Abstract
Proprietary approaches for representing annotations and image markup are serious barriers for researchers to share image data and knowledge. The Annotation and Image Markup (AIM) project is developing a standard based information model for image annotation and markup in health care and clinical trial environments. The complex hierarchical structures of AIM data model pose new challenges for managing such data in terms of performance and support of complex queries. In this paper, we present our work on managing AIM data through a native XML approach, and supporting complex image and annotation queries through native extension of XQuery language. Through integration with xService, AIM databases can now be conveniently shared through caGrid.
1. INTRODUCTION
Image annotations, including observational or computational description of image features, are essential for medical interpretations. While DICOM is the standard for medical images, there is no standard for image annotation and markup. The Annotation and Image Markup Project (AIM)1 develops a standard information model for representing and exchanging of image annotation and markup. AIM standard provides not only syntactic but also semantic interpretability with caBIG,2 DICOM and HL7 in health care and clinical trial environments. The model itself is designed using UML, including dozens of classes such as patient, observer, equipment, image, anatomic entities, image observations and image observation characteristics, geometric shapes, text annotations, calculations, and so on. The model can be implemented in XML, where data elements are deeply hierarchical. For example, the codeMeaning/codeValue elements reside in elements at the sixth level in the XML hierarchy.
XML is fast becoming the standard information exchange language for web-based applications, and is ubiquitously used for data sharing and semantic interoperability in healthcare, life sciences and many other domains.3 As a result, commercial database vendors and research institutions are researching and developing the data persistence of XML documents by building XML databases either through extension of traditional databases or new database architectures. XML databases provide significant advantages as they support standard data definition languages based on XML standards such as XML Schema, and standard XML query languages such as XPath4 and XQuery.5 Furthermore, the XML-in and XML-out approach greatly simplifies the translation of data models and query languages. XML database technology is becoming mature, and XML database products are proliferating, such as Oracle Berkeley DB XML,6 Oracle XML DB,7 IBM DB2 pureXML,8 eXist,9 Tamino,10 etc.
One immediate question is how we can effectively manage and share AIM XML documents to support the objective of AIM standard. Our goals include: i) managing complex AIM documents; ii) providing complex query support such as metadata queries, spatial queries and semantic enabled queries; iii) efficient query support; and iv) standard based sharing of AIM documents. While it is possible to map XML based annotations as relational tables and manage them through a relational database management system, the process incurs much overhead for the translation of data models and queries, and expensive joins on multiple internal association tables for retrieving the data. Storing a whole XML document as a large object (LOB) in a relational database is not desirable either due to the lack of indexing structures in LOB to support efficient queries.
In this paper, we explore the latest XML data management technologies and develop a generic XML data management system that can effectively manage AIM XML documents in terms of requirements, query support and efficiency. The AIM data can then be shared through caGrid infrastructure through xService.
The paper is organized as follows. We will give a brief overview of AIM model in Section 2, then we discuss how we manage AIM data in XML databases in Section 3. Complex query support is demonstrated in Section 4, and performance study is presented in Section 5. Section 6 shows how we can share AIM data through xService we developed, followed by Related Work and Conclusion.
2. OVERVIEW OF AIM MODEL
AIM is a caBIG In Vivo Imaging Workspace project developed by Northwestern University and Stanford University. The goal of AIM is to provide standardization for image annotation and markup, especially for clinical trials.
The AIM model describes the meaning of image pixels. Annotations become semantic information that can be used for queries and data mining, and markups can be used to outline regions of interest. The model defines a base class Annotation, and of which two classes can be instantiated: ImageAnnotation (for annotation made on images) and AnnotationOfAnnotation (for annotation derived from other annotations). Each Annotation is associated with a type attribute to define the type of annotation, such as “RECIST baseline target lesion”, “RECIST follow-up target lesion”, etc.
Annotation class has associations with User (the author), Equipment (the equipment used to create the AIM document), AnatomicEntity (the specific target in human body from which the images were generated), ImagingObservation (interpretation of an image or images, including visual features, morphologic or physiologic processes, and diseases), and Calculation (the quantitative result from mathematical or computational calculations).
ImageAnnotation class has associations with GeometricShape (with subclasses Point, MultiPoint, Polyline, Circle, and Ellipse), TextAnnotation (text displayed on images), and Segmentation (DICOM segmentation objects). ImageAnnotation class also has association with ImageReference, which could be a DICOMImageReference or a WebImageReference.
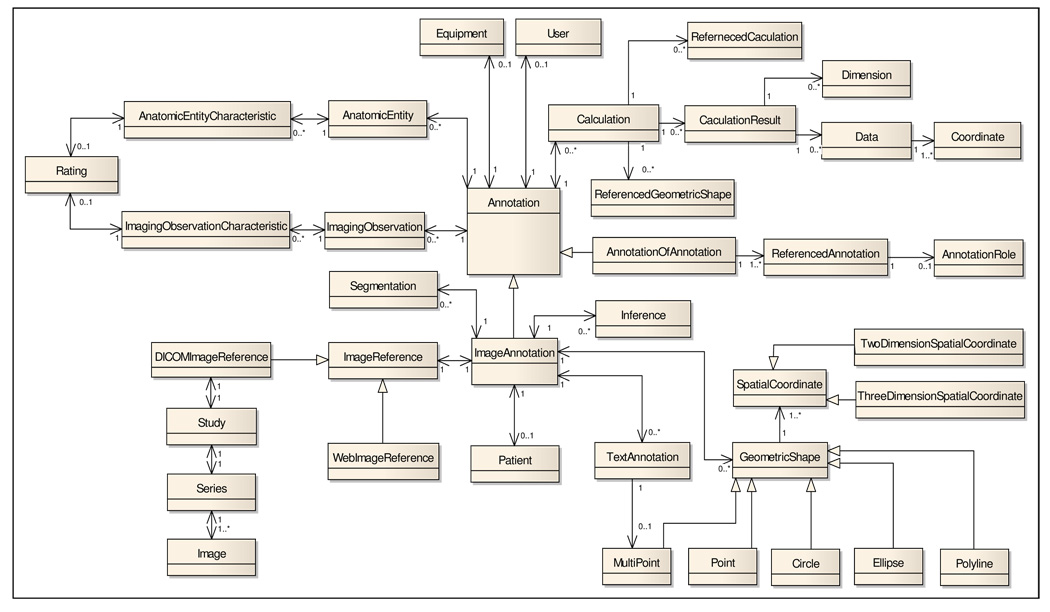
AnatomicEntity, ImagingObservation, and ImagingObservationCharacteristic classes represent essential features for an annotation, and the populated values come from certain controlled vocabulary, such as RadLex. Figure 1 shows an overview of the major classes of AIM model.
Figure 1.
Overview of AIM Data Model (V2.0)
3. MANAGING AIM DATA IN XML DATABASES
XML database systems are developed through extension of traditional relational databases or new database systems, where an XML document is the logic data object for manipulation. For example, major relational database engines such as Oracle and DB2 extend their systems with a new native data type XML or XMLType with its own backend storage and indexing support. There are many new XML database systems developed such as eXist and Tamino. Berkeley DB XML is an XML database system based on Berkeley DB engine. Currently, we support Berkeley DB XML, DB2, an eXist. XML databases conveniently support XML based data definition, and provides standard XML query languages such as XPath4 and XQuery.5 Next we discuss how we can take advantage of XML databases to manage AIM XML documents.
3.1 AIM Data Model Implementation in XML
AIM data model is first designed as a UML model. Since one essential goal for AIM is to share and exchange standardized annotation and markup documents, the XML standard is a natural choice for the intermediate representation as XML is a well established standard for data interchange and is the standard data representation for web services. The AIM UML model is therefore mapped to an XML schema, and the AIM document data are represented as XML documents, which are constructed, transmitted, manipulated, and consumed by users and applications.
There are different ways to transform a UML model to an XML schema.11 Here the caCORE SDK12 based approach is taken, with following mapping rules:
A UML class is mapped to a complex Type in XML Schema;
An attribute of a UML class is mapped as an attribute of the XML Schema complex Type that corresponds to the UML class;
A unidirectional association relationship between two classes (e.g., A → B with role name role) is mapped as enclosing elements in the XML schema, and the role name will be mapped as an intermediate element in between the two elements, e.g., <A> <role> <B> … </B> </role> </A>;
A many to many association relationship (e.g., A ↔ B with role name role1 and role2) is mapped as an enclosing elements with the role name element in between, and to preserve the backward relationship, an additional element of A is enclosed, e.g., <A> <role1> <B> <role2> <A>…</A> </role2> </B> </role1> </A>.
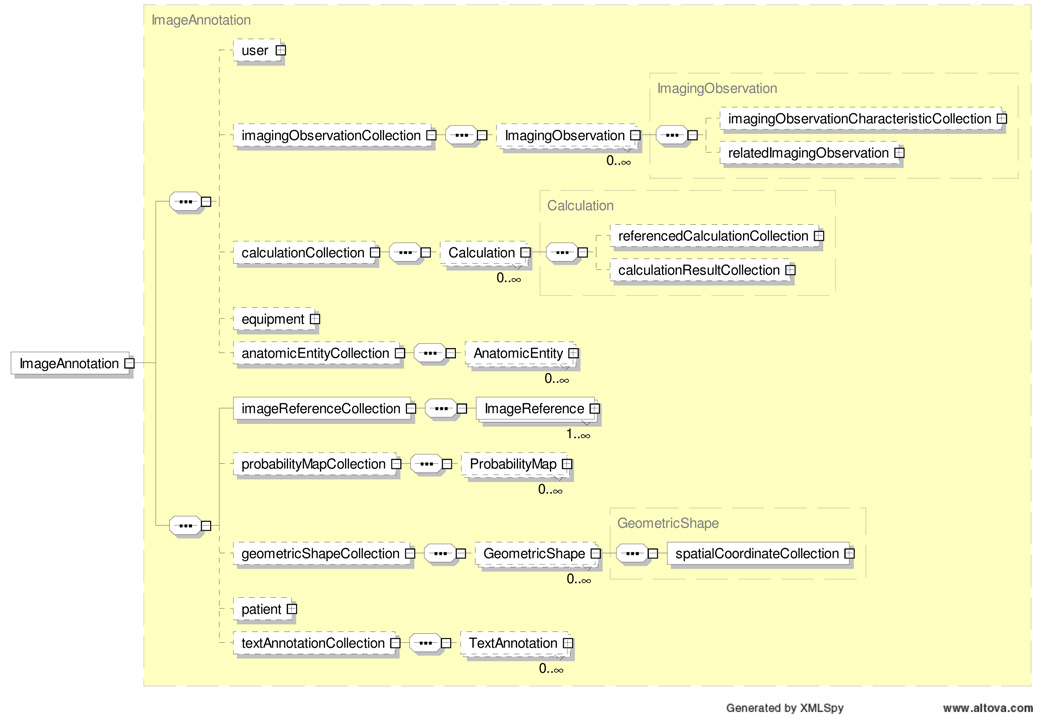
A sample AIM Schema (version 1) is shown in Figure 2.
Figure 2.
Overview of AIM XML Schema (V1)
3.2 Managing AIM XML Documents
We can classify XML databases as two major approaches: pure XML approach and hybrid relational/XML approach. For pure XML approach, only XML documents can be managed, and only XML based manipulations and APIs are available, such as Oracle Berkeley DB XML and eXist XML database; For the hybrid approach, both relational data and XML data can be managed, and the query could a combination of SQL and XML queries, such as IBM DB2 pureXML. In the pure XML approach, an AIM document is stored in an XML collection, and in the hybrid approach, an AIM document is stored in a column in XML data type. For example, for Berkeley DB XML, we can create an XML collection xmldb under folder /apps/databases/bdbxml as follows:
createContainer "/apps/databases/bdbxml/xmldb"
While in DB2, we create the following table to store AIM documents, where AIM documents are stored in the column xmlcolumn of table xmltable:
CREATE TABLE xmldb.xmltable (
docid VARCHAR(64) NOT NULL,
xmlcolumn XML,
metadata XML,
PRIMARY KEY (docid) );
3.3 Managing Metadata for AIM documents
Occasionally there are additional metadata that need to be captured about an AIM document, such as the timestamp a document is inserted, the original filename for the AIM document, a collection the AIM document belongs to, etc. Such metadata are not part of the AIM model. We develop three different solutions for the three databases. For Berkeley DB XML, it provides its own proprietary metadata management through its XmlMetaData class. Each XML document can be associated with XMLMetaData items, and each XMLMetaData item has its uri, name, and value. For DB2 and eXist, since there is internal metadata support, we associate an XML document with a metadata XML document defined in following structure:
<metadadata>
<meta>
<uri>…</uri> <name>…</name> <value>…</value>
</meta>
…
</metadata>
In eXist, a metadata XML document is managed as a regular XML document and we use its master document id as its prefix for the association. In DB2, a metadata XML column is defined for the XML table.
3.4 Managing Attached Objects
In the AIM model, there is a Segmentation class that represents DICOM segmentation objects such as binary, fractional probability, fractional occupancy and surface. The object will not be encoded into the AIM document due to its unstructured nature and large size. Instead, external objects will be linked in AIM documents.
We manage segmentation objects as files in the operating system, and provide mapping between the files and IDs in AIM documents that refer to the files through the metadata discussed above. The metadata are used to map a file’s name in the XML document to its physical storage path.
3.5 Unified XML Database Access
Due to the diversity of XML databases, to make it flexible for users to choose a database, we develop a set of unified interfaces for uploading, updating, deleting and querying XML data across different types of XML databases. Users can also quickly switch to a different database backend without change to their applications. The APIs include:
Submit. An XML document can be sumitted to the database in the form of an XML file, an XML inputstream, or an XML string;
Query. A valid XPath query can be executed on different types of XML databases;
Delete. An XML document can be deleted by its document id or an XPath query that uniquely identifies the document;
Replace. An XML document identified by its document id or a unique XPath query is replaced by a new document in the form of an XML file, an XML inputstream, or an XML string. In the future, fine-grained update of an XML document will be provided.
4. COMPLEX QUERIES ON IMAGE ANNOTATIONS AND MARKUP
XML provides two standard based query languages XPath and XQuery. XQuery depends on XPath expressions to navigate data inside an XML tree, and uses the FLOWR (FOR, LET, ORDER, WHERE and RETURN) clauses for its major expressions. XQuery5 is Turing-complete13 and natively extensible through its user defined functions.
Next we summarize a list of common AIM query scenarios and demonstrate how to express them in XML queries.
Given image UIDs, return annotation metadata such as image observation characteristics;
Given image UIDs, return geometric markup objects;
Given annotation metadata, return image or patient information;
Return all annotation data based on given image UIDs;
Return information from a group of multiple annotations based on annotation of annotations;
While most queries can be specified with XPath, queries in the last categories have to be specified in XQuery, as they require joins which are only possible through XQuery. Next we provide several example queries and show how they can be supported in XPath/XQuery. Note that different XML databases have variations on the syntax of queries. For example, to specify an XML collection, Berkeley DB XML uses collection ("xmlcollection"), while DB2 uses db2–fn:xmlcolumn ("XMLDS.XMLTABLE.XMLCOLUMN"). Here we only show the queries expressed in standard syntax.
4.1 Metadata Based Queries
Common queries are using metadata as predicates in AIM to find related information, and they can normally be expressed as XPath based queries, as shown in the following examples.
- QUERY 1 Given a series instance UID, return all image observation characteristics:
declare namespace ns1="gme://caCORE.caCORE/3.2/edu.northwestern.radiology.AIM"; /ns1:ImageAnnotation [ns1: imageReferenceCollection/ns1:ImageReference/ ns1:study/ns1:Study/ns1:series/ns1:Series/@instanceUID="1.3.6.1.4.1.9328.50.3.95047"]/ ns1:imagingObservationCollection/ns1:ImagingObservation/ imagingObservationCharacteristicCollection/ImagingObservationCharacteristic - QUERY 2. Given imaging oberservation characteristic, return study instanceUID:
declare namespace ns1="gme://caCORE.caCORE/3.2/edu.northwestern.radiology.AIM"; /ns1:ImageAnnotation [ns1:imagingObservationCollection/ns1:ImagingObservation/ ns1:imagingObservationCharacteristicCollection/ns1:ImagingObservationCharacteristic [@codeValue="2568622" and ns1:rating/ns1:Rating/@value="1.0" and ns1:rating/ns1:Rating/@name="Soft Tissue"]]/ns1:imageReferenceCollection/ ns1:ImageReference/ns1:study/ns1:Study/@instanceUID - QUERY 3. Query AIM data service using given image SOP instance UID to get all x, y coordinates on the image:
declare namespace ns1="gme://caCORE.caCORE/3.2/edu.northwestern.radiology.AIM"; /ns1:ImageAnnotation[ns1:imageReferenceCollection/ns1:ImageReference/ ns1:study/ns1:Study/ns1:series/ns1:Series/ns1:imageCollection/ns1:Image/ @sopInstanceUID="1.3.6.1.4.1.9328.50.3.122487"]/ns1:geometricShapeCollection/ ns1:GeometricShape/ns1:spatialCoordinateCollection/ns1:SpatialCoordinate - QUERY 4. Retrieve all AIM annotation documents based on given series uid:
declare namespace ns1="gme://caCORE.caCORE/3.2/edu.northwestern.radiology.AIM"; doc("aim.xml")/ns1:ImageAnnotation[ns1:imageReferenceCollection/ns1:ImageReference/ ns1:study/ns1:Study/ns1:series/ns1:Series/@instanceUID="1.3.6.1.4.1.9328.50.3.9504"]
4.2 Spatial Query Support
As markups are essentially spatial objects, it is important to support spatial queries. XQuery by itself does not support spatial operations. However, XQuery is Turing-complete and natively extensible. Thus many additional constructs needed for spatial queries can be defined in XQuery itself through user defined functions (UDF).
-
QUERY S1. Find the total area of the markups for the annotation document.
declare namespace ns1="gme://caCORE.caCORE/3.2/edu.northwestern.radiology.AIM"; for $a := collection () /ns1:ImageAnnotation[@uniqueIdentifier= "1.3.6.1.4.1.9328.50.3.122369-noduleID-13571"] /ns1:ImageAnnotation let $g := area( $a/ns1:geometricShapeCollection/ns1:GeometricShape/ ns1:spatialCoordinateCollection) return sum( $g )Here area () is a user defined function in XQuery that computes the area from coordinate collections through a recursive function.
- QUERY S2. Find the response rate of target lesions (the change rate of the sum of the longest diameter of target lesions between followup and baseline).
declare namespace ns1="gme://caCORE.caCORE/3.2/edu.northwestern.radiology.AIM"; for $baseline := collection () /ns1:ImageAnnotation[@uniqueIdentifier= "1.2.288.3.2205383238.1072.1207947057.22"] /ns1:ImageAnnotation for $followup := collection () /ns1:ImageAnnotation[@uniqueIdentifier= "1.2.288.3.2205383238.1072.1207948422.99"] /ns1:ImageAnnotation let $areabaseline := longestDiameter($baseline/ ns1:geometricShapeCollection/ns1:GeometricShape/ns1:spatialCoordinateCollection) let $areafollowup := longestDiameter($followup/ ns1:geometricShapeCollection/ns1:GeometricShape/ns1:spatialCoordinateCollection) return (sum(areafollowup)- sum(areabaseline)) / sum(areabaseline)
User-defined Spatial Functions
To support spatial queries, we define a set of user-defined functions (UDFs) in XQuery. Such UDFs could be implemented system specifically, for example, DB2 uses its table UDF to define these functions. The functions include: i) spatial relationship functions, such as contains (), within (), adjacent (), overlaps (), etc.; ii) spatial property functions, such as area (), longestdiameter (), etc.
4.3 Semantic Enabled Queries
AIM model is semantically enabled in two aspects. First, AIM data model is caBIG silver compliant – AIM data model classes and attributes are annotated through concept IDs and other information and registered at caDSR;14 Second, AIM provides the framework where its content can be authored through references to ontologies or controlled vocabularies. For example, AIM defines ImageObservation, ImageObservationCharacteristic, and AnatomicEntity classes with the following attributes: i) codingSchemeDesignator – the ontology or controlled vocabulary where the data element depends on, such as RadLex, caDSR, or EVS; ii) codeMeaning – the meaning of the concept code; and iii) codeValue – the unique identification code for the concept.
The semantic enabled data model provides two opportunities to support semantic enabled queries: semantic interoperable queries across different data sources through caDSR, or semantic enabled queries through extending XML queries in AIM databases. We will consider the latter, and show how these are achieved through considering synonyms, hypernyms, and hyponyms.
-
Query accuracy enhancement. Queries using a concept often suffer from the problem that there are synonyms for the concept, and results from other terms will be missing. To correct that, we send a request to the corresponding ontology database that the initial query term belongs to and retrieve all its synonyms. This is implemented as a function getSynonyms () that takes the initial term, and returns all the synonyms. With that synonyms, the final query would include a combination of all possible terms and return more accurate result.
For example, “parenchyma of kidney” (concept ID: RID209) has a synonym of “renal parenchyma”. An example query to return all annotations for parenchyma of kidney will be:declare namespace ns1="gme://caCORE.caCORE/3.2/edu.northwestern.radiology.AIM"; for $a in collection()/ImageAnnotation for $syn in getSynonyms("parenchyma of kidney") where $a/anatomicEntityCollection/AnatomicEntity/@codeMean/text() = $syn return $a Query expansion. When a user specifies a query with a concept, the concept itself may include subclasses of concepts, and these can generate favorable results too. For example, for a query on image observation with a concept “astrocytoma (WHO grade IV)”, since “astrocytoma (WHO grade IV)” has two subclasses “gliosarcoma” and “giant cell glioblastoma”, results from these subclasses are also acceptable for the query result. Similarly, we provide a function getHyponyms () to retrieve all hyponyms of this concept from the corresponding ontology, and include them for specifying predicates.
Query relaxation. In many cases, there may not be any result from a query using a certain term, but users may still want to look at the closest results or relaxed results by relaxing the concept to a broader scope. For example, if a query with the concept “giant cell glioblastoma” returns no result, the user may want to see if there is any result from the relaxed concept “astrocytoma (WHO grade IV)”, a super class of “giant cell glioblastoma”. Here we provide a function getHypernyms () to relax the concept to its parent concepts, and use them to adjust the predicates to possibly return relaxed result.
The implementation approaches of queries from ontology database or servers vary, as different ontology systems provide different access methods and APIs. RadLex provides RESTful services for queries. The user defined function is normally XML database dependent.
5. EFFICIENT SUPPORT OF QUERIES AND PERFORMANCE STUDY
XML databases provide different approaches on access methods through value based indexing, edge based indexing, or internal association tables. We demonstrate that such approaches significantly boost the query performance. For example, in DB2, we can create indexes through generating keys using XPath patterns. For double-slash ”//” based queries, the performance deteriorates as more index nodes need to be searched, especially for Berkeley DB XML.
Additional optimization of query performance is possible through fast disks such as RAID or solid state disks. The latter provides much better performance on random reads. The latest partitioning support for XML data in DB2 can also distribute data across multiple CPUs and storage of parallel or cluster machines to boost the performance.
To test the performance of XML based data management, we take the AIM dataset converted from LIDC,15, 16 which includes 17, 927 documents, with a total size of 155MB. The machine we use for the test is a Lenovo W500 with Core 2 Duo at 2.8GHz, 4GB of RAM, SATA 7200rpm hard drive, installed with Windows XP Professional. We have DB2 Enterprise Edition V9.5 and Oracle Berkeley DB XML V2.4.16 installed. Note that eXist is not chosen for the study, as it takes a DOM based storage model, and frequently runs out of memory.
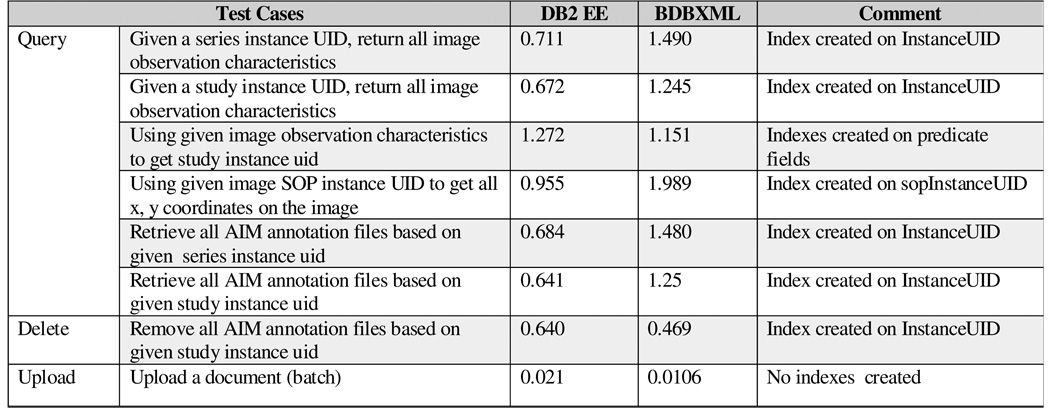
Figure 3 shows the result of several test cases. The result shows that the query support is very efficient, and DB2 performs better than Berkeley DB XML in most queries. For DB2, most queries take less than one second except the third query where two conditions are joined together. Batch uploading is very efficient even though the databases need to build the storage structure for the documents during the uploading process.
Figure 3.
Performance of AIM Queries on DB2 and Berkeley DB XML (in seconds)
6. SHARING AIM DATA ON CAGRID
caGrid is a Grid middleware infrastructure to support collaborative biomedical research studies. With a service oriented architecture, caGrid allows researchers to share both their data and analytical resources as grid services, and provides federated queries across distributed databases. caGrid takes a model driven architecture, where data models are defined in UML, and an abstract data access and query layer for the data sources is provided. From the client side, users will be able to query caGrid data sources with Common/caGrid Query Language (CQL),17 an object-oriented query language for querying data defined through UML models. For relational databases, CQL queries are translated into SQL queries through hibernate mapping created using caCORE SDK, and executed on databases of the data sources. Hibernate creates the object relational mapping and the serialization and deserialization of the query results into objects, which are subsequently encoded as XML documents for transmission to the caGrid client.
However, the current caGrid infrastructure only supports data access of relational data stored in relational databases. Existing tools and architectures will not support querying or uploading of XML data managed in XML databases such as AIM databases, which provide XML Schema based schema definition language and XPath/XQuery based query languages.
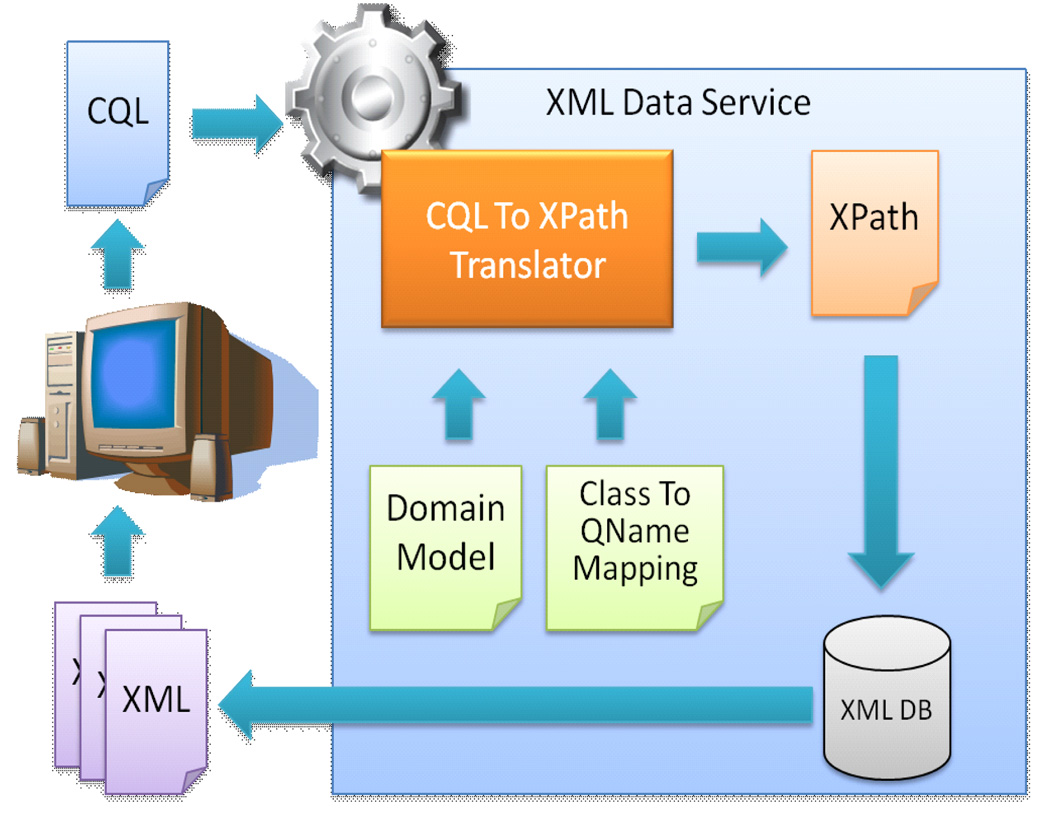
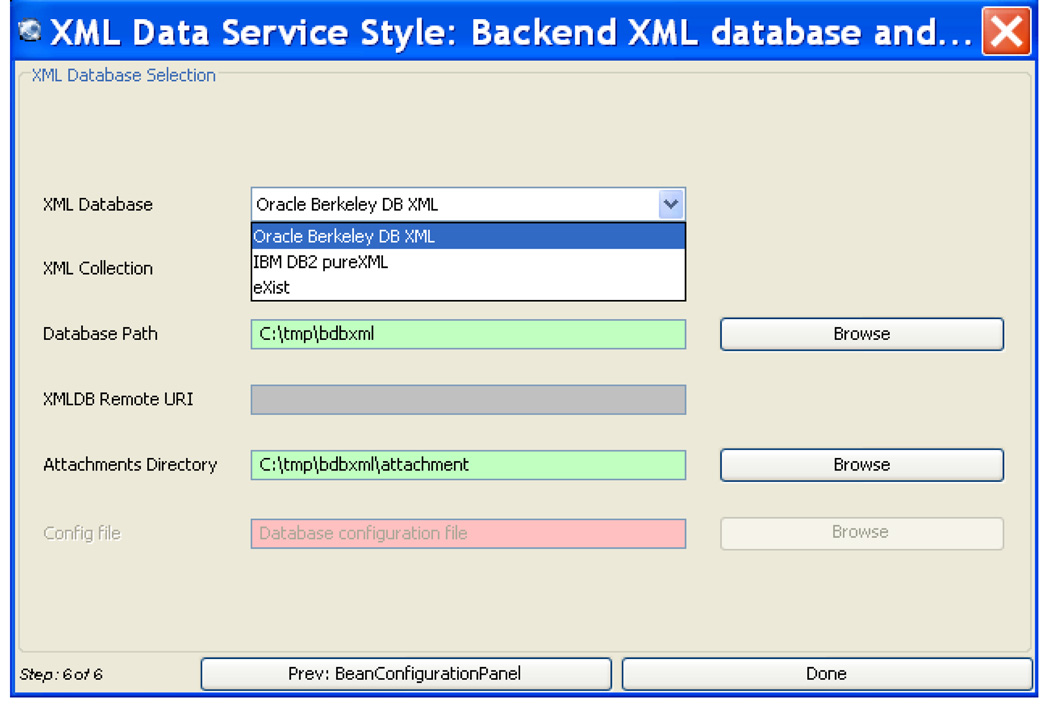
To support caGrid-based data services that support existing XML schemas, existing XML data, and/or existing XML databases, we develop aGrid XML Data Service Framework (xService).18 xService is an extension of caGrid for querying and retrieving XML documents managed in XML databases. xService provides automated data model mapping from XML schema (defined in XML Schema language) to caGrid Domain Model (represented in XMI format), query mapping from CQL to XML query language (XPath), and generic XML database interfaces for uploading, updating, querying and retrieving of XML data from diverse XML data sources discussed in this paper (Figure 4). xService also provides an extension to Introduce Toolkit for users to flexibly and rapidly create their own caGrid data services based on a predefined XML Schema. Figure 5 shows the screenshot from Introduce where a user can select which database to use for the backend XML storage.
Figure 4.
Querying Processing of caGrid with xService
Figure 5.
Sample Screenshot of Database Selection in Introduce
Through xService, caGrid clients will be able to query and upload AIM data through caGrid infrastructure. Operations provided by the AIM Data Service include: i) query, where a CQL query is taken and a CQL query result object is returned, such as count, attributes or AIM objects; ii) enumeration query, where an enumerable resource is returned for iterative next operation; iii) queryByTransfer, where query result is transported through caGrid transfer service. Transfer service uses HTTP protocol for efficient data transportation instead of SOAP protocol; iv) submit, where XML string is uploaded to the database; and v) submitWithTransfer, a caGrid transfer version of submit.
AIME19 is a collection of caGrid data services at Emory University for managing AIM documents used for multiple research projects, including TCGA Enterprise Use Case from caBIG In Vivo Imaging Workspace.
7. RELATED WORK
DICOM SR has been used to model and store image annotations and markups in DICOM,20 but DICOM SR does not provide the approach for querying the data, neither a semantic approach for representing annotations. Recently, a Semantic Web based approach has been proposed to manage ontologies and semantic metadata for medical image annotations.21
XML based approach for managing biomedical data becomes increasingly popular. One way is to provide XML based interfaces with relational based backend. For example, XNAT22 is an XML based platform for managing neuroimaging and related data, and represents data in XML at the schema level. XNAT uses relational database engine as the backend storage, and provides data and query mapping between XML and RDBMS. There is also work done to provide unique XQuery based frontend for relational based data sources.23 Project Mobius24 provides generic mapping of XML Schema elements to relational tables, although XML query support is limited.
Using XML databases is becoming popular in biomedical applications. For example, XML database has been used in25 to manage biological pathway datasets. SciPort26, 27, 28 provides an XML based approach for modeling, managing and integrating scientific experiments and data. XML is also used to integrate heterogeneous bio-molecular data.29
8. CONCLUSION
AIM is becoming the standard for representing image annotation and markup for translational research and clinical trials. In this paper, we discuss our work on using XML database based approach to manage AIM XML documents, metadata and attached segmentation objects. The XML approach shows significant benefit on supporting complex AIM queries, such as spatial queries and semantic enabled queries. Our performance study shows that the approach is efficient. Through xService, AIM data can be conveniently shared and queried through caGrid.
Acknowledgement
The project described was supported, in part, by Grant Number R24HL085343 from the National Heart, Lung, and Blood Institute. The project has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E.
REFERENCES
- 1.Channin David, Mongkolwat Pattanasak, Kleper Vladimir, Sepukar Kastubh, Rubin Daniel. The caBIG Annotation and Image Markup Project. Journal of Digital Imaging. 2009 doi: 10.1007/s10278-009-9193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The cancer Biomedical Informatics Grid. https://cabig.nci.nih.gov/
- 3.Shabo A, Rabinovici-Cohen S, Vortman P. Revolutionary impact of xml on biomedical information interoperability. IBM Syst. J. 2006;45(2):361–372. [Google Scholar]
- 4.XML Path Language. http://www.w3.org/TR/xpath.
- 5.XQuery 1.0: An XML Query Language. https://www.w3.org/TR/xquery/
- 6.Oracle Berkeley DB XML. http://www.oracle.com/database/berkeley-db/xml/
- 7.Oracle XML DB. http://www.oracle.com/technology/tech/xml/xmldb.
- 8.DB2 Express-C Overview. http://www-306.ibm.com/software/data/db2/express/
- 9.eXist XML Database. http://www.exist-db.org/
- 10.Tamino: The XML database. http://www.softwareag.com/us/products/wm/tamino/
- 11.Domínguez Eladio, Lloret Jorge, Pérez Beatriz, Rodríguez Áurea, Rubio Ángel, Zapata María. WISE. 2007. A survey of uml models to xml schemas transformations; pp. 184–195. [Google Scholar]
- 12.The caCORE Software Development Kit (SDK) http://ncicb.nci.nih.gov/infrastructure/cacoresdk.
- 13.Kepser S. Extreme Markup Languages. 2004. A Simple Proof for the Turing-Completeness of XSLT and XQuery. [Google Scholar]
- 14.Cancer Data Standards Registry and Repository (caDSR) http://ncicb.nci.nih.gov/core/caDSR.
- 15.Armato SG, III, McNitt-Gray MF, et al. The Lung Image Database Consortium (LIDC): An Evaluation of Radiologist Variability in the Identification of Lung Nodules on CT Scans. Acad Radiol. 2007;14(11):1409–1421. doi: 10.1016/j.acra.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LIDC to AIM Conversion Program. https://wiki.nci.nih.gov/display/Imaging/LIDC+to+AIM+Conversion+Program.
- 17.CQL: Common/caGrid Query Language. http://cagrid.org/display/dataservices/CQL.
- 18.caGrid XML Data Service Framework (xService) https://web.cci.emory.edu/confluence/display/xmlds/
- 19.AIM Data Service at Emory University. https://web.cci.emory.edu/confluence/display/IVI/AIMDataService.
- 20.Clunie David. DICOM structured reporting. PixelMed Publishing; 2000. [Google Scholar]
- 21.Mukherjee Saikat, Möller Manuel. Context-driven ontological annotations in dicom images: Towards semantic pacs; Proc. of Intl Joint Conference on Biomedical Engineering Systems and Technologies; 2009. [Google Scholar]
- 22.Marcus DS, Olsen TR, Ramaratnam1 M, Buckner RL. The extensible neuroimaging archive toolkit: An informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007 March;:11–33. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- 23.Bales N, Brinkley J, Lee ES, Mathur S, Re C, Suciu D. A framework for xml-based integration of data, visualization and analysis in a biomedical domain. XSym. 2005:207–221. [Google Scholar]
- 24.The Mobius Project at OSU BMI. http://www.projectmobius.org.
- 25.Jiang Keyuan, Nash Christopher. Application of xml database technology to biological pathway datasets; Proc IEEE Eng Med Biol Soc; 2006. pp. 4217–4220. [DOI] [PubMed] [Google Scholar]
- 26.Wang F, Thiel F, Furrer D, Qin C, Hackenberg G, Bourgue P, Kaltschmidt D, Wang M. An adaptable xml based approach for scientific data management and integration. SPIE Medical Imaging. 2008;volume 6919 doi: 10.1117/12.773154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Fusheng, Bourgué Pierre-Emmanuel, Hackenberg Georg, Wang Mo, Kaltschmidt David, Liu Peiya. Sciport: an adaptable scientific data integration platform for collaborative scientific research. VLDB. 2007:1310–1313. [Google Scholar]
- 28.Wang Fusheng, Liu Peiya, Pearson John, Azar Fred, Madlmayr Gerald. Experiment management with metadata-based integration for collaborative scientific research. ICDE. 2006 [Google Scholar]
- 29.Mesiti M, Jimnez-Ruiz E, Sanz I, Berlanga-Llavori R, Perlasca P, Valentini G, Manset D. Xml-based approaches for the integration of heterogeneous bio-molecular data. BMC Bioinformatics. 2009;10(S7) Suppl 12 doi: 10.1186/1471-2105-10-S12-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]