Abstract
Nested Association Mapping (NAM) has been proposed as a means to combine the power of linkage mapping with the resolution of association mapping. It is enabled through sequencing or array genotyping of parental inbred lines while using low-cost, low-density genotyping technologies for their segregating progenies. For purposes of data analyses of NAM populations, parental genotypes at a large number of Single Nucleotide Polymorphic (SNP) loci need to be projected to their segregating progeny. Herein we demonstrate how approximately 0.5 million SNPs that have been genotyped in 26 parental lines of the publicly available maize NAM population can be projected onto their segregating progeny using only 1,106 SNP loci that have been genotyped in both the parents and their 5,000 progeny. The challenge is to estimate both the genotype and genetic location of the parental SNP genotypes in segregating progeny. Both challenges were met by estimating their expected genotypic values conditional on observed flanking markers through the use of both physical and linkage maps. About 90%, of 500,000 genotyped SNPs from the maize HapMap project, were assigned linkage map positions using linear interpolation between the maize Accessioned Gold Path (AGP) and NAM linkage maps. Of these, almost 70% provided high probability estimates of genotypes in almost 5,000 recombinant inbred lines.
Keywords: Nested association mapping, Genotypic imputation
Introduction
Forward genetic approaches for relating genomic variability with phenotypic variability can be grouped as either linkage or association mapping. Because it is easy to create and maximize linkage disequilibrium in plant species the former set of methods were initially referred to as Quantitative Trait Locus (QTL) mapping, although it is now clear that association mapping also can be applied to quantitative traits. Linkage mapping is powerful but of low resolution, resulting in identifying genomic regions consisting of about 10 cM, which often consists of tens of millions of bases for most plant species. With the advent of high-throughput technologies for resequencing and genotyping, association mapping has emerged for species where it is not easy to create linkage disequilibrium. This approach exploits historical linkage and recombination accumulated over a large number of generations (Andersson and Georges 2004). Thus, it can provide high resolution information that can be used to identify the causative nucleotides underlying phenotypic variability. Depending upon the amount of linkage disequilibrium (LD) across the genome in the breeding population, association mapping can require genotyping with very high densities of molecular markers (Yu et al. 2008) and extremely large samples to achieve reasonable power (Hirschhorn and Daly 2005; Kingsmore et al. 2008).
A third approach is to combine the power of linkage mapping with the resolution of association mapping. This third approach can be thought of as an extension of the multiple family QTL approach (Jansen et al. 2003; Blanc et al. 2006), but is distinctive in that parental inbred lines are resequenced or array genotyped and this information is coupled with low-cost genotyping of their segregating progenies. The approach is conceptually equivalent to the human quantitative transmission disequilibrium test (QTDT) (Abecasis et al. 2000) combined with imputation of genotypes of relatives (Burdick et al. 2006). For the special case where the mapping population consists of multiple families of segregating progeny, usually Recombinant Inbred Lines (RILs), derived from inbred lines crossed to a single reference inbred line, the method has been called Nested Association Mapping (NAM) (Yu et al. 2008; Nordborg and Weigel 2008).
For purposes of mapping functional markers in NAM populations, parental genotypes at a large number of SNP loci need to be projected to their segregating progeny. For example, approximately 0.5 million SNPs have been genotyped in the 26 parental lines of the publicly available maize NAM population whereas only 1,106 SNP loci have been genotyped in both the parents and their 5,000 progeny. The challenge is to estimate both the genotype and genetic location of the parental genotypes in the segregating progeny. Three approaches might be considered (Yi and Shriner 2007): (1) estimate all missing genotypes by their expected values conditional on observed flanking markers (Haley and Knott 1992), (2) consider genotypes as unknowns to be predicted using an MCMC update procedure, and (3) multiple sampling of genotypes from a conditional probability distribution for each unknown locus (Sen and Churchill 2001). Given the large number of SNP loci and large number of families and progeny in NAM populations, the latter two approaches could be computationally challenging, depending upon the quality of the physical map. The first approach, however, may be accurate while computationally feasible.
Herein, we report on: (1) development of a method for imputing genotypes using an expectation approach, and (2) illustrate its use by applying it to the maize NAM population. In human family based association mapping (Burdick et al. 2006) parental SNPs are projected onto progeny in intervals with no recombinants. Herein, the method is extended to intervals with known recombination events.
Data and methods
Data
The following data sets were obtained from public information resources: (1) genotypes of 5,000 RILs representing 25 segregating families of the maize NAM mapping population (McMullen et al. 2009). These data are represented as NAM_SNP_genos_raw_20080703 at http://www.panzea.org/. (2) A composite linkage map created by McMullen et al. (2009) using the maize NAM genotypic data (http://www.panzea.org/). (3) The maize Accessioned Gold Path (AGP v1) (Wei et al. 2009), consisting of 10 chromosome pseudo-assemblies guided by the physical map, was obtained from the Arizona Genomics Institute (http://www2.genome.arizona.edu/genomes/maize). (4) the maize HapMap for the 26 founder lines of the maize NAM population. These data comprise nearly half a million SNP genotypes, and can be obtained from http://www.maizegenetics.net/maize-hap-map. Note that the maize HapMap data are continuing to be updated with new releases, so the version utilized herein will likely be outdated before publication of this manuscript.
Estimation of linkage map positions
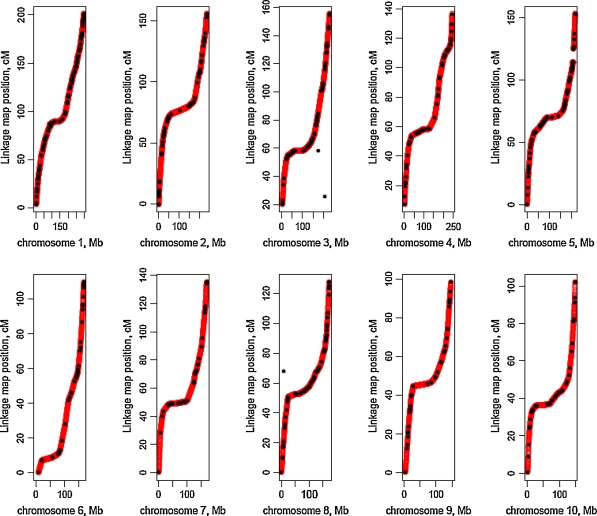
In order to detect the associations between genotypes and complex quantitative traits, it is necessary to know the linkage map positions of the polymorphic loci and to trace inheritance of these using flanking markers. The linkage map positions are unknown for the majority of the 0.5 million SNPs which are genotyped in the parental lines maize NAM families. Their linkage map positions were assigned through linear interpolation between the maize AGP v1 (Wei et al. 2009) and maize NAM linkage map (McMullen et al. 2009), as described by Kong et al. (2002). SNP loci occurring on the same BAC are assigned the same position, because the number of recombination events within BACs for 200 RILs per family is expected to be negligible (Fig. 1).
Fig. 1.
Mapped positions of physical and linkage maps obtained through linear interpolation. The dark black dots are plotted positions of BAC accessions relative to the maize NAM linkage map. The light color curves are actually individual light color dots representing high density segregating SNPs. Locations of SNP loci were obtained through linear interpolation. AC185213 and AC197480 designated as dark black dots that deviate from the curves on chromosome 3 and AC187287 on chromosome 8 were not used in linear interpolations. A break in the curve on Chromosome 5 occurs because genetic distances on the linkage map corresponds with a small physical distances on the AGP map
Imputation of parental SNPs onto segregating progeny
SNPs with known physical locations were imputed in each RIL by computing the expectation of genotypic score given flanking marker genotypic scores, as described by Haley and Knott (1992). The maize NAM population consists of RILs which were produced by self pollinating the lines for five generations after the initial cross of the parental inbred lines. Thus, not all loci are homozygous in the segregating progeny. B73 alleles were coded as −1 and the alternative alleles as 1, heterozygous genotypes as 0.
Assuming one SNP locus Q is genotyped in parental lines but not in their progeny and this locus is flanked by two SNP loci A and B which are genotyped in parental lines and their progeny within a family, the expectation of genotype score is based on the following: (1) The transition probabilities from one genotype at one locus to one genotype at another locus (P(Q = q|A = a), P(B = b|Q = q)) are obtained by Jiang and Zeng (1997). These transition probabilities are functions of the frequency of recombinants between the two flanking loci and number of selfing generations. (2) The conditional probability of genotype of SNP Q given flanking SNP loci A and B is computed as:
P(Q = q |A = a, B = b) = P(Q = q|A = a)P(B = b|Q = q)/∑qP(Q = q|A = a)P(B = b|Q = q) (Jiang and Zeng 1997). (3) The expectation for the genetic score at SNP Q is computed as (1)P(Q = 1|A = a, B = b) + (0)P(Q = 0|A = a,B = b) + (−1)P(Q = −1|A = a,B = b) = P(Q = 1|A = a,B = b) − P(Q = −1|A = a, B = b). In situations where computation is needed at terminal ends of a linkage group, SNP locus Q will have only one adjacent polymorphic SNP locus. The conditional probability is computed as P(Q = q|A = a) = P(Q = q|A = a)/∑q P(Q = q|A = a). The expectation for the genetic score is computed by (1)P(Q = 1|A = a) + (0)P(Q = 0|A = a) + (−1)P(Q = −1|A = a) = P(Q = 1|A = a) − P(Q = −1|A = a).
Results and discussion
Estimation of linkage map positions
About 90%, i.e., 444,615 of 495,091 genotyped SNPs from the maize HapMap project, were assigned linkage map positions through linear interpolation between the maize AGP and NAM linkage maps (Table 1). The mapped positions of individual SNPs are available through the GFS Sprague Population Genetics website (Table S1 http://www.agron.iastate.edu/GFSPopGen/resources.html). Approximately 10% of the SNPs were not assigned to linkage map positions because they were located in: (1) BACs that were assigned to known chromosomes, but appear to be genetically located beyond the ends of the linkage group; (2) BACs which have not been mapped consistently to the same chromosomes by the maize AGP and NAM projects (Table 2), (3) BACs which are unassigned to chromosomes and (4) three BACs whose physical and linkage locations were not consistent within chromosomes 3 and 8 (Fig. 1). With removal of these three inconsistent BACs of the latter group, all relationships between physical and linkage maps show similar smooth curves with large numbers of BACs associated with little recombination in heterochromatic regions of the genome. The continuous nature of the curves indicates that gaps in the physical map are so small that they do not seriously affect the estimation of linkage map positions of SNPs by linear interpolation. If there had been large discontinuities and changes in direction of the curves, then such interpolation for placement of SNP loci would not be justified.
Table 1.
Summary of estimated genetic locations of SNP loci in NAM parental lines obtained through linear interpolation of information from verified physical (AGP: http://www2.genome.arizona.edu/genomes/maize) and linkage (NAM: http://www.panzea.org/) maps
| Chromosome | Number of SNPs genotyped for founder lines | Number of SNPs mapped to the linkage map | Percentage |
|---|---|---|---|
| 1 | 79689 | 72744 | 91.3 |
| 2 | 59878 | 52923 | 88.4 |
| 3 | 57506 | 50383 | 87.6 |
| 4 | 52920 | 45716 | 86.4 |
| 5 | 55610 | 51390 | 92.4 |
| 6 | 40743 | 36702 | 90.1 |
| 7 | 40410 | 38441 | 95.1 |
| 8 | 41001 | 38485 | 93.9 |
| 9 | 34189 | 28496 | 83.3 |
| 10 | 33145 | 29335 | 88.5 |
| Total | 495091 | 444615 | 89.8 |
Table 2.
Inconsistent relationships between maize physical map and NAM linkage maps
| BAC designation | Linkage chromosome of SNPs on the BAC | Physical chromosome map of the BAC | Notes |
|---|---|---|---|
| AC193326 | 1 | 4 | |
| AC205979 | 1 | 5 | |
| AC210244 | 1 | 9 | |
| AC195129 | 1 | 10 | |
| AC191808 | 1 | 4 | |
| AC203181 | 1 | 5 | |
| AC182415 | 1 | 5 | |
| AC182413 | 1 | 5 | |
| AC191122 | 1 | 6 | |
| AC201963 | 2 | Not found | |
| AC189043 | 2 | Not found | |
| AC211551 | 2 | Not found | |
| AC185221 | 2 | 3 | |
| AC208466 | 2 | 4 | |
| AC199412 | 2 | 7 | |
| AC194396 | 2 | 6 | |
| AC209833 | 2 | 7 | |
| AC191668 | 2 | 4 | |
| AC185124 | 2 | 4 | |
| AC205345 | 2 | 6 | |
| AC205589 | 3 | 1 | |
| AC191661 | 3 | 4 | |
| AC206198 | 3 | 1 | |
| AC207812 | 3 | 1 | |
| AC193490 | 3 | Not found | |
| AC191299 | 3 | 8 | |
| AC200173 | 3 | 8 | |
| AC195934 | 3 | 8 | |
| AC185213 | 3 | 3 | See Fig. 1 |
| AC197480 | 3 | 3 | See Fig. 1 |
| AC208219 | 4 | 1 | |
| AC211347 | 4 | 2 | |
| AC186606 | 4 | 1 | |
| AC190571 | 4 | 5 | |
| AC195591 | 5 | 1 | |
| AC203773 | 5 | 1 | |
| AC191429 | 5 | 1 | |
| AC186432 | 5 | 1 | |
| AC191690 | 5 | 4 | |
| AC199525 | 5 | 4 | |
| AC203090 | 5 | 1 | |
| AC208986 | 5 | 4 | |
| AC204528 | 5 | 10 | |
| AC207278 | 5 | 4 | |
| AC191410–AC187045, AC194082 | 5 | 5 | See Fig. 1. Large genetic distance (10.9 cM) but small physical distance. |
| AC199708 | 6 | 8 | |
| AC194047 | 6 | 8 | |
| AC205403 | 6 | 8 | |
| AC196979 | 6 | 1 | |
| AC195845 | 7 | Not found | |
| AC202954 | 7 | 2 | |
| AC210308 | 7 | 2 | |
| AC191092 | 8 | 9 | |
| AC205129 | 8 | 6 | |
| AC197832 | 8 | 6 | |
| AC186645 | 8 | 3 | |
| AC187880 | 8 | 3 | |
| AC191611 | 8 | Not found | |
| AC203362 | 8 | 3 | |
| AC187287 | 8 | 8 | See Fig. 1 |
| AC201989 | 9 | 8 | |
| AC191402 | 9 | 3 | |
| AC208339 | 9 | 1 | |
| AC185425 | 9 | Not found | |
| AC209853 | 9 | 1 | |
| AC197895 | 9 | 1 | |
| AC190750 | 9 | 2 | |
| AC200613 | 9 | 1 | |
| AC196769 | 10 | Not found | |
| AC190844 | 10 | 2 | |
| AC206918 | 10 | 2 | |
| AC207391 | 10 | 2 | |
| AC204518 | 10 | 2 |
Imputation of SNP genotypes from parents to segregating progeny
About 444,615 SNP genotypes in the parental lines were projected onto RILs of the maize NAM population and are available for subsequent analyses at the GFS Sprague Population Genetics website (Table S2 at http://www.agron.iastate.edu/GFSPopGen/resources.html). In some families, SNP genotypes were considered missing if: (1) the genotype of either parent was missing, or (2) the genotypic score provided by the HapMap project was not equal to 0 or 1. The missing genotypes account for approximately 27% of the projected genotypes. About 5% of the projected genotypes have absolute genetic score values between 0.1 and 0.9.. The remaining 68% have absolute genetic score values in the range of 0.9 and 1.0. (Table 3).
Table 3.
Summaries of absolute expected genotic scores in segregating progeny of the maize NAM population
| Family designation | Percentage of high confidence genetic scores (0.9–1.0) | Percentage of low confidence genetic scores (0.0–0.9) | Percentage of missing scores |
|---|---|---|---|
| 1 | 69.5 | 4.8 | 25.7 |
| 2 | 69.6 | 4.8 | 25.6 |
| 3 | 69.4 | 5.6 | 25.0 |
| 4 | 69.8 | 4.7 | 25.5 |
| 5 | 68.9 | 4.4 | 26.7 |
| 6 | 69.2 | 4.7 | 26.1 |
| 7 | 68.4 | 4.5 | 27.1 |
| 8 | 75.5 | 3.3 | 21.2 |
| 9 | 68.8 | 4.4 | 26.8 |
| 10 | 68.1 | 4.6 | 27.3 |
| 11 | 66.8 | 4.7 | 28.5 |
| 12 | 65.8 | 4.6 | 29.6 |
| 13 | 69.7 | 4.8 | 25.5 |
| 14 | 65.4 | 4.3 | 30.3 |
| 15 | 70.9 | 4.8 | 24.3 |
| 16 | 69.3 | 4.7 | 26.0 |
| 17 | 62.1 | 3.8 | 34.1 |
| 18 | 69.9 | 4.1 | 26.0 |
| 19 | 67.1 | 4.7 | 28.2 |
| 20 | 71.8 | 5.1 | 23.1 |
| 21 | 71.7 | 4.5 | 23.8 |
| 22 | 70.1 | 4.3 | 25.6 |
| 23 | 69.9 | 4.8 | 25.3 |
| 24 | 67.7 | 4.3 | 27.9 |
| 25 | 61.7 | 4.7 | 33.6 |
| All families | 68.7 | 4.6 | 26.7 |
Discussion
Plant species and model organisms (e.g., mouse: Churchill et al. 2004) exhibit characteristics that favor development of NAM populations. Pure inbred lines and large segregating families are relatively easy to develop or already available, whereas large samples (minimum of 2,000 cases and controls: Hirschhorn and Daly 2005; Kingsmore et al. 2008) of unrelated, yet adapted, accessions required for association mapping are not available in most crop species. Consequently, NAM populations are being developed for Arabidopsis (Buckler and Gore 2007) as well as soybean, barley and sorghum (personal communications). Alternatively, a large number of QTL mapping studies have been completed in various crops. If the inbred parental lines, stored in germplasm repositories, are resquenced or array-genotyped, already available phenotypic data can be exploited using a multiple family QTL analysis (Jansen et al. 2003; Jannink and Wu 2003).
As shown herein, the computational challenges of imputing parental genotypes onto segregating progeny can be handled simply through linear interpolation of genetic location and subsequent calculation of expected genotypes. Such information has been shown to provide powerful, precise and accurate identification of functional markers responsible for a variety of simulated genetic architectures (Guo et al. 2010). Importantly, forward genetic approaches which require large samples for quantitative traits, are enabled by sequencing or array-genotyping of parental lines coupled with sparse genotyping of segregating progeny. This significantly reduces costs and enables genome-wide mapping through resequencing or array-genotyping of dozens of lines rather than thousands (Yu et al. 2008; Nordborg and Weigel 2008).
Acknowledgments
Funding for this research was provided by the GF Sprague endowment for population genetics in the Department of Agronomy at Iowa State University and by the Plant Sciences Institute at Iowa State University. The authors would like to thank the following individuals for providing insightful suggestions on technical aspects of this research: Dr. Shizhong Xu, University of California-Riverside. Dr. Fusheng Wei, University of Arizona, Dr. Rod Wing, University of Arizona, Dr. Doreen Ware, USDA-ARS at Cold Spring Harbor Laboratory. We also would like to thank Dr. Ed Buckler, USDA-ARS at Ithaca, New York and Dr. Michael McMullen, USDA-ARS at Columbia, Missouri for providing timely releases of data.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Baohong Guo, Email: baohong.guo@syngenta.com.
William D. Beavis, Email: wdbeavis@iastate.edu
References
- Abecasis GR, Cardon LR, Cookson WOC. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L, Georges M. Domestic-animal genomics: deciphering the genetics of complex traits. Nat Rev Genet. 2004;5:202–212. doi: 10.1038/nrg1294. [DOI] [PubMed] [Google Scholar]
- Blanc G, Charcosset A, Mangin B, Gallais A, Moreau L. Connected populations for detecting quantitative trait loci and testing for epistasis: an application in maize. Theor Appl Genet. 2006;113:206–224. doi: 10.1007/s00122-006-0287-1. [DOI] [PubMed] [Google Scholar]
- Buckler E, Gore M. An Arabidopsis haplotype map takes root. Nat Genet. 2007;39:1056–1057. doi: 10.1038/ng0907-1056. [DOI] [PubMed] [Google Scholar]
- Burdick JT, Chen WM, Abecasis GR, Cheung VG. In silico method for inferring genotypes in pedigrees. Nat Genet. 2006;38:1002–1004. doi: 10.1038/ng1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, The Complex Trait Consortium The collaborative cross: a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36d:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Guo BH, Sleper DA, Beavis WD. Nested association mapping for identification of functional markers. Genetics. 2010;186:373–383. doi: 10.1534/genetics.110.115782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Jannink JL, Wu XL. Estimating allelic number and identity in state of QTLs in interconnected families. Genet Res. 2003;81:133–144. doi: 10.1017/S0016672303006153. [DOI] [PubMed] [Google Scholar]
- Jansen RC, Jannink JL, Beavis WD. Mapping quantitative trait loci in plant breeding populations: use of parental haplotype sharing. Crop Sci. 2003;43:829–834. doi: 10.2135/cropsci2003.0829. [DOI] [Google Scholar]
- Jiang C, Zeng ZB. Mapping quantitative trait loci with dominant and missing markers in various crosses from two inbred lines. Genetica. 1997;101:47–58. doi: 10.1023/A:1018394410659. [DOI] [PubMed] [Google Scholar]
- Kingsmore SF, Lindquist IE, Mudge J, Gesler DD, Beavis WD. Genome-wide association studies: progress and potential for drug discovery and development. Nat Rev Drug Discov. 2008;7:221–230. doi: 10.1038/nrd2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong AD, Gudbjartsson F, Saint J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Maaon G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K. A high resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms D, Brown P, Browne C, Eller M, Guill K, Harjes C, Kroon D, Lepak N, Mitchell SE, Peterson B, Pressoir G, Romero S, Rosas MO, Salvo S, Yates H, Hanson M, Jones E, Smith S, Glaubitz JC, Goodman M, Ware D, Holland JB, Buckler ES. Genetic properties of the maize nested association mapping population. Science. 2009;325:737–740. doi: 10.1126/science.1174320. [DOI] [PubMed] [Google Scholar]
- Nordborg M, Weigel D. Next-generation genetics in plants. Nature. 2008;456:720–723. doi: 10.1038/nature07629. [DOI] [PubMed] [Google Scholar]
- Sen S, Churchill G. A statistical framework for quantitative trait mapping. Genetics. 2001;144:805–816. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Zhang J, Zhou S, He R, Schaeffer M, Collura K, Kudrna D, Faga BP, Wissotski M, Golser W, Rock SM, Graves TA, Fulton RS, Coe E, Schnable PS, Schwartz DC, Ware D, Clifton SW, Wilson RK, Wing RA. The physical and genetic framework of the maize genome. PLoS Genet. 2009;5(11):e1000715. doi: 10.1371/journal.pgen.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi N, Shriner D. Advances in Bayesian multiple quantitative trait loci mapping in experimental crosses. Heredity. 2007;2007:1–13. doi: 10.1038/sj.hdy.6801074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Holland JB, McMullen MD, Buckler ES. Genetic design and statistical power of nested association mapping in maize. Genetics. 2008;178:539–551. doi: 10.1534/genetics.107.074245. [DOI] [PMC free article] [PubMed] [Google Scholar]