Abstract
Aldosterone is a major regulator of epithelial Na+ absorption and acts in large part through induction of the epithelial Na+ channel (ENaC) gene in the renal collecting duct. We previously identified Dot1a as an aldosterone early repressed gene and a repressor of ENaCα transcription through mediating histone H3 Lys-79 methylation associated with the ENaCα promoter. Here, we report a novel aldosterone-signaling network involving AF9, Dot1a, and ENaCα. AF9 and Dot1a interact in vitro and in vivo as evidenced in multiple assays and colocalize in the nuclei of mIMCD3 renal collecting duct cells. Overexpression of AF9 results in hypermethylation of histone H3 Lys-79 at the endogenous ENaCα promoter at most, but not all subregions examined, repression of endogenous ENaCα mRNA expression and acts synergistically with Dot1a to inhibit ENaCα promoter-luciferase constructs. In contrast, RNA interference-mediated knockdown of AF9 causes the opposite effects. Chromatin immunoprecipitation assays reveal that overexpressed FLAG-AF9, endogenous AF9, and Dot1a are each associated with the ENaCα promoter. Aldosterone negatively regulates AF9 expression at both mRNA and protein levels. Thus, Dot1a-AF9 modulates histone H3 Lys-79 methylation at the ENaCα promoter and represses ENaCα transcription in an aldosterone-sensitive manner. This mechanism appears to be more broadly applicable to other aldosterone-regulated genes because overexpression of AF9 alone or in combination with Dot1a inhibited mRNA levels of three other known aldosterone-inducible genes in mIMCD3 cells.
The epithelial sodium channel (ENaC)2 is a heteromultimeric protein composed of three partially homologous subunits (α, β, and γ) that is expressed in the apical membrane of salt-absorbing epithelia of kidney, colon, and lung where it constitutes the rate-limiting steps in active Na+ and fluid absorption. ENaC plays a major role in the regulation of salt homeostasis and blood pressure as evidenced by the fact that ENaC mutations are associated with genetic hypertensive and hypotensive diseases, such as Liddle's syndrome (1) and pseudohypoaldosteronism type 1 (2), and the fact that it is subject to tight and complex regulation by aldosterone. Aldosterone is a major regulator of epithelial Na+ absorption and acts in large part through ENaC induction in the renal collecting duct (3, 4). Aldosterone administration or hyperaldosteronism induced by a low-Na+ diet increases ENaCα gene transcription, without increasing β- or γ-subunit expression (5-9), and without a separate effect on ENaCα mRNA turnover (10) in this segment. Although ENaCα synthesis is believed to be the rate-limiting step in Na+ channel formation in the collecting duct, only limited information exists regarding the specific mechanisms governing transcriptional regulation of this gene, in particular epigenetic mechanisms exerting such controls.
Traditional models of aldosterone trans-activation of target genes, including ENaCα, have emphasized interaction of the liganded miner-alocorticoid receptor or glucocorticoid receptor (GR) with hormone response elements on target genes (11). The 5′-flanking regions of human, mouse, and rat ENaCα contain a highly conserved imperfect glucocorticoid-response element (GRE) that is essential for GR- or mineralocorticoid receptor-mediated trans-activation in the presence of glucocorticoid or aldosterone (10,12,13). Cell-specific expression and corticosteroid-mediated regulation of murine ENaCα requires the proximal 1.56-kb of 5′-upstream sequence that harbors the conserved GRE and multiple GRE half-sites (12). Before interacting with the cognate hormone response element, the ligand-receptor complex must have accessibility to the DNA, which is compacted in chromatin.
Recently, we characterized the murine disruptor of telomeric silencing alternative splice variant “a” (Dot1a), a histone H3 Lys-79 methyltransferase, and its expression in the mouse kidney and mouse inner medullary collecting duct cell line mIMCD3 (14). We further demonstrated that Dot1a is a novel aldosterone-regulated histone modification enzyme that is basally associated with chromatin at the ENaCα promoter and hypermethylates histone H3 Lys-79 associated with the ENaCα promoter, thereby repressing ENaCα transcription (15). We reported that aldosterone treatment down-regulates Dot1a expression and histone H3 Lys-79 methylation at the ENaCα promoter, thereby relieving the Dot1a-mediated repressive effect on ENaCα transcription (15). The effect of Dot1a was substantial, because RNA interference knockdown of Dot1a resulted in at least a tripling of endogenous ENaCα mRNA expression and of the activity of a stably integrated ENaCα promoter-reporter gene (15). Because mIMCD3 cells retain many of the phenotypic properties of the inner medullary collecting duct of kidney in vivo (16), respond to aldosterone (Ref. 15, and data hereafter), and express all of the components involved in the novel aldosterone signaling network reported here (see Fig. 1 and Refs 14,15, and 17), we have continued to use mIMCD3 cells as our model.
FIGURE 1. Isoform-specific interaction between Dot1 and AF9 in yeast two-hybrid assay.
Different versions of murine Dot1a and Dot1b were expressed as GAL4-DB fusions (1-14) and tested for their ability to interact with various GAL4-AD-AF9 fusions (15-17) in yeast strain AH109. Interaction between any two fusions was identified by the activation ofthethreereporters,resultingintheAde+ His+ and Mel1+ phenotype. Presence(+)or absence (−) of interaction is shown, with +, ++, and +++ indicating the weak, modest, and strong interactions, respectively. 1–12, Dot1a fusions: full-length (1), deletion of the putative leucine zipper, aa 576–597 (2), 2–95 (3), 96–1222 (4), 1223–1540 (5), 417–1222 (6), 417–478 (7), 479–659 (8), 660–828 (9), 829–972 (10), 1112–1222 (11), and 417–828 (12). 2–308 of Dot1b corresponding to 96–416 of Dot1a deleted 322–335 (13). Dot1b corresponding to 96–1222 of Dot1a deleted 322–335 (14). 15–17: AF9 fusions: full-length (15), 339–557 (16), 397–557 (17). Gray, aa 322–335 of Dot1a. Black, leucine zipper motifs. ND, not determined.
Because sequence-specific DNA binding activity has not been defined in any known Dot1 proteins, including Dot1a, a central mechanistic question in understanding this novel regulatory network is how Dot1a becomes associated with specific regions of the ENaCα promoter, and specifically, whether it binds to it or is recruited by a specific DNA-binding protein. The level of histone H3 Lys-79 methylation is low at some specific loci and high at other loci (18). Moreover, human Dot1L has been shown to interact with AF10, a mixed lineage leukemia (MLL) fusion partner involved in acute myeloid leukemia. The interaction between human Dot1L and AF10 results in mistargeting of human Dot1L to leukemia-relevant genes such as Hoxa9 through the DNA-binding domain of MLL retained in the MLL-AF10 fusion (19). These observations suggest that Dot1-interacting partners may directly or indirectly target Dot1-catalyzed histone H3 Lys-79 methylation to specific genomic loci. Accordingly, we sought to identify protein partners of Dot1a that might allow its association at the ENaCα promoter.
In this study, we report that AF9, a putative transcription factor, physically and functionally interacts with Dot1a to form a nuclear repressor complex, which, via direct or indirect binding to specific sites in the ENaCα promoter, regulates histone H3 Lys-79 methylation at these sites and represses basal transcription of ENaCα. Aldosterone down-regulates the Dot1 a-AF9 complex, at least in part by inhibiting Dot1 and AF9 expression, leading to targeted histone H3 Lys-79 hypomethylation and transcriptional activation of ENaCα. Furthermore, overexpression of AF9 or Dot1a in mIMCD3 cells inhibited expression of three other aldosterone-induced genes, connecting tissue growth factor, period homolog, and preproendothelin. Taken together, these data not only define a novel aldosterone signaling network governing ENaCα transcription, but also point to new aspects of function and regulation of AF9 and Dot1a-mediated histone H3 Lys-79 methylation that may be more broadly applied to other aldosterone target genes.
MATERIALS AND METHODS
Reagents
Antibodies against dimethyl histone H3 Lys-79, dimethylated histone H3 Lys-9, and acetylated H3 Lys-9 (Upstate), α-tubulin (Santa Cruz), EGFP (Clontech), and FLAG (Sigma), Lipofectamine™ 2000 reagent (Invitrogen), and aldosterone (Sigma) were purchased and used according to the manufacturer's instructions. The chicken anti-AF9 antibody (20), EGFP-Dot1a, (14), pcDNA-Dot1a, pGL3Zeocin-1.3 ENaCα, and anti-Dot1 antibody (15), and anti-H+ ,K+ -ATPase α2 sub-unit antibody (21) have been described. A peptide corresponding to mouse AF9 aa 421–441 was used to generate a new anti-AF9 antibody in rabbit (New England Peptide Inc., Gardner, MA). The antibody specifically recognized recombinant (data not shown) and endogenous AF9 from mIMCD3 cells or mouse kidneys (Fig. 1B and hereafter). Details of its characterization will be given in a related manuscript.3 This antibody was used in immunoprecipitation and chromatin immunoprecipitation (ChIP) assays, and the chicken AF9 antibody was used only in immunoblot assays. PCR fragments containing different versions of Dot1a or Dot1b were cloned into pGBKT7 (Clontech) at NdeI-EcoRI or pCMV-BD (Stratagene) at EcoRI-NotI for expression of GAL4 BD-Dot1a in yeast or mammalian cells, respectively. Various AF9 segments were amplified using the expressed sequence tag clone BC021420 (ATCC) as template and cloned into pGADT7 (Clontech), pGEX6P-1 (Amersham Biosciences), or pRFP-V5 (a modified pDsRed-monomer-C1, Clontech) at EcoRI-XhoI to generate GAL4 AD-AF9, GST-AF9, or red fluorescence protein-tagged fusions. A similar EcoRI-NotI PCR fragment containing the AF9 coding region was cloned into pCMV-AD (Stratagene) for mammalian expression of GAL4 AD-AF9 or into pME18S-HDAC2 (22,23) to replace the HDAC2 portion for expressing FLAG-AF9 fusions. pRFP-V5 was obtained by subcloning the annealing oligonucleotides (WZ814, TCGACCGCGGAGGCTTGAATTCCGGCTCGAGCGGCCGCGGTAC and WZ815, CGCGGCCGCTCGAGCCGGAATTCAAGCCTCCGCGG) into pDsRed-monomer-C1 at XhoI-KpnI to replace the multiple cloning site regions. To clone AF9 into pCDNA3.1(+), we used the annealing oligonucleotides (WZ748, GATCTCTTAAGATGGCTTTAGAATTCCGGCTCGAGCGGCCG CG; and WZ749, TCGACGCGGCCGCTCGAGCCGGAATTCTAAAGCCATCTTAAGA) to replace the AflII-SalI fragment of pCDNA3.1(+) to generate pcDNA3.1(+)-V1, which contains a modified multiple cloning site region and a Kozak sequence. Next, a fragment containing the AF9 coding region was amplified and cloned into pCDNA3.1-V1 at AflII-NotI to produce pcDNA-AF9. Two AF9-specific target sequences (RNAi number 1, GGTAGAAGAGTCCGGGTAC, encoding aa 72–78; RNAi number 2, GGCCCCCTGCTTCAGATTC, encoding aa 272–278) were annealed and cloned into pSilencer-2.1-U6-Hygro (Ambion) at BamHI-HindIII according to the manufacturer's instructions. The resulting plasmids were designated pAF9-RNAi#1 and pAF9-RNAi#2, respectively. BLAST search against mouse genome sequences confirms that these RNAi target sequences perfectly match with only AF9. All inserts in the constructs were verified by DNA sequencing.
Cell Culture, Aldosterone Treatment, Transient and Stable Transfections, and RNA Interference
mIMCD3 cells were maintained with Dulbecco's modified Eagle's medium/F-12 plus 10% fetal bovine serum. For aldosterone time course studies, cells were cultured in Dulbecco's modified Eagle's medium/F-12 plus 10% charcoal-stripped fetal bovine serum for at least 50 h before adding 1 μm aldosterone or 0.01% ethanol as vehicle control at different time points. All cells were then harvested at the same time point. Transient transfections were performed using the Lipofectamine™ 2000 reagent as described (14). To knockdown AF9 mRNA levels by RNA interference, pAF9RNA#1 and pAF9RNA#2 were transfected separately into mIMCD3 cells to generate stably integrated cell lines following selection with hygromycin (800 μg/ml). 15 to 20 colonies from each transfection were examined by immunoblot analysis with the chicken AF9 antibody for the efficiency of silencing AF9 expression, and those with the lowest AF9 expression were chosen for further study. pSilencer-2.1-U6-Hygro was employed similarly as a negative control.
Yeast and Mammalian Two-hybrid Screen, Immunoblot, Immunoprecipitation, GST Pulldown
The yeast two-hybrid screen was performed as described in our earlier work (24). Mammalian two-hybrid assays were conducted with the corresponding system according to the manufacturer's (Stratagene) instructions. Immunoprecipitation, immunoblot, and GST pulldowns were performed according to our published protocols (14, 23, 24) with the exception of a modified cell lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, pH 8.0, 1% Triton X-100,1% SDS, and a mixture of protease inhibitors) used to prepare whole cell lysates of mIMCD3 cells or mouse kidneys. The lysates were stored at −80°C for 16h, centrifuged at 14,000 × g for 10 min to remove the cell debris and possibly the majority of SDS in an Eppendorf centrifuge (Brinkmann Instruments, Inc.), and then diluted 10-fold for immunoprecipitation or GST pulldown assays. To demonstrate coimmunoprecipitation of the endogenous AF9 and Dot1 in the mIMCD3 or mouse kidney lysates, 4 ml of lysates (~1.2 mg of protein) were pre-cleared with 50 μl of protein A/G plus beads. The cleared lysates were then incubated in the presence of 50 μl of fresh protein A/G plus beads and 9 μg of rabbit antibody specific for AF9, Dot1, or H+ ,K+ -ATPase α2 subunit (as negative control) at 4 °C for 16 h. The beads were gently washed with 6 ml of phosphate-buffered saline for 5 min for 6 times. To prevent loss of the beads, all incubations, washes, and bead collections were carried out on ice by using Poly-prep chromatography columns or Bio-spin disposable chromatography columns (Bio-Rad). The immunoprecipitated proteins were eluted from the beads by incubation with 50 μl (200 ng/μl) of the corresponding peptide used to generate the AF9 or Dot1 antibodies overnight at 4 °C. To match the reaction conditions, a mixture of these two peptides at the equal molar ratio was used in the samples when anti-H+ ,K+ -ATPase α2 subunit antibody was used.
Deconvolution Immunofluorescence Microscopy
mIMCD3 cells were cultured in Dulbecco's modified Eagle's medium/F-12 plus 10% fetal bovine serum in 4-chamber polystyrene vessel tissue culture-treated glass slides (BD Falcon), transfected with pEGFP-Dot1a, pRFP-AF9, or in combination with Lipofectamine 2000. 24 h after transfection, cells were briefly washed with 1× phosphate-buffered saline twice, fixed in 1% paraformaldehyde, and stained with 4′,6-diamidino-2-phenylindole/Vectashield (Vector Laboratories). Image analysis was performed at the Multi-User Fluorescence Imaging and Microscopy Core Facility, Department of Pathology and Laboratory Medicine, University of Texas Medical School, Houston, TX. The protocols for image analysis were detailed in an earlier publication (25).
ChIP, Real-time Quantitative PCR (qPCR), or RT-qPCR, and Statistical Analysis
ChIP assays were performed using the Chromatin Immunoprecipitation Assay Kit (Upstate) according to the manufacturer's instruction with the following modifications: 1) the protein A-agarose-antibody-protein complex was washed additionally twice with modified lysis buffer (described above) to increase the specificity; 2) as in the immunoprecipitation assays described above, all incubation, washings, and bead collections were performed by using the disposable columns to avoid bead loss; and 3) the eluted DNA from the washed agarose-antibody-protein complex with elution buffer (1% SDS, 0.1 m NaHCO3) was reverse cross-linked and purified with the PCR Purification Kit (Qiagen), rather than by phenol/chloroform extraction as suggested by the manufacturer. Real-time qPCR of RT-PCR products for analysis of mRNA expression or of DNA isolated in ChIP assays, as well as statistical analysis were performed essentially the same as we previously detailed (15).
RESULTS
Dot1a Specifically Interacts with AF9 in Yeast and Mammalian Two-hybrid Assays
We previously cloned Dot1a, the murine homolog of human Dot1L, demonstrated its methyltransferase activity specific for histone H3 Lys-79, and identified it as an integral component of the aldosterone signaling network controlling ENaCα transcription (15). To identify Dot1a protein partners that may participate in this novel signaling network, we screened a mouse kidney cDNA library ligated to the GAL4 activation domain using Dot1a as bait in a yeast two-hybrid assay. These studies identified a specific interaction between Dot1a and the protein encoded by AF9 (accession number BC021420; Fig. 1). The cDNA clone isolated from yeast encodes only aa 339–557 of AF9, which was fused in-frame with the GAL4 activation domain. Interestingly, AF9 was originally identified from human leukemia patients containing a t(9,11)(p22;q23) translocation (27), resulting in MLL-AF9 fusion harboring hAF9 aa 376–568 (corresponding to mouse AF9 aa 365–557). To test more rigorously the Dot1a-AF9 interaction and to further define the AF9 domain responsible for the interaction, GAL4 AD fusions containing full-length AF9 or aa 397–557 were generated and tested for interaction with GAL4 BD-Dot1a fusion in the yeast two-hybrid assay. As expected, full-length AF9 interacted with Dot1a (Fig. 1). Moreover, AF9 396–557 displayed an even stronger interaction with Dot1a than did the full-length of AF9 in this assay, possibly because of the presence of a repression activity in AF9 outside this C-terminal fragment.
Previous studies established that human Dot1L 1–416 and mouse Dot1a 1–478 possess histone H3-Lys-79-specific methyltransferase activity (28, 14). However, no functions have been assigned to the long C-terminal portion. To map the domains in Dot1a responsible for interacting with AF9, we divided Dot1a into 13 fragments, some of which were determined by their presence in Dot1a, but absence in Dot1b. As AF9 397–557 exhibited a stronger interaction with Dot1a, it was used to test all of these Dot1a fragments. These allowed us to identify that the methyltransferase domain (aa 1–416), the putative leucine zipper domain (aa 576–597), and the very C-terminal part (aa 1112–1540) of Dot1a apparently are not necessary for the interaction. On the contrary, Dot1a aa 479–659 and 829–972 are capable, but less competent compared with the full-length of Dot1a, to mediate the interaction with AF9 (Fig. 1). These results therefore define for the first time an AF9-interacting function in the internal portion of Dot1a.
To demonstrate further the specific interaction in vivo of Dot1a with AF9, we carried out mammalian two-hybrid assays. As in the yeast two-hybrid assay, AF9 was expressed as a GAL4-AD fusion and examined for its ability to activate a GAL4-dependent luciferase construct in the presence or absence of a GAL4-BD fusion construct harboring Dot1a. Coexpression of both Dot1a and AF9 fusions resulted in a greater than 200-fold activation of the luciferase activity compared with each alone (Fig. 2A). The above results, in combination with the GST pulldown, coimmunoprecipitation, and colocalization assays shown in Fig. 2, allow us to conclude that Dot1a interacts specifically with AF9 in vitro and in vivo.
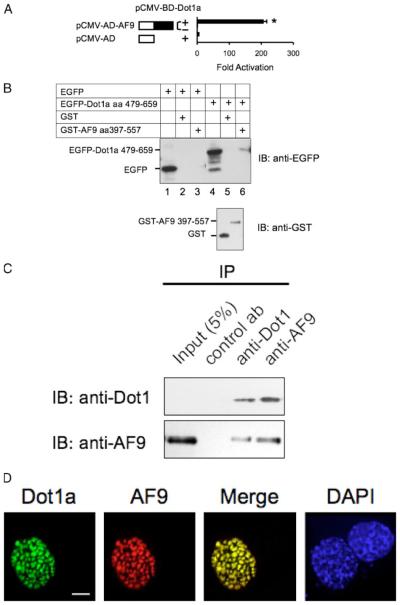
FIGURE 2. Dot1a and AF9 interact in vitro and in vivo.
A, mammalian two-hybrid assay confirming the in vivo interaction of Dot1a and AF9. AF9 was expressed as GAL4 activation domain fusion, and tested to activate a GAL4-dependent luciferase reporter in the presence (+) or absence (−)of GAL4 binding domain fused to Dot1a in mIMCD3 cells. The bars represent the average -fold of activation from three independent experiments. *, p<0.05 versus other bars. B, GST pulldown assay showing specific domains in Dot1a and AF9 responsible for the interaction. GST and GST-AF9-(397–557) fusion were purified from E. coli and incubated with whole cell lysates of mIMCD3 cells expressing EGFP or EGFP-Dot1a-(479–659), as indicated. Input of the lysates (10%, lanes 1 and 4) and proteins bound to glutathione-Sepharose 4B beads wereexamined by immunoblot (IB) analysis with the anti-EGFP antibody. The inputs of GST and GST-AF9 fusion were analyzed similarly with the anti-GST antibody (lower panel). C, co-immunoprecipitation assay demonstrating that the endogenous Dot1 and AF9 proteins in mIMCD3 cells are present in the same protein complex. Whole cell lysates of mIMCD3 cells were immunoprecipitated (IP) with rabbit antibodiesspecific for Dot1, AF9, or H+,K+ -ATPase α2 subunit (as a negative control antibody) as detailed under “Materials and Methods.” Immunoprecipitated proteins were eluted from the protein A/G-agarose beads and subjected to immunoblot analysis in parallel with the antibodies as indicated. D, deconvolution microscopy revealing nuclear colocalization of Dot1a and AF9. mIMCD3 cells were cotransfected with constructs encoding EGFP-Dot1a and red fluorescent protein-tagged AF9 (RFP-AF9) and examined by deconvolution microscopy as detailed under “Materials and Methods.” DAPI, 4′,6-diamidino-2-phenylindole. Scalebar,3 μm.
Dot1a Binds AF9 in the GST Pulldown Assay
To confirm biochemically the findings revealed in the yeast two-hybrid assay that Dot1a interacts with AF9, and that aa 397–557 in AF9 and aa 479–659 in Dot1a are important for the interaction, we performed GST pulldown assays. We initially intended to express and purify the GST fusion containing full-length AF9 from Escherichia coli and test its ability to retain an EGFP-tagged fusion harboring full-length Dot1a from cell lysates of pEGFP-Dot1a-transfected mIMCD3 cells. This approach was unsuccessful because we never obtained the GST-AF9 fusion, possibly because of its toxicity, insolubility, or both. Furthermore, whereas EGFP-tagged full-length Dot1a was easily detected by epifluorescence or deconvolution microscopy in mIMCD3 cells (Fig. 2D), the fusion protein was frequently detected as multiple smaller fragments by immunoblot analysis, presumably resulting from post-translational cleavage or extensive degradation during extraction. Therefore, the GST pulldown assay was performed with GST-AF9-(397–557) purified from E. coli and whole cell lysates derived from mIMCD3 cells transiently transfected with a construct encoding EGFP-Dot1a-(479–659). GST and EGFP empty vectors were included as negative controls. As shown in Fig. 2B, whereas EGFP failed to interact with the AF9 fusion (lane 3), EGFP-Dot1a-(479–659) bound the AF9 fusion (lane 6), but not GST alone (lane 5). The observed results were not because of differences in the expression levels of EGFP and EGFP-Dot1a fusions (Fig. 2B, lanes 1 and 4) nor the amounts of GST and GST-AF9 fusions used in the assay (Fig. 2B, lower panel), as they were all comparable.
Dot1a Coimmunoprecipitates and Colocalizes with AF9
Next, we determined that Dot1a and AF9 interact at the endogenous protein level. Three aspects were considered in performing the coimmunopre-cipitation experiments. 1) Our previous studies demonstrated that Dot1a mRNA was the principal isoform expressed in mIMCD3 cells, mouse kidney, and all other tissues examined. Therefore, mIMCD3 cells and mouse kidneys were used to prepare the whole lysates. 2) Because the existing AF9 antibody (raised in chicken (20)) did not work well in immunoprecipitation assays, we developed a new AF9 antibody with a peptide corresponding to mouse AF9 aa 421–441. This antibody specifically recognizes a recombinant GST-AF9 fusion expressed in E. coli (data not shown) and the endogenous AF9 from mIMCD3 cells or mouse kidneys (Fig. 1B and hereafter). 3) AF9 has an expected molecular mass of ~60 kDa and comigrates with IgG heavy chain at ~50 kDa in immunoblots. To increase the specificity and eliminate signals from IgG, we eluted the immunoprecipitated proteins from the protein A/G beads with the corresponding peptide used to generate the AF9 or Dot1 antibodies. A mixture of the two peptides at an equal molar ratio was applied to the samples when the negative control antibody (rabbit anti-H+ ,K+ -ATPase α2 subunit (21)) was used. In this way, all reactions were carried out under the same or very similar conditions except for the use of the antibodies. As shown in Fig. 2C, although Dot1 protein was undetectable in the mIMCD3 cell lysate (first lane, upper panel), enrichment of the Dot1a protein, however, by immunoprecipitation with the Dot1 antibody allowed the detection of the Dot1 protein. Dot1 protein was also coimmunoprecipitated by the AF9 antibody. Reciprocally, AF9 was immunoprecipitated by the AF9 antibody and coimmunoprecipitated by the Dot1 antibody. In either case, neither Dot1 nor AF9 were immunoprecipitated by the same amount of rabbit anti-H+ ,K+ -ATPase α2 subunit antibody under the mild washing conditions as detailed under “Materials and Methods.” Similar results were obtained when mouse kidney lysate was used (data not shown).
Interacting proteins with physiological relevance should colocalize within the cell. Unfortunately, the distribution and colocalization of the endogenous AF9 and Dot1 proteins in mIMCD3 cells was not successfully determined because the chicken and rabbit anti-AF9 antibodies were not effective in immunostaining this cell line. Accordingly, EGFP-Dot1a was co-expressed with red fluorescent protein-tagged AF9 by transient transfection of the corresponding constructs into mIMCD3 cells. As shown in Fig. 2D, Dot1a and AF9 displayed nuclear colocalization and colocalized at large discrete foci. The neighboring, unsuccessfully transfected cell identified by 4′,6-diamidino-2-phenylindole staining of the nucleus showed no fluorescence for the two proteins (Fig. 2D). Furthermore, cells transfected with a single construct only displayed the corresponding fluorescence (data not shown), demonstrating the specificity of each fluorescence fusion protein. Similar results were obtained independently with 293T or Mz-ChA-1 cholangiocarcinoma cells derived from human gall bladder (data not shown).
Overexpression of AF9 with Dot1a Synergistically Inhibits Expression of ENaCα and Other Aldosterone Up-regulated Genes in mIMCD3 Cells
Our previous studies demonstrated that Dot1a represses ENaCα transcription by mediating histone H3 Lys-79 methylation associated with the ENaCα promoter in a methyltransferase-dependent manner, and that aldosterone relieves this Dot1a-mediated repression by down-regulating Dot1a expression, leading to ENaCα trans-activation (15). Because Dot1a, the major isoform of Dot1 in mIMCD3 cells interacts with AF9, we first sought to test directly the hypothesis that AF9 also participates in the repression of ENaCα transcription. mIMCD3 cells were transiently transfected with a control plasmid or pFLAG-AF9 and examined by real-time RT-qPCR for expression of ENaCα mRNA, with housekeeping genes β-actin (not shown) or GAPDH as a control. Expression of FLAG-AF9 was monitored by immunoblot analysis with the anti-FLAG antibody and anti-α-tubulin as control (Fig. 3A). Similar to Dot1a overexpression (15), AF9 overexpression suppressed ENaCα mRNA levels to ~30% (p < 0.05) of that of the vector control (Fig. 3A).
FIGURE 3. Overexpressed AF9 down-regulates ENaCα expression by mediating histone H3 Lys-79 methylation of the ENaCα promoter in mIMCD3 cells.
A, real-time RT-qPCR showing that overexpression of AF9 decreases the expression of endogenous ENaCα and other aldosterone up-regulated genes. Total RNAs from mIMCD3 cells transiently transfected with pCMV500 (Vec)orpFLAG-AF9(AF9)wereanalyzed by real-time RT-qPCRto determine the mRNAlevelsofthegenesindicated and normalizedto that of GAPDH from the same sample. GAPDH levels were invariant under the different conditions. FLAG-AF9 expression and equal loading were monitored by immunoblot analysis with the anti-FLAG and anti-α-tubulin antibodies. n = 3. B, luciferase assay demonstrating that overexpression of AF9 and Dot1a synergistically repressed the expression of the ENaCα promoter-luciferase construct. An mIMCD3-derived cell line harboring a stably transfected pGL3Zeocin-1.3ENaCα was transiently transfected with pcDNA3.1 (Vec), pcDNA-AF9, and/or pcDNA-Dot1a along with Renilla luciferase reporter pRL-SV40 as an internal control. Firefly luciferase activity of eachsamplewas normalized to its Renilla luciferaseactivity. The firefly luciferaseactivity of the vector-transfected cells was designated as 1 and utilized to determine the relative level and the significance of the other samples. *, p < 0.05 versus Vec, n = 6. C, diagram of the ENaCα promoter. Fragments designated Ra and R0–R3 were shown along with their relative position to the major transcription start site (+1) of ENaCα. The filled circle and rectangles represent the putative GRE site (−811) and GRE half-sites (−983, −416, −325, −241 and −234),respectively. D, ChIP assay showing that AF9 was associated with and promoted histone H3 Lys-79 methylation of the ENaCα promoter. Similarly transfected mIMCD3 cells as in A were subjected to ChIP assay with the indicated antibodies. Immunoprecipitated DNA was analyzed by real-time qPCR with primers designed to specifically amplify Ra and R0–R3 as shown in C. The relative H3 Lys-79 methylation was defined as ChIP efficiency, i.e. the immunoprecipitated amount of material to that of the input sample, as determined by real-time qPCR. The relative H3 Lys-79 methylation of R0 from the vector-transfected cells was set to 1 and used to calculate those of all other samples accordingly. *, p < 0.05 versus vector control; n = 3. Representative agarose gel analyses of the final qPCR products were shown to verify the quantification determined by real-time qPCR and specificity for each sample.
It has been reported that connecting tissue growth factor, period homolog, and preproendothelin were also up-regulated upon aldosterone addition in mIMCD3 cells, as evidenced by multiple assays using GAPDH as an internal control (17). To test whether AF9 represses these aldosterone-inducible genes, the same cDNA samples examined above were further analyzed for expression of these genes. As shown in Fig. 3A, these aldosterone-inducible genes, but not GAPDH, were also down-regulated about 2–5-fold at the mRNA level when AF9 was overex-pressed. Overexpression of EGFP-Dot1a also decreased expression of these genes to various degrees (1–9-fold, data not shown) in mIMCD3 cells.
To verify independently that AF9 represses ENaCα expression by modulating its promoter activity, we cloned AF9 into a modified pcDNA3.1 vector to generate pcDNA-AF9 for expression of untagged AF9. Transfection of pcDNA-AF9 or pcDNA3.1 as control into a mIMCD3 cell line harboring the stably transfected ENaCα promoter-luciferase construct (15) was then performed, followed by luciferase assay. As shown in Fig. 3B, overexpression of AF9 resulted in an ~35% reduction in the luciferase activity compared with the vector control. Moreover, cotransfection of AF9 and Dot1a elicited an even more dramatic effect (~80% reduction) on luciferase activity than either alone (Fig. 3B). These results indicate that Dot1a and AF9 function together to repress ENaCα expression by down-regulating ENaCα promoter activity.
AF9 Is Associated with the ENaCα Promoter and Modulates Histone H3 Lys-79 Methylation at the ENaCα Promoter
We have reported that reduction of Dot1a expression by either aldosterone treatment or Dot1a-specific RNA interference inhibits the level of histone H3 Lys-79 methylation associated with the R0, R1, and R3 subregions in the ENaCα promoter (15), whereas overexpression of Dot1a results in hypermethylation in these subregions (15). Accordingly, we hypothesized that AF9 directly or indirectly binds the ENaCα promoter, and through its interaction with Dot1a, represses ENaCα transcription by increasing the local level of chromatin-associated histone H3 Lys-79 methylation at the ENaCα promoter. To test directly this hypothesis in vivo, mIMCD3 cells were transiently transfected with the same control plasmid or FLAG-AF9 as above, and examined by ChIP coupled with real-time qPCR using methods we previously reported (15). As shown in Fig. 3C, overexpressing AF9 caused a statistically significant 2–3-fold increase in histone H3 Lys-79 methylation in each of the R0–R3 regions examined compared with the vector control. However, no significant change in histone H3 Lys-79 methylation was detected in Ra, which is 5′ upstream of R0. Therefore, it is less likely that overexpression of AF9 elicited a global change in H3 Lys-79 methylation. Furthermore, ChIP assays with antibodies recognizing histone H3-acety-lated Lys-9 or the N-terminal tail of histone H3 run in parallel experiments uncovered no obvious differences under the various transfection conditions over the entire vicinity of the ENaCα promoter examined (Fig. 3D), suggesting that the changes in histone H3 Lys-79 methylation were specific and not the result of a generalized effect on histones. Furthermore, ChIP with IgG yielded background signals that were undetectable by agarose gel analysis (Fig. 3D).
To test directly whether AF9 associates with any of the Ra and R0–R3 subregions of the ENaCα promoter, ChIP with the anti-FLAG antibody was also performed, followed by real-time qPCR. Consistent with an increase in H3 Lys-79 methylation in R0–R3, but not in Ra following AF9 overexpression, FLAG-AF9 was efficiently immunoprecipitated by the anti-FLAG antibody and found in the chromatin associated with all four subregions (R0–R3), but not with Ra. Moreover, only background signals from the vector-transfected cells were detected, demonstrating the specificity of the anti-FLAG antibody (Fig. 3D). These data along with additional ChIP presented below strongly suggest that AF9 is associated with the ENaCα promoter. Moreover, the data indicate that AF9 cooperates in regulating the local chromatin structure by modulating histone H3 Lys-79 methylation through its interaction with Dot1a, thereby favoring ENaCα transcription.
RNA Interference-mediated Knockdown of AF9 Expression Increases Expression of Endogenous ENaCα and the ENaCα Promoter-luciferase Construct
To confirm further that AF9 represses ENaCα expression by modulating histone H3 Lys-79 methylation through its interaction with Dot1a, we employed RNA interference to knockdown specifically AF9 mRNA and monitored its effects on expression of endogenous ENaC and the ENaCα promoter-luciferase construct. Two AF9 RNAi constructs (pAF9-RNAi#1 and pAF9-RNAi#2) were constructed and transfected into mIMCD3 cells to establish stable cell lines as detailed under “Materials and Methods” and schematically represented in Fig. 4A. Immunoblot analysis demonstrated that AF9 protein levels were significantly reduced to 45 and 12%, respectively, in the cell lines derived from cells transfected with pAF9-RNAi#1 or pAF9-RNAi#2, compared with the control cells transfected with a similar construct harboring a nonspecific RNAi sequence (Fig. 4B). The expression of the housekeeping gene α-tubulin was not measurably affected by the transfections, providing evidence of the specificity of the RNAi targeting (Fig. 4B). Real-time RT-qPCR revealed that ENaCα mRNA expression was 7-fold greater in the cells transfected with pAF9-RNAi#1 than that in the control cells. ENaCα mRNA abundance was more dramatically increased (more than 8-fold) by more efficient knockdown of AF9 in pAF9-RNAi#2-transfected cells (Fig. 4C). In either case actin mRNA levels were relatively constant (Fig. 4C). Furthermore, transient transfection of the ENaCα promoter-luciferase construct into these cell lines also resulted in 2–3-fold higher levels of luciferase activity in AF9-targeted cells than the non-targeted cells (Fig. 4D).
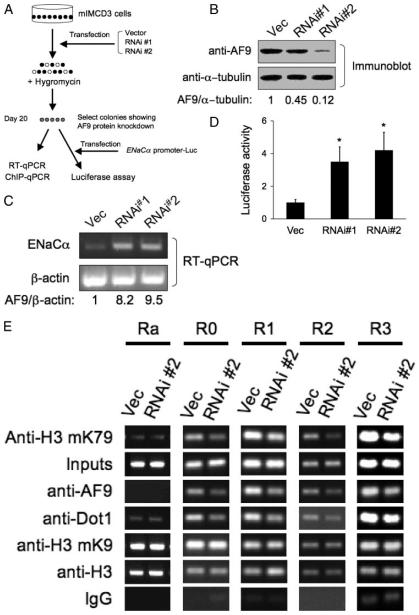
FIGURE 4. RNA interference knockdown of AF9 expression impairs AF9, Dot1, and histone H3 Lys-79 methylation associated with the ENaCα promoter and increases expression of endogenous ENaCα and the activity of the ENaCα promoter-luciferase construct.
A, diagram of the experiment procedure. B, immunoblots showing knockdown of AF9. Three independently mIMCD3 cell lines stably transfected with pSilencer 2.1-U6-hygro negative control (Vec), pAF9-RNAi#1 (RNAi#1), or pAF9-RNAi#2 (RNAi#2) were examined by immunoblotting with the chicken AF9 antibody or the anti-α-tubulin antibody. The normalized AF9 level (to the α-tubulin abundance) is shown with the AF9 of the vector-transfected cells arbitrarily setto 1. C, RT-qPCR showing that knockdown of AF9 increases ENaCα mRNA levels. The same cell lines as in B were examined with realtime RT-qPCR for expression of ENaCα or β-actin as control. n = 3. D, the same cells as in B were transiently transfected with pGL3Zeocin-1.3ENaCα, followed by luciferase assay as described in the legend to Fig. 3B. *, p < 0.05 versus Vec, n = 3. E, ChIP assay showing that knockdown of AF9 decreased occupancy of AF9, Dot1, and histone H3 Lys-79 methylation in the ENaCα promoter. The same cells as in B were subjected to ChIP assay as in Fig. 3D, except for the use of the rabbit antibodies specific for Dot1 or AF9. n = 3.
AF9 Knockdown Is Correlated with Reduction of AF9, Dot1, and Histone H3 Lys-79 Methylation Associated with the ENaCα Promoter
To determine whether the increased ENaCα mRNA expression in AF9 knockdown cells is accompanied with corresponding changes in the abundance of the endogenous AF9 and Dot1 proteins and methylated histone H3 Lys-79 associated with the ENaCα promoter, we repeated ChIP assay with the mIMCD3 cells that exhibited a more efficient knockdown of AF9 by RNAi#2. As shown in Fig. 4E, compared with the control cells, RNAi#2-transfected cells showed a 2–5-fold decrease of AF9, Dot1, and thus histone H3 Lys-79 methylation associated with all R0-R3 subregions, but not in Ra. Only minimal Dot1a association with Ra was observed, possibly because Dot1a, like its counterpart hDot1L (29), possesses a low level of non-sequence-specific DNA binding activity. However, minimal association of Dot1a with Ra was not changed by overexpression of AF9, which apparently was not associated with the subregion. Furthermore, no measurable changes were observed for the total histone H3 or dimethylated H3 Lys-9 in all five regions examined between the two cell lines, indicating that knockdown of AF9 elicited a specific rather than a global effect on chromatin structure. In brief, the data strongly support the argument that AF9 represses ENaCα expression and does so at least partially through interacting with Dot1 to mediate histone H3 Lys-79 methylation associated with the ENaCα promoter in mIMCD3 cells.
Aldosterone Down-regulates AF9 mRNA and Protein Expression in mIMCD3 Cells
Our previous work (15) indicated that aldosterone down-regulates Dot1a mRNA levels and reduces histone H3 Lys-79 methylation in bulk histones in mIMCD3 cells. Because AF9 interacts with Dot1a and represses ENaCα expression at least partially through its interaction with Dot1a, we examined whether aldosterone also regulates AF9 expression in these cells. mIMCD3 cells were starved in charcoal-stripped serum for at least 50 h, followed by addition of vehicle (ethanol) or 1 μM aldosterone at different time points (Fig. 5A) before harvest. As a result, a time course of aldosterone treatment over 1–7 h was obtained with all cells collected at the same time point. Cell lysates from these cells were used to prepare identical immunoblots probed with the anti-AF9 or anti-α-tubulin antibodies as a loading control. Representative immunoblots are shown in Fig. 5A. Aldosterone-treated cells exhibited lower levels of AF9 protein at 1 h of treatment, compared with that of the corresponding vehicle-treated cells (Fig. 5A, lane 1 versus 2). This response was progressively attenuated and eventually disappeared at the later time points. Quantitative analysis of the corresponding bands by densitometry indicated that aldosterone significantly (p < 0.05) lowered the AF9 level at both the 1- and 1.5-h time points (data not shown). As further evidence of this response to aldosterone, real-time RT-qPCR revealed that aldosterone-treated mIMCD3 cells exhibited almost three times lower AF9 mRNA levels at the 1.5-h time point than the control (Fig. 6B). Taken together, these results along with our previous study (15) demonstrate that aldosterone coordinately down-regulates Dot1a and AF9 expression.
FIGURE 5. Aldosterone down-regulates AF9 expression at both mRNA and protein levels.
A, immunoblots showing that aldosterone dynamically regulated AF9 protein expression in mIMCD3 cells. mIMCD3 cells were cultured in Dulbecco's modified Eagle's medium/F-12 plus 10% charcoal-stripped fetal bovine serum for at least 50 h, then treated with vehicle (ethanol, –) or 1 μM aldosterone (+) for the indicated times. The whole cell lysates were analyzed by immunoblot assay with the antibodies specific for AF9 antibody or α-tubulin. B, real-time RT-qPCR demonstrating that aldosterone decreased AF9 mRNA expression. Total RNAs from the mIMCD3 cells treated as in A for 1.5 h were analyzed by real-time RT-qPCR for expression of AF9 as described in the legend to Fig. 3A. n = 3.
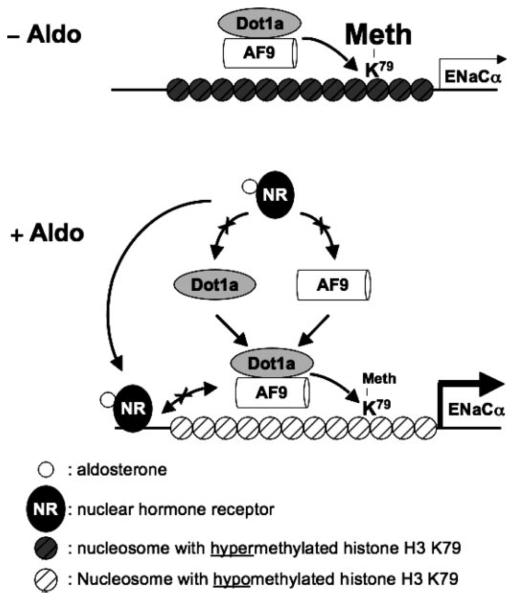
FIGURE 6. Hypothetical model for aldosterone-sensitive repression of ENaCα transcription by Dot1a-AF9 complex modulating histone H3 Lys-79 hypermethylation associated with the ENaCα promoter.
Under basal conditions, Dot1a and AF9 form a nuclear complex that directly or indirectly binds specific sites of the ENaCα promoter, leading to hypermethylation of histone H3 Lys-79 and repression of ENaCα (A). Aldosterone (blank circles) stimulates ENaCα transcription by regulating two different complexes. In the classical pathway, aldosterone binds and activates the nuclear receptors (NR) that are either GR and/or mineralocorticoid receptor homo- or heterodimer to bind GRE for transactivation of ENaCα. In a parallel pathway, aldosterone down-regulates the amount of Dot1a-AF9 complex by reducing Dot1a (15) and AF9 expression, presumably via NR-dependent (shown) or NR-independent mechanisms (not shown) to limit their availability, leading to histone H3 Lys-79 hypomethylation at the specific subregions and release of repression of ENaCα. In either case, AF9-free Dot1a binds DNA nonspecifically and catalyzes histone H3 Lys-79 methylation throughout the genome at the basal level (not shown). The putative counter-interaction between the NR-aldosterone complex and the Dot1a-AF9 complex is also shown and may tune the ultimate level of ENaCα transcription. Similar mechanisms may be applicable to other aldosterone-regulated genes.
DISCUSSION
Aldosterone is known to enhance ENaCα transcription and thereby increase renal tubular Na+ reabsorption. The signaling cascade linking aldosterone to transcriptional activation of ENaCα is incompletely defined, but available data indicate the operation of both hormone response element-dependent and -independent pathways. Our recent report (15) indicated that the actions of aldosterone on the chromatin-modifying enzyme Dot1a and the association of this enzyme with the ENaCα promoter mediate transcriptional regulation. In this report, we have added AF9, with its ability to associate with the ENaCα promoter and to bind Dot1a and to be down-regulated in a coordinate manner with Dot1a in response to aldosterone, as a key mechanistic component to our model. Specifically, we have shown that, by multiple independent assays (yeast and mammalian two-hybrid assays, GST pulldown assay, coimmunoprecipitation and colocalization analyses, and ChIP assays), Dot1a interacts specifically with AF9 in vitro and in vivo in renal collecting duct cells. Knockdown of AF9 mRNA expression by RNA interference increased expression of endogenous ENaCα and the activity of an ENaCα promoter-luciferase construct in mIMCD3 cells, a phenotype identical with that of aldosterone treatment, which down-regulates both Dot1a and AF9 expression. In contrast, overexpression of AF9 decreased ENaCα mRNA levels and expression of the ENaCα promoter-luciferase reporter. We found in ChIP assays that FLAG-tagged AF9 associated with four of five defined subregions of the ENaCα 5′-flanking region and enhanced histone H3 Lys-79 methylation associated with these sites in vivo. In the region (Ra) where AF9 association was apparently absent, modulating AF9 expression by transient transfection of the FLAG-AF9 constructor RNAi-mediated AF9 knockdown did not elicit corresponding changes in histone H3 Lys-79 methylation. Moreover, endogenous Dot1 and AF9 were associated with the ENaCα 5′-flanking region. Finally, overexpression of Dot1a or AF9 resulted in significant reductions of mRNA levels of three other aldosterone-inducible genes, but not GAPDH. Our findings that AF9 associates with some subregions of the ENaCα promoter and is correlated with histone H3 Lys-79 hypermethylation in these subregions directly links Dot1a-AF9-mediated repression of ENaCα to the changes in the chromatin structure of its promoter. Therefore, we conclude that AF9 is a novel non-histone protein that physically and functionally interacts with Dot1a, potently represses ENaCα transcription, directs local histone H3 Lys-79 methylation through its interactions with Dot1a and the ENaCα promoter, and is down-regulated by aldosterone. Based on these observations and findings reported in the literature, we propose a new aldosterone signaling network important for governing transcription of ENaCα and other aldosterone target genes (Fig. 6).
AF9 was originally identified from human leukemia samples containing a t(9,11)(p22;q23) translocation (27). It isabundantly expressed in kidney and multiple other tissues (30). To date, more than 30 genes have been identified as fusion partners of the mixed lineage leukemia (MLL) gene to generate in-frame fusion proteins that are thought to cause leukemogenesis (31). MLL-AF9 is one of the most common forms (32). AF9 interacts with AF4, another MLL fusion partner (20), specific isoforms of BCL-6 corepressor (BCoR) (32), and MPc3, a member of the Polycomb group multiprotein complexes involved in gene silencing by modifying chromatin structure (30). BCoR suppresses AF9 trans-activation activity (32), and is known to associate with histone deacetylases. MPc3 can simultaneously bind AF9 and RING1 (30), and in turn RING1 may interact with other Polycomb group proteins even when bound to MPc3. Thus AF9 may acquire a dominant-negative function by recruitment of, or to, corepressors or chromatin modifying proteins. Apart from these observations, little is known about the AF9 function in normal cells.
Histone H3 Lys-79 methylation has long been considered to be a conserved hallmark of active chromatin regions (18), although even in yeast, the organism most extensively studied in this regard, the role of histone H3 Lys-79 methylation in association with the Sir (silent information regulator) proteins, and thus in gene silencing is still controversial (18,33,34). Our previous (15) and current data support the alternative view that hypermethylation of histone H3 Lys-79 can result in repression, rather than activation of transcription in mammalian cells. Whether histone H3 Lys-79 methylation results in gene activation or repression may depend on the patterns of other histone modifications leading to either the association or the dissociation of distinct regulatory proteins. For example, Dot1-mediated methylation of histone H3 Lys-79 in vivo strongly depends on the intact nucleosomal structure and Rad6-dependent ubiquitination of histone H2B at Lys-123 (35), an example of trans-histone regulation between modifications on different histones. Rad6 has been shown to function, through its H2B ubiquitination activity, as a transcriptional repressor in silent chromatin at the repressible ARG1 gene (36-40). In addition, Dot1 has been shown to interact with the silencing protein Sir2, an NAD-dependent histone deacetylase that is thought to maintain the very low level of histone acetylation characteristic of silenced loci (36). SIRT1, the mammalian homolog of yeast Sir2, is widely expressed among tissues, including kidney, and has been demonstrated to deacetylate Lys-9 and Lys-14 of histone H3 and Lys-16 of H4 in the context of synthetic, acetylated peptides (41). In addition to its role as a histone deacetylase, SIRT1 deacetylates non-histone substrates, and directly interacts with p300/CBP-associated factor and CBP (CREB-binding protein)/p300 to inhibit the acetylation status and activity of these enzymes. SIRT1 deacetylation of p53, NF-κB p65, p300, and forkhead transcription factors inhibits transcription regulated by these factors (42-44). Murine SIRT1 is also involved in the epigenetic silencing of chromatin states mediated by the Polycomb group multiprotein complexes and is physically associated with a complex containing the E(Z) histone methyltransferase (44). Given the ability of Dot1 and AF9 each to bind others regulatory proteins, it is intriguing to speculate that AF9 binds in a sequence-specific manner to specific regions of the ENaCα promoter and there serves as a nidus for the assembly of a repressor complex that includes directly bound Dot1a and a specific spatial arrangement of co-regulatory proteins known to associate with AF9 (such as MPc3 or BCoR) or Dot1a (SIRT1), leading to Dot1a-mediated histone H3 Lys-79 methylation, and SIRT1-mediated histone deacetylation at this locus and transcriptional repression of ENaCα. Our current efforts are directed at examining this hypothesis.
The present study also indicates that Dot1a-mediated histone H3 Lys-79 methylation can occur in a targeted manner. Dot1 has been generally considered to methylate histone H3 Lys-79 in a non-targeted manner (18, 33, 38), despite the fact that a non-random pattern of histone H3 Lys-79 methylation was observed throughout the genome in both yeast and mammalian cells (18, 33). It has been proposed that yeast DNA-binding proteins such as Rap1 recruit silencing proteins (Sir) to the regions containing undermethylated histone H3 Lys-79 and Sir binding then blocks histone H3 Lys-79 methylation. On the contrary, regions with hypermethylated histone H3 Lys-79 are restricted from Sir protein binding, which subsequently leads to increased histone H3 Lys-79 methylation and even weaker Sir binding (18). However, the pattern and regulation of histone H3 Lys-79 methylation in mammalian cells are largely unclear. Dot1a may be preferentially recruited or retained at particular gene control regions recognized by AF9 or other Dot1-interacting proteins, leading to local hypermethylation. In the case of ENaCα, AF9 complexed with Dot1a might bind R0–R3 regions of the ENaCα promoter directly or indirectly at specific sites, nucleate the hypermethylation of histone H3 Lys-79, and then spread it into the neighboring nucleosomes. It should be noted that whether AF9 possesses sequence-specific DNA binding activity and whether R0-R3 regions of ENaCα promoter contain any AF9 binding sites remains to be determined. Many genes, including AF9, fused in-frame to MLL are identified in translocations associated with both acute lymphoblastic and acute myelogenous leukemia (31). These genes encode proteins that collectively do not share a common structural motif or biochemical function. However, we identified three MLL fusion partners that specifically interacted with Dot1a in our yeast two-hybrid screen, including AF9 described here and AF10 that has been reported (19). The sequence regions responsible for the interactions are all present in the corresponding MLL fusions, suggesting that mis-targeting of the Dot1 methyltransferase may represent a shared mechanism for these MLL fusions to give rise of leukemia.
Identifying Dot1-interacting partners and defining their DNA binding activity and specificity should provide new clues about how Dot1-mediated histone modifications are regulated in normal cells. In addition, no functions have been assigned to the extended C-terminal fragment that is presented in mouse and human Dot1 proteins (14,28), butabsent in the yeast counterpart (38). Because the domains important for the interaction (Dot1aaa 479–659 and 829–972) are also present in human Dot1L, but almost missing in yeast Dot1, and BLAST searches with the AF9 sequence against the yeast genome do not yield any significant matches, it is unlikely that a similar Dot1-AF9 interaction exists in yeast. The sequence differences between mammalian and yeast Dot1 proteins may provide a molecular basis for distinct mechanisms regulating histone H3 Lys-79 methylation.
Acknowledgment
We thank Brian J. Poindexter for help performing the deconvolution microscopy.
Footnotes
This work was supported by National Institutes of Health Grants K01 DK70834 (to W. Z.), R01 DK075065 (to B. C. K.), and R01 DK47981 (to B. C. K.), and endowment funds from The James T. and Nancy B. Willerson Chair (to B. C. K.).
The abbreviations used are: ENaC, epithelial sodium channel; Dot1, disruptor of telo-meric silencing; GR, glucocorticoid receptor; GRE, glucocorticoid-response element; mIMCD3, mouse inner medullary collecting duct cell line-3; MLL, mixed lineage leukemia; NF-κB, nuclear factor for immunoglobulin κ chain in B cells; BCoR, BCL-6 core-pressor; Sir, silencing information repressor; EGFP, epidermal growth factor protein; aa, amino acid(s); ChIP, chromatin immunoprecipitation; RNAi, RNA interference; GST, glutathione S-transferase; qPCR, quantitative PCR; RT, reverse transcriptase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
W. Zhang, X. Xia, M. R. Reisenauer, C. S. Hemenway, and B. C. Kone, unpublished data.
REFERENCES
- 1.Shimkets RA, Warnock DG, Bositis CM, Nelson-Williams C, Hansson JH, Schambelan M, Gill JR, Jr., Ulick S, Milora RV, Findling JW, Canessa CM, Rossier BC, Lifton RP. Cell. 1994;79:407–414. doi: 10.1016/0092-8674(94)90250-x. [DOI] [PubMed] [Google Scholar]
- 2.Chang SS, Grunder S, Hanukoglu A, Rosler A, Mathew PM, Hanukoglu I, Schild L, Lu Y, Shimkets RA, Nelson-Williams C, Rossier BC, Lifton RP. Nat. Genet. 1996;12:248–253. doi: 10.1038/ng0396-248. [DOI] [PubMed] [Google Scholar]
- 3.Eaton DC, Malik B, Saxena NC, Al-Khalili OK, Yue G. J. Membr. Biol. 2001;184:313–319. doi: 10.1007/s00232-001-0098-x. [DOI] [PubMed] [Google Scholar]
- 4.Thomas CP, Itani OA. Curr. Opin. Nephrol. Hypertens. 2004;13:541–548. doi: 10.1097/00041552-200409000-00010. [DOI] [PubMed] [Google Scholar]
- 5.Asher C, Wald H, Rossier BC, Garty H. Am. J. Physiol. 1996;271:C605–C611. doi: 10.1152/ajpcell.1996.271.2.C605. [DOI] [PubMed] [Google Scholar]
- 6.Escoubet B, Coureau C, Bonvalet JP, Farman N. Am. J. Physiol. 1997;272:C1482–C1491. doi: 10.1152/ajpcell.1997.272.5.C1482. [DOI] [PubMed] [Google Scholar]
- 7.Debonneville C, Flores SY, Kamynina E, Plant PJ, Tauxe C, Thomas MA, Munster C, Chraibi A, Pratt JH, Horisberger JD, Pearce D, Loffing J, Staub O. EMBO J. 2001;20:7052–7059. doi: 10.1093/emboj/20.24.7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mink S, Haenig B, Klempnauer KH. Mol. Cell. Biol. 1997;17:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stokes JB, Sigmund RD. Am. J. Physiol. 1998;274:C1699–C1707. doi: 10.1152/ajpcell.1998.274.6.C1699. [DOI] [PubMed] [Google Scholar]
- 10.Mick VE, Itani OA, Loftus RW, Husted RF, Schmidt TJ, Thomas CP. Mol. Endocrinol. 2001;15:575–588. doi: 10.1210/mend.15.4.0620. [DOI] [PubMed] [Google Scholar]
- 11.Stockand JD. Am. J. Physiol. 2002;282:F559–F576. doi: 10.1152/ajprenal.00320.2001. [DOI] [PubMed] [Google Scholar]
- 12.Kohler S, Pradervand S, Verdumo C, Merillat AM, Bens M, Vandewalle A, Beermann F, Hummler E. Biochim. Biophys. Acta. 2001;1519:106–110. doi: 10.1016/s0167-4781(01)00228-7. [DOI] [PubMed] [Google Scholar]
- 13.Zentner MD, Lin HH, Deng HT, Kim KJ, Shih HM, Ann DK. J. Biol. Chem. 2001;276:29805–29814. doi: 10.1074/jbc.M103153200. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Hayashizaki Y, Kone BC. Biochem. J. 2004;377:641–651. doi: 10.1042/BJ20030839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W, Xia X, Jalal DI, Kuncewicz T, Xu W, Lesage GD, Kone BC. Am. J. Physiol. 2006;290:C936–C946. doi: 10.1152/ajpcell.00431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rauchman MI, Nigam SK, Delpire E, Gullans SR. Am. J. Physiol. 1993;265:F416–F424. doi: 10.1152/ajprenal.1993.265.3.F416. [DOI] [PubMed] [Google Scholar]
- 17.Gumz ML, Popp MP, Wingo CS, Cain BD. Am. J. Physiol. 2003;285:F664–F673. doi: 10.1152/ajprenal.00353.2002. [DOI] [PubMed] [Google Scholar]
- 18.Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 20.Erfurth F, Hemenway CS, De Erkenez AC, Domer PH. Leukemia. 2004;18:92–102. doi: 10.1038/sj.leu.2403200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang W, Xia X, Zou L, Xu X, LeSage GD, Kone BC. Am. J. Physiol. 2004;286:F1171–F1177. doi: 10.1152/ajprenal.00043.2003. [DOI] [PubMed] [Google Scholar]
- 22.Yao YL, Yang WM, Seto E. Mol. Cell. Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Kone BC. Am. J. Physiol. 2002;283:F904–F911. doi: 10.1152/ajprenal.00156.2002. [DOI] [PubMed] [Google Scholar]
- 24.Kuncewicz T, Balakrishnan P, Snuggs MB, Kone BC. Am. J. Physiol. 2001;281:F326–F336. doi: 10.1152/ajprenal.2001.281.2.F326. [DOI] [PubMed] [Google Scholar]
- 25.Poindexter BJ. J. Burns and Wounds. 2005;4:101–111. 128. [PMC free article] [PubMed] [Google Scholar]
- 26.Deleted in proof
- 27.Iida S, Seto M, Yamamoto K, Komatsu H, Akao Y, Nakazawa S, Ariyoshi Y, Takahashi T, Ueda R. Jpn. J. Cancer Res. 1993;84:532–537. doi: 10.1111/j.1349-7006.1993.tb00172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 29.Min J, Feng Q, Li Z, Zhang Y, Xu RM. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 30.Hemenway CS, de Erkenez AC, Gould GC. Oncogene. 2001;20:3798–3805. doi: 10.1038/sj.onc.1204478. [DOI] [PubMed] [Google Scholar]
- 31.Ayton PM, Cleary ML. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 32.Srinivasan RS, de Erkenez AC, Hemenway CS. Oncogene. 2003;22:3395–3406. doi: 10.1038/sj.onc.1206361. [DOI] [PubMed] [Google Scholar]
- 33.van Leeuwen F, Gafken PR, Gottschling DE. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 34.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng HH, Xu RM, Zhang Y, Struhl K. J. Biol. Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 36.Huang H, Kahana A, Gottschling DE, Prakash L, Liebman SW. Mol. Cell. Biol. 1997;17:6693–6699. doi: 10.1128/mcb.17.11.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. J. Biol. Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 38.Alvarez de la Rosa D, Li H, Canessa CM. J. Gen. Physiol. 2002;119:427–442. doi: 10.1085/jgp.20028559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun ZW, Allis CD. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 40.Turner SD, Ricci AR, Petropoulos H, Genereaux J, Skerjanc IS, Brandl CJ. Mol. Cell. Biol. 2002;22:4011–4019. doi: 10.1128/MCB.22.12.4011-4019.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 42.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG. J. Biol. Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 44.Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, Reinberg D. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]