Abstract
Thermoinhibition, or failure of seeds to germinate when imbibed at warm temperatures, can be a significant problem in lettuce (Lactuca sativa L.) production. The reliability of stand establishment would be improved by increasing the ability of lettuce seeds to germinate at high temperatures. Genes encoding germination- or dormancy-related proteins were mapped in a recombinant inbred line population derived from a cross between L. sativa cv. Salinas and L. serriola accession UC96US23. This revealed several candidate genes that are located in the genomic regions containing quantitative trait loci (QTLs) associated with temperature and light requirements for germination. In particular, LsNCED4, a temperature-regulated gene in the biosynthetic pathway for abscisic acid (ABA), a germination inhibitor, mapped to the center of a previously detected QTL for high temperature germination (Htg6.1) from UC96US23. Three sets of sister BC3S2 near-isogenic lines (NILs) that were homozygous for the UC96US23 allele of LsNCED4 at Htg6.1 were developed by backcrossing to cv. Salinas and marker-assisted selection followed by selfing. The maximum temperature for germination of NIL seed lots with the UC96US23 allele at LsNCED4 was increased by 2–3°C when compared with sister NIL seed lots lacking the introgression. In addition, the expression of LsNCED4 was two- to threefold lower in the former NIL lines as compared to expression in the latter. Together, these data strongly implicate LsNCED4 as the candidate gene responsible for the Htg6.1 phenotype and indicate that decreased ABA biosynthesis at high imbibition temperatures is a major factor responsible for the increased germination thermotolerance of UC96US23 seeds.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-010-1425-3) contains supplementary material, which is available to authorized users.
Introduction
The genus Lactuca L. belongs to the Compositae or Asteraceae family and is represented by approximately 100 species distributed in warm and temperate regions of Europe, Asia, North America, Africa and Australia (Dziechciarkova et al. 2004; Lebeda et al. 2004). Lettuce (Lactuca sativa L.) is an important crop species with an annual farm-gate value of over $2.7 billion in the US in 2007 (NASS 2008). Thermoinhibition (inhibition of seed germination by high temperature) in lettuce can delay or prevent germination in warm season plantings, resulting in reduced field emergence and stand establishment (Valdes et al. 1985). Thermoinhibitory conditions often occur in major winter lettuce production areas of California and Arizona where temperatures permissive of germination are exceeded during late summer and fall plantings. Delays in germination and asynchronous emergence can diminish yield (Cantliffe et al. 1981), necessitate multiple harvests, and create other management problems through non-uniformity of crop development, resulting in losses in quality and profitability (Benjamin 1990).
The maximum temperatures for lettuce seed germination are genetically heritable (Argyris et al. 2005; Guzman et al. 1992) and are influenced by environmental factors, such as the temperature and light environment during seed maturation and germination (Contreras et al. 2008, 2009; Gray et al. 1988; Harrington and Thompson 1952; Hayashi et al. 2008; Kozarewa et al. 2006; Kristie and Fielding 1994; Saini et al. 1989; Sung et al. 1998). An intact endosperm envelope that encloses the lettuce embryo, but not the outer fused testa and pericarp, is essential for the imposition of thermoinhibition (Borthwick and Robbins 1928; Dunlap et al. 1990; Globerson et al. 1974; Sung et al. 2008).
Physiologically, the maximum germination temperature of lettuce seeds appears to be determined primarily through the antagonistic interactions of gibberellins (GA) and abscisic acid (ABA) (Gonai et al. 2004; Toh et al. 2008) and the regulation of their synthesis and degradation by phytochrome (Sawada et al. 2008a, b; Seo et al. 2006). Seeds of ABA-deficient and -insensitive mutants (aba1 and abi3) in Arabidopsis thaliana germinate at supra-optimal temperatures, supporting a major role for ABA in the thermoinhibition mechanism (Tamura et al. 2006). The expression of genes involved in ABA biosynthesis in Arabidopsis (e.g., AtNCED genes encoding 9-cis-epoxycarotinoid dioxygenases) is up-regulated at high imbibition temperatures, with AtNCED9 playing the major role in thermoinhibition, while the expression of genes involved in GA synthesis (e.g., AtGA3ox genes encoding GA 3β-hydroxylases) is down-regulated (Toh et al. 2008). In cultivated lettuce, ABA content is maintained at elevated levels when seeds are imbibed at high temperatures, but decreases rapidly when imbibition occurs at optimal temperatures for germination (Argyris et al. 2008b; Toh et al. 2008; Yoshioka et al. 1998). Gibberellins promote ABA catabolism and thus increase the maximum temperature of lettuce seed germination (Gonai et al. 2004). Ethylene application also promotes germination at high temperatures, and endogenous ethylene production is greater in seeds capable of germinating at high temperature (Kozarewa et al. 2006; Prusinski and Khan 1990; Saini et al. 1986). Large scale transcriptional profiling of genes involved in these hormonal biosynthesis and deactivation pathways showed that genetic variation in maximum germination temperature was associated with reciprocal changes in expression of ABA versus GA and ethylene biosynthesis, metabolism, and response genes (Argyris et al. 2008b).
As in many other crop species, molecular markers have been utilized in Lactuca species for taxonomic and biodiversity studies, germplasm identification, improving disease resistance, dissecting quantitative traits and marker-assisted selection (MAS) for crop improvement (Argyris et al. 2005; Dziechciarkova et al. 2004; Grube et al. 2005; Lebeda et al. 2007; Michelmore et al. 1991). Molecular marker identification has been enhanced through the development of an expressed sequence tag (EST) database of over 135,000 ESTs from multiple Lactuca species through the Compositae Genome Project (CGP) (http://cgpdb.ucdavis.edu/). This database has aided the construction of an integrated genetic map using six inter- and intra-specific crosses of Lactuca spp. and comprises over 2,700 markers spanning 1,505 cM in nine linkage groups (LGs) corresponding to the nine chromosomes of Lactuca (Truco et al. 2007). More recently, marker density of the integrated genetic map has been greatly augmented using a tiled microarray representing ~29,000 unigenes for massively parallel genetic mapping, allowing over 10,000 polymorphic loci to be mapped (van Leeuwen et al. 2009). These resources have facilitated the identification of quantitative trait loci (QTLs) and the genetic mapping of candidate genes associated with seed germination, dormancy and thermotolerance in a recombinant inbred line (RIL) population derived from L. sativa cv. Salinas and L. serriola accession UC96US23 (Argyris et al. 2005, 2008a). For example, a major QTL for high temperature germination (Htg6.1) collocated with LsNCED4, a key regulated gene in the ABA biosynthetic pathway (Argyris et al. 2008b). The expression of this gene and seed ABA content were inversely correlated with the maximum germination temperature.
The dissection of the genetic architecture of quantitative traits is accomplished effectively through backcrossing and the development of near-isogenic lines (NILs). A gene, chromosomal segment, or QTL can be isolated in a different and uniform genetic background (that of the recurrent parent [RP]) and its additive effects validated (MacKay et al. 2009). NILs have broad applications as immortal mapping populations with greater mapping power as compared to RILs (Keurentjes et al. 2007). In lettuce, NILs and backcross-inbred line populations have been used to study herbicide resistance, to detect markers associated with disease resistance, and to examine potentially beneficial traits derived from wild germplasm (Jeuken and Lindhout 2004; Mallory-Smith et al. 1990; Paran et al. 1991). NILs are particularly critical for studies of seed traits such as dormancy, as the status of an individual seed is scored as either germinated or not (qualitatively), while populations of seeds exhibit quantitative variation in dormancy characteristics (Bradford 1996). To this end, NILs have been used effectively in validating seed dormancy QTLs in Arabidopsis and rice (Oryza sativa L.) (Bentsink et al. 2006; Gu et al. 2006). In lettuce, the Htg6.1 QTL for high temperature germination conferred significant increases in the maximum germination temperature in advanced backcross (BC) progeny (Argyris et al. 2008a).
The present study builds on the genetic resources developed in Lactuca and extends previous work identifying the role of Htg6.1 and different allelic forms of LsNCED4 in regulating thermoinhibition in RILs and advanced BC progeny (Argyris et al. 2005, 2008a, b). Here we report on several candidate gene/QTL co-locations associated with seed traits and in detail on the collocation of LsNCED4 with Htg6.1. We also describe the development and analysis of NILs introgressed with the Htg6.1 QTL associated with germination thermotolerance.
Materials and methods
Genetic mapping of candidate germination/dormancy genes and QTL analyses
Development of the RIL mapping population derived from a cross between L. sativa cv. Salinas and L. serriola UC96US23 has been described previously (Argyris et al. 2005; Truco et al. 2007). Utilizing an EST-based mapping approach with sequences retrieved from the CGP database (http://www.cgpdb.ucdavis.edu/), unigene contigs with high protein sequence homology to known germination/dormancy-related candidates (reviewed in Bentsink et al. 2007) were identified through keyword or BLASTX searches (Altschul et al. 1997). Identification of polymorphic EST candidate germination and dormancy contigs and primer design followed the methodology of Lai et al. (2005) (Online Resource Table S1). Polymorphisms in polymerase chain reaction (PCR) amplicons were screened in parental lines Salinas and UC96US23 of the RIL population using agarose or single-stranded conformational polymorphism (SSCP) gel electrophoresis according to the methods of Bodenes et al. (1996) with the following modifications: gels were fixed and stained in a single step with a combination 1% acetic acid/0.15% silver nitrate solution for 10 min, rinsed in deionized H2O, then developed in 1 M NaOH + 0.037% formaldehyde for 8–10 min. Gels were then rinsed thoroughly for 3 min, air-dried and scored. PCR products for genes of interest were sequenced to confirm their identities. The population of 90 RILs in F7 generation were genotyped with these markers and linkage analysis performed using JoinMap v. 2.0 (Stam 1995) to place candidates on the linkage map. A complementary approach used candidate gene sequences representing L. sativa deposited in GenBank and primer sets designed at intervals along the coding sequence to generate 250 bp PCR amplicons and subsequent analysis of these amplicons from both parent lines by SSCP for polymorphism detection.
For germination/dormancy genes from L. sativa found in GenBank that were either not found in the CGP database, not polymorphic, or not able to be mapped using the above approaches, a sequencing approach was adopted. Oligonucleotide primers were designed to the ends of full length coding regions and PCR amplicons for Salinas and UC96US23 were purified with ExoSAP-IT® reagent (USB, Cleveland, OH). Sequencing reactions were performed using a Big Dye Terminator v.3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and sequencing analysis conducted by the genomics facility at UC Davis (http://www.cgf.ucdavis.edu/home/) on an ABI Prism-3730 DNA analyzer (Applied Biosystems, Foster City, CA). PCR products were sequenced up to 8× redundancy to ensure accurate single nucleotide polymorphism (SNP) calls. Primer sets were then designed around putative SNPs detected between the Salinas and UC96US23 sequences, and PCR products were screened on SSCP gels to ensure SNPs were co-dominant and scorable. Screening in the RIL mapping population and genetic mapping were then performed as above.
A subset of 473 markers and mapped germination/dormancy candidate genes approximately 5–7 cM apart was chosen as the framework map (Online Resource Fig. S1) for analysis of phenotypic data for germination at 25°C (low temperature germination, LTG) and 35°C (high temperature germination, HTG), as well as germination at 20°C in darkness following exposure to far-red light (GFR). Data from F7:8 RIL populations consisting of 108 individuals grown in three environments (Davis, CA; Yuma, AZ; and De Lier, The Netherlands) in the summer of 2002 (Argyris et al. 2005) were reanalyzed using Windows QTL Cartographer V. 2.5 (Basten et al. 2001). Details of how these traits were measured are described in Argyris et al. (2005). Listwise deletion was employed to eliminate individuals missing phenotypic data. Subsequent QTL analyses were performed with data sets ranging from 94 to 74 RILs. Germination percentages were probit transformed and QTL effects identified by composite interval mapping (CIM) utilizing standard model 6 and parameters outlined previously (Argyris et al. 2005). QTL were verified by 1,000 permutations of phenotypic data and declared significant (p < 0.05) above the permutated log of the odds (LOD) threshold value. Multilocus and least square means analyses were conducted to estimate additive and interaction effects of QTL as described previously (Argyris et al. 2005; Gandhi et al. 2005).
Following the initial QTL analysis, markers from the high density Affymetrix GeneChip® mapping microarray (van Leeuwen et al. 2009) were combined with the framework markers comprising LG6 from the Lactuca integrated map (Truco et al. 2007) to obtain a more densely populated genetic map for the refinement of the Htg6.1 QTL interval. Single position polymorphisms (SPPs) were obtained for unigenes by analysis of 113 RILs hybridized in duplicate to the lettuce Affymetrix gene chip (van Leeuwen et al. 2008). SPPs were ordered using MadMapper (http://cgpdb.ucdavis.edu/XLinkage/MadMapper) and a SPP was selected from each distinct genetic bin (unique recombinations between adjacent markers) covering LG6 (van Leeuwen et al. 2009). The ordered markers without genetic distances were entered into QTL Cartographer to identify relevant sets of SPP genetic markers that were encompassed by the QTL interval. BLASTX (Altschul et al. 1997) was performed for all unigenes in the QTL interval against the Arabidopsis genome (1 × e−20 threshold) to obtain putative candidate gene identities. Markers encompassing the QTL interval plus the framework markers from the integrated map were entered into JoinMap V 4.0 (Stam 1995). The resulting maps with genetic distances were then reanalyzed in QTL Cartographer to determine the newly defined confidence intervals and effects of the Htg6.1 QTL.
NIL development and testing
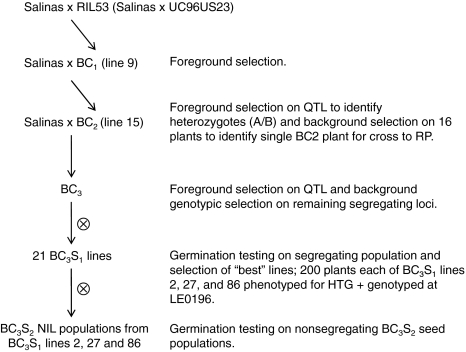
To create NILs for QTL Htg6.1 that contained LsNCED4, MAS using foreground (linked to the QTL) and background (unlinked to the QTL) markers was performed during three generations of backcrossing with L. sativa cv. Salinas as the RP as outlined in Fig. 1. Foreground and background MAS was performed with PCR-based markers designed from EST sequences in the CGP database (http:/www.cgpdb.ucdavis.edu/) (Online Resource Table S2). Markers were selected in areas of the donor parent (DP) genome at the outset of the backcrossing/MAS process to monitor gains in RP genome and QTL introgression based on a genetic linkage map composed of over 1,700 amplified fragment length polymorphism (AFLP), simple sequence repeat (SSR), and EST markers coalescing into 9 LGs (corresponding to chromosome number) and spanning ~1,213 cM. Details of map construction were as described previously (Truco et al. 2007). Selected markers were co-dominant and could be easily resolved in backcross populations. To genotype individuals during backcrossing, genomic DNA was extracted as described previously (Kesseli et al. 1994) and PCR-amplified using primer sequences for markers with designations from LE0000-LE3999 (Online Resource Table S2) and an annealing temperature of 57°C. Polymorphisms were visualized using SSCP gels.
Fig. 1.
Pedigree of NILs developed for the introgression of the Htg6.1 QTL containing LsNCED4 using backcrossing and marker-assisted selection
The DP of the Htg6.1 QTL allele was RIL53 of the F7 generation of the RIL population (Fig. 1). While several RILs possessed the UC96US23 chromosomal segment containing the entire confidence interval of Htg6.1 on LG6, RIL53 possessed the greatest number of RP alleles in comparison to other RILs. The progeny of the cross between Salinas and RIL53 was designated BC1. MAS was performed using six foreground markers within and flanking the QTL interval on LG6. Selection on an additional 25 markers dispersed over the remaining eight LGs was performed concurrently to identify individuals with the highest proportion of the RP. Nine BC1 lines were confirmed as heterozygous at marker LE0196 that was closest to the maximum QTL LOD peak at approximately 65 cM from the end of LG6 (the collocation of LsNCED4 with Htg6.1 was not determined until later, so this gene was not used in MAS). LE0196 corresponds to contig CLS_S3_Contig6125 on the high density map which subsequently mapped at 43.0 cM while LsNCED4 mapped at 43.5 cM (Supplementary Table S3). An individual BC1 plant was backcrossed again to the Salinas RP to yield 120 BC2 plants (Fig. 1). Among these, 16 retained the entire Htg6.1 QTL confidence interval, of which BC2 NIL15 had the highest percentage of RP alleles. This line was therefore crossed to Salinas to yield 100 BC3 NILs on which foreground and background selections were repeated. All BC3 NILs were allowed to self-pollinate and seed populations segregating at the Htg6.1 locus were tested for germination as detailed in Argyris et al. (2008a). BC3 NILs 2, 27 and 86 were determined to have the Htg6.1 introgression (heterozygous), a high percentage of RP genome homozygous, different segregating RP background loci, and their progeny had the greatest HTG percentages among the BC3 NILs tested. Sister NILs derived from each of these three BC3 NILs by one generation of selfing were used for the evaluation of the effect of Htg6.1 on increasing the maximum germination temperature.
From each of the BC3 NILs 2, 27 and 86, 400 BC3S1 seeds were sown in Petri dishes at 33°C. From these, germinated seedlings were separated and recorded as displaying a HTG phenotype and the remaining seeds were transferred to 25°C until they germinated. Both HTG and non-HTG seedlings were transplanted to flats in a greenhouse at 25/15°C with ambient light duration and intensity when day length exceeded 14 h. Plants were maintained for 1 week at these temperatures and a 14/10 h light/dark diurnal cycle with a supplemental light intensity of 400 W/m2 when daylength was <14 h. From the original group of 400 seeds, 200 seedlings (including all those exhibiting a HTG phenotype) of each of the three BC3 NIL lines, as well as Salinas and UC96US23, were transferred to 10-cm pots on 10 November 2006 (Fig. 1). DNA was extracted from all BC3S1 NIL plants. The BC3S1 NILs were allowed to self-pollinate to generate BC3S2 seed. Seeds were hand-harvested beginning 5 April 2007 and continuing until 20 June 2007. Following harvest, seeds of each BC3S2 NIL were cleaned and stored in paper envelopes at 9°C and ~25% relative humidity (RH) until tested.
Germination testing
Approximately 30 BC3S2 NIL seed lots that had been genotyped at LsNCED4 (15 homozygous for Salinas alleles [A/A] and 15 homozygous for UC96US23 alleles [B/B] of each the BC3 NILs 2, 27 and 86) as well as parental lines were germinated (two replications of 25 seeds each) on a thermogradient table at 1°C intervals from 27 to 33°C. To test the effect of the LsNCED4 genotype on germination in larger populations, tests at 30°C were performed on three replicates of 25 seeds. Seeds were distributed on two absorbent blotter paper discs (VWR Scientific Products, West Chester, PA) saturated with 4 mL de-ionized water in 5 cm-diameter Petri dishes (Gelman Sciences, Ann Arbor, MI). Seeds were placed into a germination cabinet (Hoffman Manufacturing, Albany, OR) under continuous fluorescent light and constant temperature conditions and evaluated for germination (radicle emergence) at 72 h.
Gene expression assays
The expression of LsNCED4 was measured in 100 seeds comprising 20 seeds from each of five BC3S2 NIL seed lots derived from of each of the three BC3 NILs. These combined samples for seeds for each NIL, pooled separately by genotypic class (either A/A or B/B genotypes), were divided into three replications of each genotype and imbibed at 31°C for 24 h. At this time, germinated seedlings were removed, and the remaining seeds were frozen in liquid nitrogen. RNA was extracted, cDNA synthesized, and quantitative PCR analysis performed as described previously (Argyris et al. 2008b).
Results
QTL and candidate gene mapping
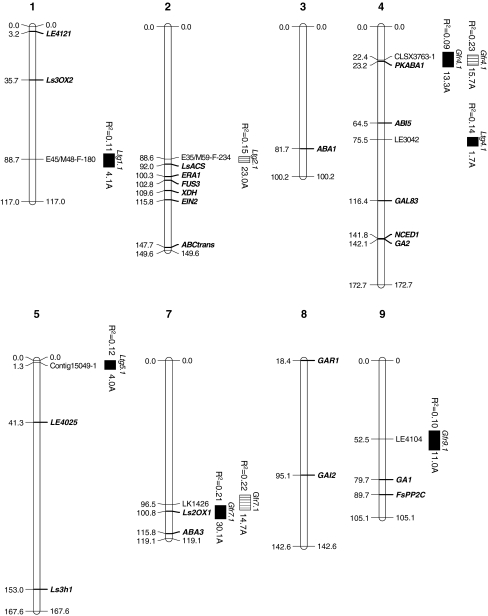
Twenty-three lettuce genes identified in the CGP database and GenBank that could potentially be involved in germination or dormancy were mapped (Table 1). These candidates included genes encoding homologs of proteins involved in hormonal biosynthesis, signaling, or metabolism (9 for ABA, 8 for GA, and 2 for ethylene), a transcription factor involved in seed development (FUSCA3), a protein phosphatase 2C (FsPP2C), and a component of the sucrose non-fermenting (SNF)-related protein kinases (GAL83). At least one candidate gene mapped to each of the nine lettuce chromosomes, with five genes clustering within a 23 cM interval near the bottom of LG2 (Fig. 2). This cluster included two ethylene-related genes encoding ACC SYNTHASE (LsACS1) and ETHYLENE INSENSITIVE 2 (LsEIN2), two ABA-related genes encoding ENHANCED RESPONSE TO ABA (LsERA1) and XANTHINE DEHYDROGENASE (LsXDH) as well as FUSCA3. A second region of interest occurred on LG4, where 9-cis-EPOXYCAROTENOID DIOXYGENASE 1 (LsNCED1) and ent-KAURENE SYNTHASE (LsKS or GA2) were separated by only 0.3 cM. LsNCED4 (GI:84579411) co-segregated with the maximum LOD peak identifying Htg6.1, as well as with QTL for germination sensitivity to ABA and GA, as reported previously (Argyris et al. 2008b). Subsequently, segregation analysis of SPP markers using the Affymetrix GeneChip provided an ultra-dense map averaging more than one marker per cM containing the Htg6.1 QTL on LG6 (Fig. 3). All genes represented on the microarray that were polymorphic for SPPs and mapped within ~10 cM of LsNCED4 are listed in Online Resource Table S3. Other than LsNCED4, no obvious functional annotations associated with seed germination, plant hormones or temperature sensitivity were evident.
Table 1.
Candidate genes encoding proteins potentially involved in germination or dormancy mapped in a Lactuca sativa cv. Salinas × L. serriola UC96US23 RIL population
| Markera | ESTa or GenBank ID | Gene | Description | LG | Positionb |
|---|---|---|---|---|---|
| ABA related | |||||
| LE4128 | QGF5H03.yg.ab1 | XDH | Xanthine dehydrogenase | 2 | 109.6 |
| LE4015 | QG CA Contig2867 | ABA1 (ZEP) | Zeaxanthin epoxidase | 3 | 81.7 |
| LE4019 | QGG18L02.yg.ab1 | ABA3 | Molybdenum cofactor sulfurase | 7 | 115.8 |
| LE4010 | QG CA Contig7543 | ABI5 | ABA insensitive 5 | 4 | 64.5 |
| LE4245 | AB120115 | LsAO1 | Abscisic acid aldehyde oxidase | 6 | 21.1 |
| LE4131 | QG CA Contig6166 | LsNCED1 | 9-cis-Epoxycarotenoid dioxygenase 1 | 4 | 141.8 |
| LE4250 | AB120110 | LsNCED4 | 9-cis-Epoxycarotenoid dioxygenase 4 | 6 | 65.1 |
| LE4129 | QGG28D16.yg.ab1 | ERA1 | Enhanced response to ABA | 2 | 100.3 |
| LE4021 | QG CA Contig3084 | SnRK2.2 | Protein kinase responsive to ABA | 4 | 23.2 |
| GA related | |||||
| LE4209 | AB031204 | LsCPS1 | Ent-copalyl diphosphate synthase | 9 | 79.7 |
| LE4022 | QGF25D22.yg.ab1 | LsKS | Ent-kaurene synthase | 4 | 142.1 |
| LE4201 | AB031206 | LsGA2ox1 | Gibberellin 2-oxidase 1 | 7 | 100.8 |
| LE4251 | AB012205 | LsGA3ox1 | Gibberellin 3β-hydroxylase 1 | 5 | 153.0 |
| LE4220 | AB012206 | LsGA3ox2 | Gibberellin 3β-hydroxylase 2 | 1 | 35.7 |
| LE4025 | QGF7L19.yg.ab1 | GA20ox | Gibberellin 20-oxidase | 5 | 41.3 |
| LE4134 | QG CA Contig7236 | GAR1 | Suppressor of gibberellin insensitive | 8 | 18.4 |
| LE4130 | QG CA Contig5082 | GAI2 | Gibberellin insensitive 2 | 8 | 95.1 |
| Ethylene related | |||||
| LE4034 | QGG10D02.yg.ab1 | LsACS1 | ACC synthase | 2 | 92.0 |
| LE0358 | QGC20F11.yg.ab1 | LsEIN2 | Ethylene insensitive 2 | 2 | 115.8 |
| Miscellaneous | |||||
| LE4132 | QG CA Contig5234 | FsPP2C | Protein phosphatase 2C | 9 | 89.7 |
| LE4133 | QGB11D05.yg.ab1 | FUSCA3 | DNA binding/transcription factor | 2 | 102.8 |
| LE4051 | QGB28M14.yg.ab1 | GAL83 | Docking protein of SNF1 kinase | 4 | 116.4 |
a http://www.cdgdb.ucdavis.edu
bDecember 2006 map: http://cgpdb.ucdavis.edu/database/genome_viewer/viewer/
Fig. 2.
Candidate genes in the lettuce consensus map composed of nine linkage groups and collocations with QTL (LG6 is shown in Fig. 3). Mapped candidate genes are in bold italics. Genomic locations of QTL for seed germination traits detected with data reanalyzed for seeds grown in two environments are shown: solid bars Davis, CA, horizontal open bars Yuma, AZ. Lengths of bars denote 1-LOD confidence intervals. Phenotypic variances and additive genetic effects on germination percentage are adjacent to bars (A Salinas, B UC96US23)
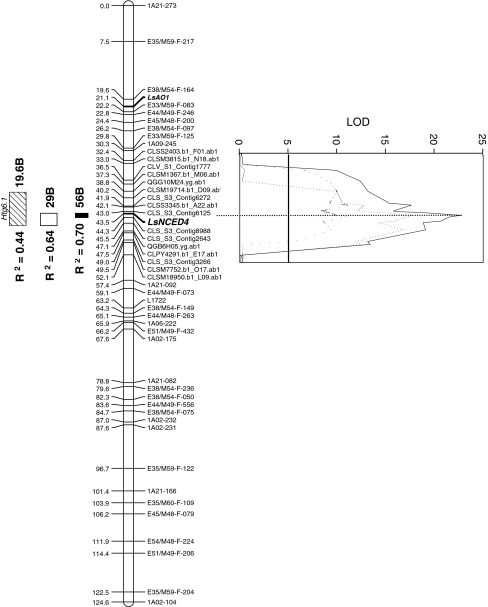
Fig. 3.
Genetic map of LG6 and positions, LOD thresholds and genetic effects of Htg6.1 using markers from the lettuce integrated map and SPP markers from a mapping microarray. Numbers indicate locations (in cM) of genetic markers used in CIM analysis. Solid bar Davis, CA 2002, open bar De Lier, The Netherlands 2002; diagonal lined bar Davis, CA 2006. Phenotypic variances and additive genetic effects are shown adjacent to bars. Germination/dormancy candidate genes located on LG6 are in bold italics. Solid, dotted and dashed lines in LOD graph represent Davis 2002, Netherlands 2002 and Davis 2006 populations, respectively. Solid vertical line at LOD 5 is average permutated threshold value for declaring QTL significant
When a new QTL analysis was performed using the ultra high-density genetic map within the QTL region of LG6 and germination data from seeds produced in 2002, the Htg6.1 QTL was detected in the Salinas × UC96US23 RIL populations grown at Davis and in The Netherlands, as reported previously (Argyris et al. 2005). Htg6.1 was again detected in the same population from seeds produced in 2006 at Davis, strengthening the evidence for a major genetic component for HTG variation being determined by the Htg6.1 QTL. The maximum 1-LOD peaks (ranging from 11 to 23) at 43.5 cM aligned with the map position of LsNCED4 in all three cases and the 2-LOD Htg6.1 QTL confidence intervals were approximately 1, 2.5 and 7 cM for seeds grown in Davis 2002, Netherlands 2002, and Davis 2006, respectively (Fig. 3). The proportion of phenotypic variance for HTG explained by the QTL ranged from 44 to 70% while the additive genetic effect of replacement of the Salinas haplotype with the UC96US23 haplotype at Htg6.1 resulted in 19.6–56% mean increases in germination percentage at 35°C.
In addition to Htg6.1 on LG6, five QTL for three traits in two environments collocated with candidate genes on three different LGs (Fig. 2). Four QTL were detected for germination in the dark after exposure to far-red light (GFR), two each from Davis- and Yuma-grown populations on LGs 4 and 7, respectively. Gfr4.1 accounted for 9 and 23% of the phenotypic variation for GFR in the Davis and Yuma environments, respectively, and was localized to a 10 cM 1-LOD confidence interval. The QTL alleles from Salinas resulted in a 14% mean increase in germination. Gfr4.1 co-segregated with a homolog of SnRK2.2, an ABA-responsive protein kinase in Arabidopsis demonstrated to be involved in ABA signal transduction associated with seed dormancy (Fujii et al. 2007; Nakashima et al. 2009). The second pair of GFR QTL on LG7 (Gfr7.1) was of larger magnitude, accounting for 21 and 22% of the phenotypic variation for GFR in the Davis and Yuma environments, respectively, and resulting in a mean additive effect of a 22% increase in germination due to the Salinas haplotype. These QTL co-segregated with a gene potentially encoding GIBBERELLIN 2-OXIDASE 1 (LsGA2ox1) that is responsible for the metabolic inactivation of active GAs (Nakaminami et al. 2003; Wang et al. 2004). An analysis of the two-way additive effects of Gfr-linked marker loci revealed significant interactions within individual environments (p < 0.05 and p < 0.001 in Davis and Yuma environments, respectively) but not for data combined across environments. However, the presence of Salinas haplotypes at both Gfr4.1 and Gfr7.1 resulted in 42.9% mean germination as compared to 6.1% for UC96US23 haplotypes at the same QTL across both production locations, providing some evidence for epistatic interaction between these QTL in stimulating germination following exposure to far-red light.
A single QTL of large magnitude for germination at 25°C (LTG) accounting for 15% of the phenotypic variation was detected on LG2 for seeds grown in the Yuma environment (Fig. 2). The Salinas QTL haplotype resulted in a 23% increase in germination. A gene encoding ACC SYNTHASE (LsACS1) regulating the committed step of ethylene biosynthesis (Yang and Hoffman 1984) at 92 cM mapped just outside the 2-LOD confidence interval for Ltg2.1 (87–91 cM) (Fig. 2).
Analysis of HTG phenotypes in BC3S2 NILs fixed for Salinas or UC96US23 alleles of LsNCED4 at Htg6.1
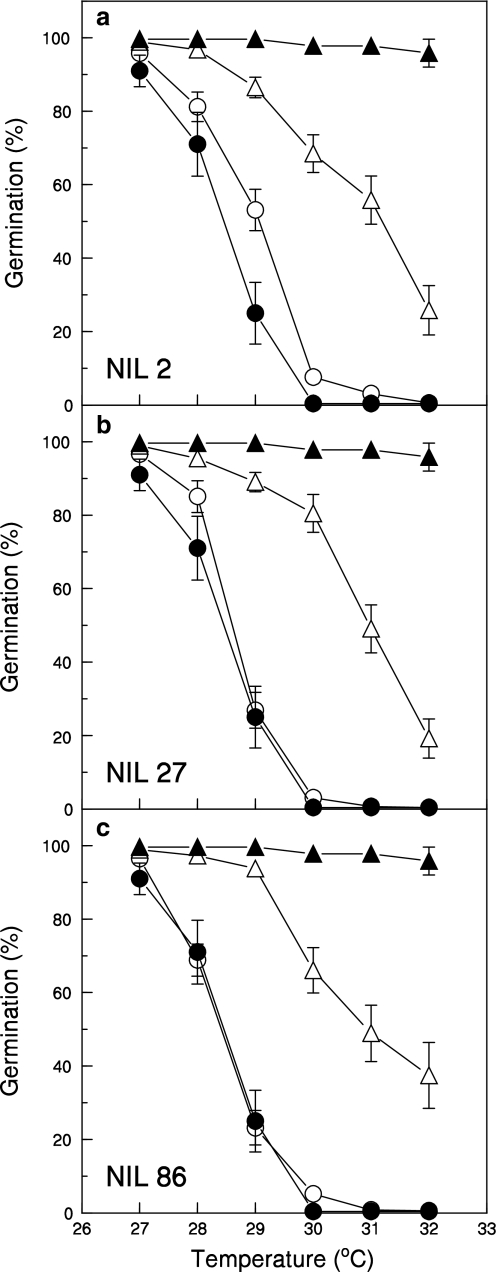
LsNCED4 was sequenced from BC3S2 sister NILs generated from each of the three BC3 NILs to identify BC3S2 seed lots homozygous for each parental allele. Seed lots that were homozygous for the UC96US23 LsNCED4 allele (B/B) had significantly higher mean germination (p < 0.001) over a temperature gradient from 28 to 32°C as compared to sister NIL seed lots homozygous for the Salinas allele (A/A) (Fig. 4). Combined mean germination of the three sets of BC3S2 sister NIL populations of the B/B class was ≥86% up to 29°C before declining to 28% at 32°C. In contrast, the combined mean germination of A/A seeds lots began to decline sharply at 28°C, and was <10% at 30°C and <1% at 32°C due to thermoinhibition. The difference between the two groups was maximal at 30°C, where the combined mean germination of B/B seed lots exceeded that of A/A seeds by 66.2% (71.6% germination in the former as compared to 5.4% germination in the latter). The germination phenotype of A/A seed lots closely paralleled that of the Salinas parental line; however, germination of the UC96US23 parental line was greater than 95% up to 32°C, which was significantly higher than that of the B/B BC3S2 NILs (Fig. 4).
Fig. 4.
Germination across a temperature gradient of Salinas (filled circles), UC96US23 (filled triangles) and 15 BC3S2 sub-NIL seed lots derived from NILs 2 (a), 27 (b), and 86 (c) having either Salinas (A/A open circles) or UC96US23 alleles (B/B open triangles) at LsNCED4. Error bars are ±SD. Differences in germination between BC3S2 NIL genotypic classes were significant (p < 0.001) between 29 and 32°C
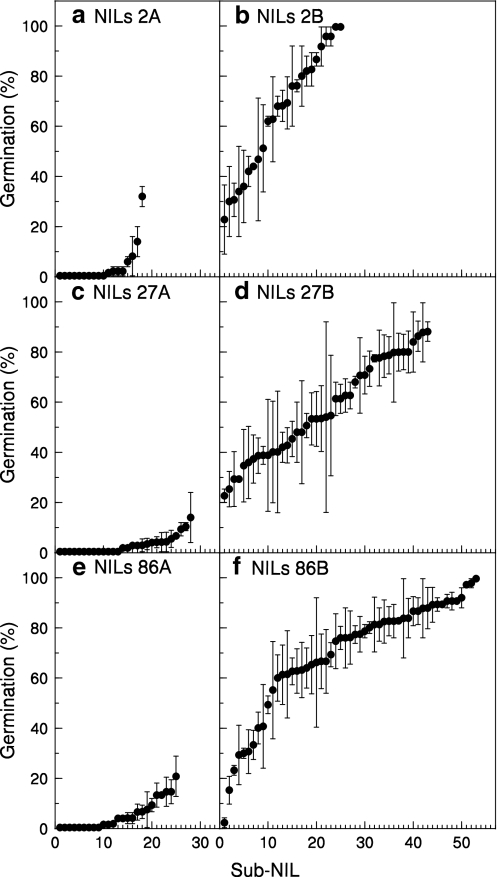
Germination of individual BC3S2 NIL seed lots homozygous for the B allele at LsNCED4 varied widely when tested at 30°C, the temperature at which differences in germination between A/A and B/B genotypes were maximized (Fig. 5b, d, f). The overall mean germination for each set of B/B BC3S2 NIL populations ranged from 58.3 to 65.5%, and was highest in seeds derived from NIL 86. However, germination ranged from >20 to 100% in the 125 B/B BC3S2 NIL seed lots tested from the three sets of sister NILs. In nearly all cases, this was significantly higher than germination of the 70 BC3S2 NIL seed lots homozygous for the A allele at LsNCED4, which showed less variation and ranged from 0 to <15% germination in most lines (Fig. 5a, c, e). With the exception of four seed lots (2 derived from NIL 2 and 2 derived from NIL 86), there was a clear separation between the two allelic classes for germination at 30°C.
Fig. 5.
Germination phenotypes at 30°C of BC3S2 NIL seed lots homozygous at LsNCED4 for either Salinas (A/A a, c, e) or UC96US23 alleles (B/B b, d, f) derived from NIL 2 (a, b), NIL 27 (c, d) and NIL 86 (e, f). The sub-NILs are ordered according to increasing germination percentages. The mean germination percentages averaged across sub-NILs were a, 5.1% (n = 17), b 60.7% (n = 26), c 2.7% (n = 28), d 58.3% (n = 46), e 5.4% (n = 25), and f 65.5% (n = 53). Error bars indicate ±SD
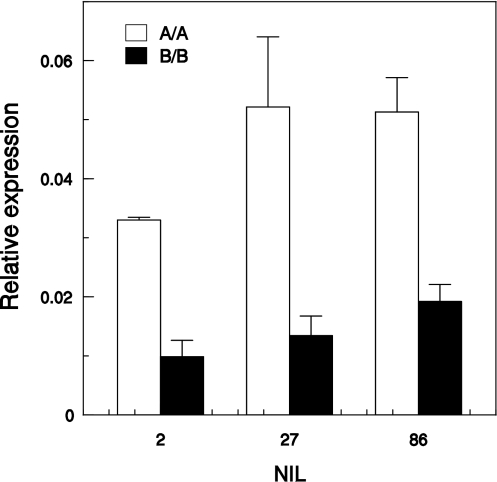
The expression of LsNCED4 following imbibition at 30°C in pooled seed lots homozygous for the B allele and exhibiting a HTG phenotype was significantly lower in all BC3S2 sister NILs when compared with seed lots homozygous for the A allele and exhibiting thermoinhibition (Fig. 6). Mean relative expression levels were similar in all B/B BC3S2 NILs and were 2–2.5-fold lower than in A/A BC3S2 NILs.
Fig. 6.
Expression of LsNCED4 at 30°C in seed lots pooled according to genotype at LE0196 (Htg6.1; 0.5 cM from LsNCED4). A/A Salinas genotype, B/B UC96US23 genotype. Error bars show SE of three biological replicates
Discussion
Mapping of candidate germination/dormancy genes and collocations with QTLs
Genetic mapping of putative candidate genes in lettuce can determine whether they collocate with QTLs influencing germination and dormancy. Those genes that do not collocate can be ruled out as major genetic determinants of the phenotypes observed, although they may still be under the control of loci within a QTL. Although QTLs typically encompass large genomic intervals and many loci, those candidate genes that do collocate with QTL can suggest identity and function. We mapped 22 candidate genes known to be associated with seed germination and dormancy in a RIL population in which QTLs for germination traits had previously been characterized and identified possible functional associations underlying three QTL.
The NCED genes in Arabidopsis, particularly AtNCED9, have been shown to play a major role in thermoinhibition of germination (Toh et al. 2008). In lettuce, a major QTL associated with the alleviation of thermoinhibition (Htg6.1) was identified on LG6 (Argyris et al. 2005). We previously reported the collocation of LsNCED4 (which has highest homology to AtNCED6) with Htg6.1, but the number of markers available at that time was insufficient to fine map the locus (Argyris et al. 2008b). The high-density SPP map developed using a custom Affymetrix GeneChip for lettuce allowed the construction of a high-density linkage map (van Leeuwen et al. 2009) that localized Htg6.1 to substantially smaller genomic intervals (1–7 cM depending on the seed production environment) than the 15 cM previously reported (Argyris et al. 2005). The proportion of phenotypic variance and additive genetic effects explained by the UC96US23 QTL allele were larger than previously reported; this was largely due to the fact that the QTL analysis was done only with LG6, so all of the phenotypic variance was attributed to this linkage group.
Although not conclusively identifying LsNCED4 as Htg6.1, the QTL interval has been reduced to as little as 1 cM centered on LsNCED4 (Fig. 3), and no other polymorphic genes known to map to this region encode proteins with obvious connections to germination (Online Resource Table S3). In addition, several ABA-, GA- and ethylene-related candidate genes can be ruled out as being responsible for the primary effect of the significantly increased maximum germination temperature because they segregate independently of Htg6.1 (Table 1). These findings strongly point to LsNCED4 as a key determinant of thermoinhibition in lettuce, consistent with its elevated expression and a higher ABA content in thermoinhibited seeds (Fig. 6; Argyris et al. 2008b) and with similar results for NCED genes in Arabidopsis (Gonai et al. 2004; Tamura et al. 2006; Toh et al. 2008). These data also demonstrate the powerful potential of SPP arrays for rapidly fine mapping and identifying candidate genes closely associated with QTL.
Salinas seeds are capable of germinating to high percentages in the dark at 20°C, while UC96US23 seeds require light for germination at all temperatures (Argyris et al. 2008b). Two pairs of QTL for germination in darkness at 20°C following far-red light exposure (GFR) on LGs 4 and 7 were associated with candidate genes for GA catabolism (LsGA2ox1) and ABA signaling (SnRK2.2 or PKABA1) (Fig. 2). The QTL were detected in multiple environments and showed evidence of epistatic interactions; therefore, potentially identifying areas of the lettuce genome involved in differentially regulating sensitivity to light in Salinas as compared to UC96US23 seeds. GA metabolism via GA 2-oxidases may be involved in regulating GA content and action during germination (Argyris et al. 2008b; Nakaminami et al. 2003; Wang et al. 2004). Two GA2ox genes have been identified in lettuce, one of which (LsGA2ox2) was expressed primarily in seeds and showed some responsiveness to red light, although not to GA1 (Nakaminami et al. 2003). The expression (mRNA abundance) of LsGA2ox1 was higher in both Salinas and UC96US23 seeds imbibed in the dark when compared with seeds imbibed in the light, while LsGA2ox2 mRNA was detected only briefly following imbibition and at low levels as compared to LsGA2ox1 in both light and dark (Argyris et al. 2008b). Up-regulation of LsGA2ox1 in the dark would be expected to reduce active GA levels and suppress germination.
SnRK2.2 mediates ABA signaling during seed germination in Arabidopsis, with snrk2.2 mutants showing a reduced level of seed dormancy (Fujii et al. 2007; Nakashima et al. 2009). The full length SnRK2.2 homolog in lettuce has not been cloned nor its expression studied, but up-regulation of SnRK2.2 would be expected to enhance ABA action and suppress germination. Recently, a putative family of ABA receptor proteins (RCAR or PYR/PYL proteins) have been linked to modification of PP2C phosphatase and SnRK2 kinase activities in the ABA signal transduction pathway (Cutler et al. 2010; Fujii et al. 2009; Ma et al. 2009; Park et al. 2009), adding significance to this collocation of a lettuce SnRK2.2 homolog with Gfr4.1.
Hayashi et al. (2008) identified two significant QTLs for HTG and four for the GFR trait in a cross between lettuce cultivars Diplomat and Margarita. Overlapping QTLs for HTG and GFR phenotypes were associated with a gene for white versus dark brown seed color that Hayashi et al. (2008) termed br. However, br was identified for brown versus black seed color and is located on chromosome 2 (O’Brien 2007; Waycott et al. 1999), while the w locus on chromosome 7 is associated with white versus brown seed color (Kesseli et al. 1994; Waycott et al. 1999). Because the testa color phenotypes in Diplomat and Margarita are dark brown and white, respectively, the locus identified by Hayashi et al. (2008) is likely associated with w rather than br. As w is located on chromosome 7, the HTG and GFR QTLs identified by Hayashi et al. may be associated with the Gfr7.1 locus reported here. As AFLP markers were used to develop their map, it is not currently possible to compare the locations of these QTLs between the two populations nor to identify candidate genes associated with their QTLs. An additional QTL for HTG was identified on LG9 in a RIL population derived from a cross between Salinas and L. sativa accession PI251246 (O’Brien 2007). We are currently developing this population further to fine map this QTL relative to candidate genes. These multiple sources for tolerance of high germination temperatures and reduced light requirements offer the possibility of combining alleles to achieve higher maximum germination temperatures and less dependence on exposure to light in cultivated lettuce seeds.
Phenotypic effects of Htg6.1 in BC3S2 NILs
Introgression of the Htg6.1 QTL containing LsNCED4 reduced thermoinhibition and elevated the maximum germination temperature by 2–3°C in BC3S2 NIL seed lots of three sets of sister NILs derived from a single BC2 plant (BC2 NIL15) (Figs. 4, 5). Those lines homozygous for the UC96US23 allele at LsNCED4, which mapped at the maximum LOD peak of Htg6.1 (Fig. 3), showed significantly higher germination at supra-optimal temperatures (28–29°C) and at higher temperatures where Salinas and BC2S2 NIL seed lots with the Salinas allele at LsNCED4 were completely thermoinhibited (30–32°C). NIL seed lots carrying the UC96US23 allele at Htg6.1 also exhibited a greater response to seed priming, a hydration/dehydration process that increases the maximum germination temperature of lettuce seeds (Schwember and Bradford 2010). The effect of the UC96US23 allele at LsNCED4 in significantly increasing germination at high temperature in near-isogenic backgrounds, where effects of other loci influencing HTG are minimized, supports earlier evidence for the gene as a major genetic determinant of maximum germination temperature in the Salinas × UC96US23 RIL population (Argyris et al. 2008a).
The effect of the Htg6.1 QTL was maximal at 30°C (Fig. 4), although large variation in germination among individual BC3S2 NILs was observed (Fig. 5). This variance may have resulted from differences in genetic background due to the segregation of the few remaining polymorphic segments. In addition, characteristics of lettuce seed germination are influenced strongly by light and temperature conditions during seed maturation (Contreras et al. 2008, 2009; Hayashi et al. 2008); therefore, much of the variance observed between sister BC3S2 RILs may have been due to maternal environment during seed development. At a minimum, the effect of the UC96US23 QTL haplotype was significant in increasing thermotolerance as compared to BC3S2 NILs lacking the introgression and some NILs may have possessed additional loci that allowed germination at the higher temperatures. Further fine mapping, mutant and transgenic analyses are in progress to conclusively demonstrate the role of LsNCED4 in determining the maximum germination temperature of lettuce seeds.
Conclusions
Natural variation, as an alternative to mutational approaches, can provide additional genetic resources to identify genes that are involved in the control of seed dormancy (Bentsink et al. 2006; Chiang et al. 2009; Gu et al. 2008, 2006). We have exploited the variation present for thermoinhibition between cultivated lettuce and an accession of its wild progenitor species, L. serriola, to map the Htg6.1 QTL in a RIL population and introgress this QTL into BC3S2 NILs where it was subsequently confirmed to extend the range of germination thermotolerance. The mapping of candidate genes for seed germination and dormancy identified collocation of specific genes with QTL for HTG and germination in darkness and provided evidence that LsNCED4 may be responsible for the effect of the Htg6.1 QTL allele from L. serriola. The multiple BC NIL populations developed in this work can serve as informative mapping populations to identify other QTLs for HTG, for fine mapping Htg6.1, and as advanced germplasm for breeding applications. Thermoinhibition is a common production problem, but breeding efforts have not been focused on the improvement of seed germination characteristics, in part due the lack of tools to select efficiently for improved phenotypes. However, exploitation of the variation present for seed thermoinhibition in natural Lactuca populations and the identification of candidate genes and genetic markers associated with it have provided insight into the physiological factors controlling its imposition and release and should lead to the development of lettuce cultivars with improved high temperature tolerance.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by United States Department of Agriculture Initiative for Future Agriculture and Food Systems grant 2000-04292, by National Science Foundation Plant Genome Program Grants 0421630 and 0820451, and by the National Research Initiative competitive Grant no. 2008-02509 from the USDA National Institute of Food and Agriculture. We thank Dr. Andrés Schwember for producing and phenotyping seeds of the RIL population in 2006 and developing Table S3 and Drs. David Still and Eiji Hayashi for the data used to map the GFR QTL.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyris J, Truco MJ, Ochoa O, Knapp SJ, Still DW, Lenssen GM, Schut JW, Michelmore RW, Bradford KJ. Quantitative trait loci associated with seed and seedling traits in Lactuca. Theor Appl Genet. 2005;111:1365–1376. doi: 10.1007/s00122-005-0066-4. [DOI] [PubMed] [Google Scholar]
- Argyris J, Dahal P, Truco MJ, Ochoa O, Still DW, Michelmore RW, Bradford KJ. Genetic analysis of lettuce seed thermoinhibition. Acta Hortic. 2008;782:23–33. [Google Scholar]
- Argyris JM, Dahal P, Hayashi E, Still DW, Bradford KJ. Genetic variation for lettuce seed thermoinhibition is associated with temperature-sensitive expression of abscisic acid, gibberellin, and ethylene biosynthesis, metabolism, and response genes. Plant Physiol. 2008;148:926–947. doi: 10.1104/pp.108.125807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basten CJ, Weir BS, Zeng ZB. QTL Cartographer. Raleigh: Department of Statistics, North Carolina State University; 2001. [Google Scholar]
- Benjamin LR. Variation in time of seedling emergence within populations: a feature that determines individual growth and development. Adv Agron. 1990;44:1–25. doi: 10.1016/S0065-2113(08)60816-0. [DOI] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Soppe W, Koornneef M. Genetic aspects of seed dormancy. In: Bradford KJ, Nonogaki H, editors. Seed development, dormancy and germination. Oxford: Blackwell; 2007. pp. 113–132. [Google Scholar]
- Bodenes C, Laigret F, Kermer A. Inheritance and molecular variations of PCR-SSCP fragments in pedunculate oak (Quercus robur L.) Theor Appl Genet. 1996;93:348–354. doi: 10.1007/BF00223175. [DOI] [PubMed] [Google Scholar]
- Borthwick HA, Robbins WW. Lettuce seed and its germination. Hilgardia. 1928;3:275–305. [Google Scholar]
- Bradford KJ. Population-based models describing seed dormancy behaviour: implications for experimental design and interpretation. In: Lang A, editor. Plant dormancy: physiology, biochemistry and molecular biology. Wallingford: CAB International; 1996. pp. 313–339. [Google Scholar]
- Cantliffe DJ, Shuler KD, Guedes AC. Overcoming seed thermodormancy in a heat-sensitive romaine lettuce by seed priming. HortScience. 1981;16:196–198. [Google Scholar]
- Chiang GCK, Barua D, Kramer EM, Amasino RM, Donohue K. Major flowering time gene, FLOWERING LOCUS C, regulates seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2009;106:11661–11666. doi: 10.1073/pnas.0901367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras S, Bennett MA, Metzger JD, Tay D. Maternal light environment during seed development affects lettuce seed weight, germinability, and storability. HortScience. 2008;43:845–852. [Google Scholar]
- Contreras S, Bennett MA, Metzger JD, Tay D, Nerson H. Red to far-red ratio during seed development affects lettuce seed germinability and longevity. HortScience. 2009;44:130–134. [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61:651–679 [DOI] [PubMed]
- Dunlap JR, Scully BT, Reyes DM. Seed coat-mediation of lettuce germination responses to heat and sodium chloride. J Rio Grande Valley Hortic Soc. 1990;43:55–61. [Google Scholar]
- Dziechciarkova M, Lebeda A, Dolezalova I, Astley D. Characterization of Lactuca spp. germplasm by protein and molecular markers—a review. Plant Soil Environ. 2004;50:47–58. [Google Scholar]
- Fujii H, Verslues PE, Zhu J-K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park S-Y, Cutler SR, Sheen J, Rodriguez PL, Zhu J-K. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi SD, Heesacker AF, Freeman CA, Argyris J, Bradford K, Knapp SJ. The self-incompatibility locus (S) and quantitative trait loci for self-pollination and seed dormancy in sunflower. Theor Appl Genet. 2005;111:619–629. doi: 10.1007/s00122-005-1934-7. [DOI] [PubMed] [Google Scholar]
- Globerson D, Kadmanza A, Ginzburg C. Genetic method for studying the role of seed coats in germination of lettuce. Ann Bot. 1974;38:201–203. [Google Scholar]
- Gonai T, Kawahara S, Tougou M, Satoh S, Hashiba T, Hirai N, Kawaide H, Kamiya Y, Yoshioka T. Abscisic acid in the thermoinhibition of lettuce seed germination and enhancement of its catabolism by gibberellin. J Exp Bot. 2004;55:111–118. doi: 10.1093/jxb/erh023. [DOI] [PubMed] [Google Scholar]
- Gray D, Wurr DCE, Ward JA, Fellows JR. Influence of post-flowering temperature on seed development, and subsequent performance of crisp lettuce. Ann Appl Biol. 1988;113:391–402. doi: 10.1111/j.1744-7348.1988.tb03315.x. [DOI] [Google Scholar]
- Grube RC, Wintermantel WM, Hand P, Aburomia R, Pink DAC, Ryder EJ. Genetic analysis and mapping of resistance to lettuce dieback: a soilborne disease caused by tombusviruses. Theor Appl Genet. 2005;110:259–268. doi: 10.1007/s00122-004-1825-3. [DOI] [PubMed] [Google Scholar]
- Gu X-Y, Kianian SF, Foley ME. Isolation of three dormancy QTLs as Mendelian factors in rice. Heredity. 2006;96:93–99. doi: 10.1038/sj.hdy.6800757. [DOI] [PubMed] [Google Scholar]
- Gu X-Y, Turnipseed EB, Foley ME. The qSD12 locus controls offspring tissue-imposed seed dormancy in rice. Genetics. 2008;179:2263–2273. doi: 10.1534/genetics.108.092007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman VL, Nagata RT, Datnoff LE, Raid RN. ‘Florida 202’ and ‘Everglades’: new butterhead lettuce cultivars adapted to Florida. HortScience. 1992;27:852–853. [Google Scholar]
- Harrington JD, Thompson RC. Effect of variety and area of production on subsequent germination of lettuce seed at high temperatures. Proc Am Soc Hortic Sci. 1952;59:445–450. [Google Scholar]
- Hayashi E, Aoyama N, Still DW. Quantitative trait loci associated with lettuce seed germination under different temperature and light environments. Genome. 2008;51:928–947. doi: 10.1139/G08-077. [DOI] [PubMed] [Google Scholar]
- Jeuken MJW, Lindhout P. The development of lettuce backcross inbred lines (BILs) for exploitation of the Lactuca saligna (wild lettuce) germplasm. Theor Appl Genet. 2004;109:394–401. doi: 10.1007/s00122-004-1643-7. [DOI] [PubMed] [Google Scholar]
- Kesseli RV, Paran I, Michelmore RW. Analysis of a detailed genetic linkage map of Lactuca sativa (lettuce) constructed from RFLP and RAPD markers. Genetics. 1994;136:1435–1446. doi: 10.1093/genetics/136.4.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJB, Bentsink L, Alonso-Blanco C, Hanhart CJ, Vries HBD, Effgen S, Vreugdenhil D, Koornneef M. Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics. 2007;175:891–905. doi: 10.1534/genetics.106.066423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozarewa I, Cantliffe DJ, Nagata RT, Stoffella PJ. High maturation temperature of lettuce seeds during development increased ethylene production and germination at elevated temperatures. J Am Soc Hortic Sci. 2006;131:564–570. [Google Scholar]
- Kristie DN, Fielding A. Influence of temperature on the Pfr level required for germination in lettuce cv. Grand rapids. Seed Sci Res. 1994;4:19–25. doi: 10.1017/S0960258500001951. [DOI] [Google Scholar]
- Lai Z, Livingstone K, Zou Y, Church SA, Knapp SJ, Andrews J, Rieseberg LH. Identification and mapping of SNPs from ESTs in sunflower. Theor Appl Genet. 2005;111:1532–1544. doi: 10.1007/s00122-005-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeda A, Dolezalova I, Ferakova V, Astley D. Geographical distribution of wild Lactuca species (Asteraceae, Lactuceae) Bot Rev. 2004;70:328–356. doi: 10.1663/0006-8101(2004)070[0328:GDOWLS]2.0.CO;2. [DOI] [Google Scholar]
- Lebeda A, Ryder EJ, Grube R, Doležalová I, Křístková E. Genetic resources, chromosome engineering, and crop improvement series. In: Singh R, editor. Lettuce (Asteraceae; Lactuca spp) Boca Raton: CRC Press; 2007. pp. 377–472. [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- MacKay T, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- Mallory-Smith CA, Thill DC, Dial MJ, Zemetra RS. Inheritance of sulfonylurea herbicide resistance in Lactuca spp. Weed Technol. 1990;4:787–790. [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis—a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami K, Sawada Y, Suzuki M, Kenmoku H, Kawaide H, Mitsuhashi W, Sassa T, Inoue Y, Kamiya Y, Toyomasu T. Deactivation of gibberellin by 2-oxidation during germination of photoblastic lettuce seeds. Biosci Biotechnol Biochem. 2003;67:1551–1558. doi: 10.1271/bbb.67.1551. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- NASS (2008) National Agricultural Statistics Service, USDA
- O’Brien LK (2007) Lettuce (Lactuca sativa) seed and seedling traits and Sclerotinia disease resistance: a quantitative trait loci analysis of a recombinant inbred line population. MS Thesis, University of California, Davis, CA, 169 p
- Paran I, Kesseli RV, Michelmore R. Identification of restriction fragment length polymorphism and random amplified polymorphic DNA markers linked to downy mildew resistance genes in lettuce using near-isogenic lines. Theor Appl Genet. 1991;85:985–993. doi: 10.1139/g91-157. [DOI] [PubMed] [Google Scholar]
- Park S-Y, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, T-fF Chow, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu J-K, Schroeder JI, Volkman BF, Cutler SR. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinski J, Khan AA. Relationship of ethylene production to stress alleviation in seeds of lettuce cultivars. J Am Soc Hortic Sci. 1990;115:294–298. [Google Scholar]
- Saini HS, Consolacion ED, Bassi PK, Spencer MS. Requirement for ethylene synthesis and action during relief of thermoinhibition of lettuce seed germination by combinations of gibberellic acid, kinetin, and carbon dioxide. Plant Physiol. 1986;81:950–953. doi: 10.1104/pp.81.4.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini HS, Consolacion ED, Bassi PK, Spencer MS. Control processes in the induction and relief of thermoinhibition of lettuce seed germination: actions of phytochrome and endogenous ethylene. Plant Physiol. 1989;90:311–315. doi: 10.1104/pp.90.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, Aoki M, Nakaminami N, Mitsuhashi W, Tatematsu K, Kushiro T, Koshiba T, Kamiya Y, Inoue Y, Nambara E, Toyomasu T. Phytochrome- and gibberellin-mediated regulation of abscisic acid metabolism during germination of photoblastic lettuce seeds. Plant Physiol. 2008;146:1386–1396. doi: 10.1104/pp.107.115162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, Katsumata T, Kitamura J, Kawaide H, Nakajima M, Asami T, Nakaminami K, Kurahashi T, Mitsuhashi W, Inoue Y, Toyomasu T. Germination of photoblastic lettuce seeds is regulated via the control of endogenous physiologically active gibberellin content, rather than of gibberellin responsiveness. J Exp Bot. 2008;59:3383–3393. doi: 10.1093/jxb/ern192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwember AR, Bradford KJ. A genetic locus and gene expression patterns associated with the priming effect on lettuce seed germination at elevated temperatures. Plant Mol Biol. 2010;73:105–118. doi: 10.1007/s11103-009-9591-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, North H, Marion-Poll A, Sun T-p, Koshiba T, Kamiya Y, Yamaguchi S, Nambara E. Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 2006;48:354–366. doi: 10.1111/j.1365-313X.2006.02881.x. [DOI] [PubMed] [Google Scholar]
- Stam P (1995) JoinMap 2.0 deals with all types of plant mapping populations. Plant Genome III Abstracts http://www.intl-pag.org/3/abstracts/47pg3.html
- Sung Y, Cantliffe DJ, Nagata RT. Seed developmental temperature regulation of thermotolerance in lettuce. J Am Soc Hortic Sci. 1998;123:700–705. [Google Scholar]
- Sung Y, Cantliffe DJ, Nagata RT, Nascimento WM. Structural changes in lettuce seed during germination at high temperature altered by genotype, seed maturation temperature, and seed priming. J Am Soc Hortic Sci. 2008;133:300–311. [Google Scholar]
- Tamura N, Yoshida T, Tanaka A, Sasaki R, Bando A, Toh S, Lepiniec L, Kawakami N. Isolation and characterization of high temperature-resistant germination mutants of Arabidopsis thaliana. Plant Cell Physiol. 2006;47:1081–1094. doi: 10.1093/pcp/pcj078. [DOI] [PubMed] [Google Scholar]
- Toh S, Imamura A, Watanabe A, Nakabayashi K, Okamoto M, Jikumaru Y, Hanada A, Aso Y, Ishiyama K, Tamura N, Iuchi S, Kobayashi M, Yamaguchi S, Kamiya Y, Nambara E, Kawakami N. High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 2008;146:1368–1385. doi: 10.1104/pp.107.113738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truco MJ, Antonise R, Lavelle D, Ochoa O, Kozik A, Witsenboer H, Fort SB, Jeuken MJW, Kesseli RV, Lindhout P, Michelmore R, Peleman J. A high-density integrated genetic linkage map of lettuce (Lactuca spp.) Theor Appl Genet. 2007;115:735–746. doi: 10.1007/s00122-007-0599-9. [DOI] [PubMed] [Google Scholar]
- Valdes VM, Bradford KJ, Mayberry KS. Alleviation of thermodormancy in coated lettuce seeds by seed priming. HortScience. 1985;20:1112–1114. [Google Scholar]
- van Leeuwen H, Stoffel K, Mathrakott S, Chen F, Tan X, Yao J, Wong G, Cui X, Kozik A, Truco MJ, Michelmore RW, Van Deynze A. Analysis of single feature polymorphism markers with a lettuce Affymetrix GeneChip for massively parallel marker discovery, genotyping and mapping. San Diego: Plant and Animal Genome XVI; 2008. p. 31. [Google Scholar]
- van Leeuwen H, Stoffel K, Kozik A, Cui X, Ashrafi H, McHale L, Lavelle D, Wong G, Chen F, Truco MJ, Van Deynze A, Michelmore RW. High-density mapping of the lettuce genome with SFP markers in over 15, 000 unigenes. San Diego: Plant and Animal Genome XVII; 2009. [Google Scholar]
- Wang H, Caruso LV, Downie AB, Perry SE. The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell. 2004;16:1206–1219. doi: 10.1105/tpc.021261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waycott W, Fort SB, Ryder EJ, Michelmore RW. Mapping morphological genes relative to molecular markers in lettuce (Lactuca sativa L.) Heredity. 1999;82:245–251. doi: 10.1038/sj.hdy.6884730. [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol. 1984;35:155–189. doi: 10.1146/annurev.pp.35.060184.001103. [DOI] [Google Scholar]
- Yoshioka T, Endo T, Satoh S. Restoration of seed germination at supraoptimal temperatures by fluridone, an inhibitor of abscisic acid biosynthesis. Plant Cell Physiol. 1998;39:307–312. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.