Abstract
Staphylococcus aureus is a significant human pathogen that causes skin-structure, invasive, and hospital-associated infections worldwide. The complement system is vital to innate defense against many bacterial infections. As shown with other pathogens, mechanisms for circumventing complement attack may include recruitment of the complement regulatory protein factor H (fH). In the present study, we show that S. aureus binds fH in a dose-dependent and time-dependent manner. Interestingly, this interaction does not require complement activation nor C3-fragment presence and occurs efficiently in the absence of other serum components suggesting a mechanism other than bridging between intermediary molecules. However, fH binding is greater when incubated with normal human serum compared to heat-inactivated serum, which suggests that complement activation may enhance fH binding. S. aureus-bound fH was found to inhibit the alternative pathway through disruption of the alternative pathway C3 convertase as shown by an increase in Bb release and a decrease in total C3-fragment deposition. Furthermore, S. aureus-bound fH retains cofactor activity for factor-I mediated cleavage of C3b. These studies show that the acquisition of fH to the S. aureus surface inhibits complement-mediated opsonization via disruption of the alternative pathway convertase; thus, we report an immune-evasion mechanism not previously described for S. aureus.
Keywords: Staphylococcus aureus, factor H, complement, immune evasion
1. Introduction
Staphylococcus aureus continues to be one of the most frequent causes of community and hospital-associated bacterial infections, with methicillin-resistant S. aureus (MRSA) becoming increasingly common (CDC, 2004; Rosenthal et al., 2010). S. aureus is responsible for a multitude of superficial and invasive infections resulting in significant morbidity and mortality worldwide (Chang et al., 2003; Hakim et al., 2007; Haupt et al., 2008; Nizet, 2007). As antibiotic resistance continues to rise, novel therapies for treating and preventing S. aureus infection are essential. As such, elucidating the mechanisms by which S. aureus interacts with and evades the host immune response provides a means to identify novel therapeutic targets.
As a successful pathogen, S. aureus can survive and replicate within its host, and in doing so, must subvert the host immune response. One of the primary targets of staphylococcal immune evasion is the complement system, a vital component of the innate immune defense against bacterial infections (Hair et al., 2008; Ricklin et al., 2009). Complement activation results in the opsonization of the bacterial cell surface, thereby facilitating bacterial uptake and their subsequent destruction via phagocytes (Foster, 2005). To circumvent complement-mediated opsonization, S. aureus secretes potent molecules that directly target the alternative pathway C3-convertase (C3bBb). These include the staphylococcal complement inhibitor (SCIN), the staphylococcal extracellular complement-binding protein (Ecb), and the extracellular fibrinogen-binding protein (Efb) (Chen et al., 2010; Jongerius et al., 2010; Ricklin et al., 2009). While both Ecb and Efb prevent the formation of C3bBb by binding to C3b (Chen et al., 2010; Jongerius et al., 2010), SCIN inactivates this convertase by fixating C3bBb (Ricklin et al., 2009). Complement evasion by S. aureus also occurs via the recruitment of the soluble complement regulator factor I to the bacterial surface, which has previously been demonstrated by our lab (Cunnion et al., 2004b; Hair et al., 2010; Hair et al., 2008).
Host cells are protected from complement attack via both membrane-associated and soluble complement regulatory proteins. The plasma protein factor H is a 155 kDa major fluid-phase complement regulator that disrupts the alternative pathway C3 convertase by displacing factor Bb and also acts as a cofactor for factor I-mediated cleavage of C3b (Liszewski et al., 2008; Zipfel et al., 2002). Factor H-like protein 1 (FHL-1) is a 42 kDa splice variant of fH, with similar functionality to fH (Johnsson et al., 1998; Zipfel et al., 2002). These regulatory proteins are composed of short-consensus repeats (SCRs) of which the first seven SCRs are common to both proteins (Jozsi and Zipfel, 2008; Zipfel et al., 2002). Factor H related protein 1 (FHR-1), the product of a different gene, is present in two forms: FHR1-α (37 kDa) and FHR1-β (43 kDa). FHR-1 contains five SCRs that are homologous to those found in fH; however, its function is not well described (Friberg et al., 2008; Zipfel et al., 2002). Pathogens such as Streptococcus pneumoniae, S. pyogenes, and Borrelia burgdorferi are known to acquire fH as an immune evasion tactic to inhibit activation of the alternative pathway of complement (Jarva et al., 2002; Jarva et al., 2003; Kenedy et al., 2009; Kraiczy et al., 2001; Pandiripally et al., 2002; Perez-Caballero et al., 2000).
A previous study identifies the secreted S. aureus protein Sbi (Staphylococcus aureus binder of IgG) as a fH-binding protein which binds fH via the Sbi domains III and IV; however, this binding requires the presence of C3b or C3d, with little or no binding evident in the absence of these molecules (Haupt et al., 2008). In the present study, we show that intact S. aureus binds considerable amounts of fH whether in serum or in purified form irrespective of complement activation or C3-fragment presence. We also demonstrate, for the first time, that fH is functionally active on the S. aureus surface, disrupting the alternative pathway C3 convertase and inhibiting opsonization.
2. Materials and methods
2.1. Bacteria
S. aureus were grown to mid-logarithmic phase (OD600 0.8 – 1.5) in Columbia 2% NaCl broth at 37ºC. Six laboratory strains (Newman, Wood, Lowenstein, Reynolds, Wright, Becker) and ten clinical isolates (5 MSSA, 5 MRSA) were used for initial fH-binding studies. The clinical isolates were obtained as de-identified, discarded specimens from a clinical microbiology laboratory (Eastern Virginia Medical School IRB 06-04-WC-0040). Strains Reynolds, Newman, and a clinical isolate named R7 were used for additional studies, as indicated. R7 is a community associated methicillin resistant S. aureus (CA-MRSA) determined to be a USA300 strain by pulsed-field gel electrophoresis (data not shown).
2.2. Buffers
The following buffers were used: GVBS ++ (veronal-buffered saline [VBS] with 0.1% gelatin, 0.15 mM CaCl2, and 1.0 mM MgCl2), GVBS (VBS with 0.1% gelatin), GVBS EDTA (VBS with 0.1% gelatin and 0.01 M EDTA), and VBS-Ni (VBS with 1mM NiCl2). GVBS++ buffer and VBS-Ni allow the activation of the complement system due to the presence of cations; alternatively, EDTA chelates these cations and, therefore, inhibits the complement cascade.
2.3. Serum
Normal human serum (NHS) was made as previously described (Cunnion et al., 2001) from the blood of four healthy human volunteers in accordance with an Institutional Review Board approved protocol (EVMS IRB 02-06-EX-0216). The serum was pooled, aliquoted, and frozen at −80ºC. Heat-inactivated serum was generated by heating NHS at 56ºC for 30 minutes. Cobra-venom-factor treated serum (CVS) was produced by the addition of 20 μg of cobra-venom factor (CompTech) to 1 mL of NHS, and heated at 37°C for 1 hour. C3-depleted serum was purchased commercially (CompTech).
2.4. Cell-wall preparations
Cell-wall extracts were prepared as previously described (Cheung and Fischetti, 1988). Briefly, S. aureus cells were sedimented from 20 mL cultures, washed twice with GVBS EDTA and resuspended in 30% raffinose buffer to stabilize the bacterial protoplasts; protease inhibitors (complete mini, Roche) and DNase were also added. Cell-wall proteins were solubilized by incubating with 10 μg lysostaphin (Sigma) at 37°C for 1 hour, with rotation. The protoplasts were sedimented and cell-wall proteins were recovered in the supernatant.
2.5. fH-binding assays
S. aureus (1 × 108 organisms) were incubated with various concentrations of serum for 30 mins at 37°C to assay the binding of fH under these conditions. Additionally, the binding of purified fH (CompTech) was assessed using 2.5 × 108 organisms that were incubated with various amounts of fH for 1 hr at 37°C. The assays were conducted using the buffers indicated. Following incubation with sera or pure proteins, cells were washed thoroughly with GVBS EDTA and bound proteins were stripped using 2% SDS at 95°C for 5 mins or extracted with 0.1M glycine (pH 2.5) for 20 mins at 37°C, as described by Friberg et al (Friberg et al., 2008). Among a variety of methods tested, 2% SDS yielded the maximum recovery of fH from S. aureus.
A variation of this assay involved the assessment of fH retention and release by S. aureus strain Reynolds. Bacteria were incubated with 20% NHS in GVBS++ buffer for 30 mins at 37°C followed by washing in GVBS EDTA and subsequent resuspension in 3% BSA/PBS. Aliquots of 1 × 108 organisms/50uL of buffer were assessed for retention and release at various time points. Released fH from the cell surface was captured in the supernatant and retained fH was stripped in 50 uL of 2% SDS at 95°C for 5 mins.
2.6. Whole-cell ELISA
Binding of fH to immobilized S. aureus strains Reynolds and R7 was performed as described by Pandiripally et al. (Pandiripally et al., 2002), with modifications. Bacteria were washed in PBS then resuspended in carbonate buffer. Flat-bottom Immulon 2 plates (Thermo Labsystems) were coated with 1 × 107 organisms in 50 uL of carbonate buffer and incubated overnight at 4°C. Wells were washed to remove unbound cells with wash buffer (PBST [phosphate buffered saline with 0.05% tween] containing 0.5% gelatin) then blocked with wash buffer for 90 mins at 37°C. Various amounts of purified fH (in wash buffer) were added to wells and allowed to incubate at room temperature for 1 hr. Control wells were incubated with wash buffer only. Following the removal of unbound fH, the presence of fH was assessed. The primary and secondary probes used were chicken anti-fH (Accurate Chem.) and goat anti-chicken HRP (Genway Biotech, Inc.), respectively. Probes were added in wash solution and allowed to incubate for 1 hr at room temperature with wash steps in between incubations. Plates were developed with TMB substrate (Thermo Scientific), stopped with 1N H2SO4, and read at 450 nm. Absorbance values were analyzed for fH presence with values from wells not incubated with fH subtracted as background.
2.7. C3b cleavage to iC3b
Washed mid-logarithmic S. aureus were incubated with 10% heat-inactivated serum for 30 mins at 37°C in GVBS to bind serum fH, based on an assay described by Friberg et al. (Friberg et al., 2008). Bacteria were washed 3 × with GVBS. To aliquots of 1 × 108 organisms, various combinations of factor I, C3b or C3 (1 μg each) were added and allowed to incubate for 1 hr at 37°C. Controls included bacteria not incubated with serum, and samples of purified fH and factor I with purified C3 or C3b and no bacteria. Purified proteins were purchased commercially from CompTech. The bacteria were sedimented and the supernatant was analyzed for evidence of iC3b via Western Blotting, under reducing conditions.
2.8. Opsonization with purified complement components
C3b molecules were deposited as described by Horstmann et al. (Horstmann et al., 1985), with modifications. Washed mid-logarithmic S. aureus Reynolds (4.0 × 109 organisms in VBS-Ni buffer) were incubated with 50 μg C3 in 1 mL VBS-Ni to which 100 uL of fluid phase C3bBb(Ni) enzyme was added and incubated at 20°C for 30 min. The enzyme was preformed with 30 μg C3b, 20 μg fB, 5 μg fD incubated in 50 uL VBS-Ni for 5 min at 20°C. The cells were washed 3 × in VBS-Ni then split evenly into two groups: one group was incubated with 8 μg fH, the other without fH (control), for 30 min at 37°C. The cells were sedimented then resuspended in VBS-Ni; aliquots of 5 × 108 organisms were further incubated with 8 μg C3, 5 μg fB, 2 μg properdin and 0.2 μg fD in 125 μL VBS-Ni at 4°C for various time points. The cells were then sedimented and shed Bb was recovered in the supernatant. The cells were washed 3 × in GVBS EDTA then incubated in 50 μL of 25 mM methylamine at 37°C for 1 hr to release C3 fragments bound to the cell surface via ester linkage into the supernatant; the bacteria were removed via centrifugation (Cunnion et al., 2004b).
2.9. ELISA
Flat-bottom Immulon 2 plates (Thermo Labsystems) were coated with 50 μL of either goat anti C3 (10 μg/mL), goat anti fH (1:1000 dilution), or goat anti fB (1:1000 dilution) in carbonate buffer (4°C, overnight), for C3, fH or Bb ELISA, respectively. Washed wells were blocked with 3% BSA/PBST (2 hr, room temperature) then washed again. Samples were added to the wells in block buffer (1 hr, room temperature). Primary and secondary probes were as follows: chicken anti-C3 (Sigma) and goat anti-chicken HRP (Sigma) for C3 ELISA; mouse monoclonal anti fH (Serotec Ltd., Oxford, United Kingdom) and goat anti-mouse HRP (Sigma) for fH ELISA; mouse anti-Bb neoantigen (Quidel) and goat anti-mouse HRP for Bb ELISA. Probes were added in block buffer and allowed to incubate for 1 hr at room temperature with wash steps in between incubations. Plates were developed with TMB substrate (Thermo Scientific), stopped with 1N H2SO4, and read at 450 nm. Purified C3 and fH (CompTech) were used as standards for quantitation of protein. Absorbance values were analyzed for Bb presence with control wells subtracted as background.
2.10. Molecules of fH per bacterium calculation
Given fH = 155 kDa = 155,000 g/mol: μg/mL fH × extraction volume (mL) = g fH; g fH × mol × (6.022 × 10 23 molecules/mol) / (155,000 g) = molecules of fH; (molecules of fH)/(number of bacteria in sample) = molecules of fH per bacterium.
2.11. Statistical analysis
ELISA data were analyzed by a two-tailed paired Student’s t test. Calculated P values of ≤ 0.05 were considered statistically significant.
3. Results
3.1. fH binds to S. aureus in a time- and dose-dependent manner
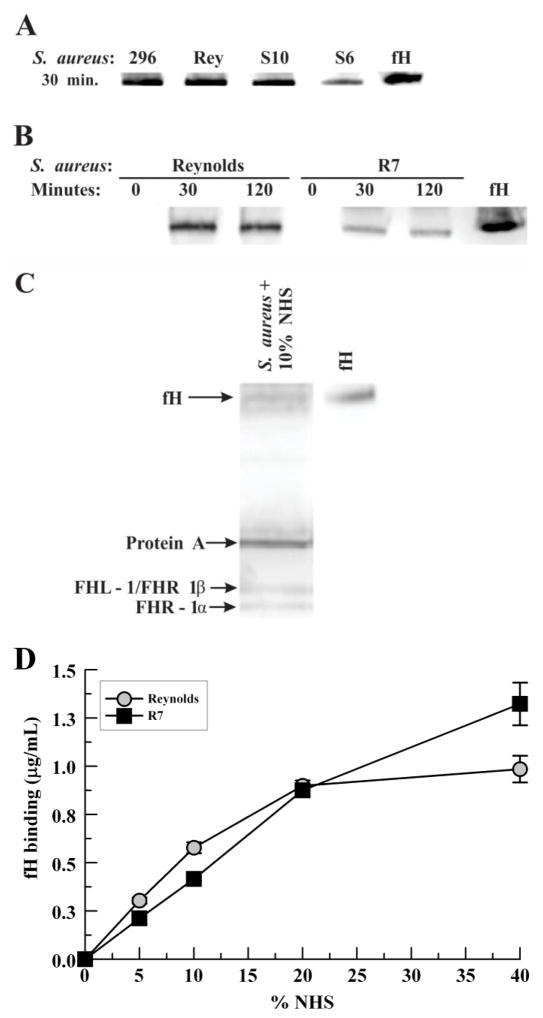
To examine the commonality for binding serum fH by S. aureus, six laboratory strains and ten clinical isolates were incubated with 10% NHS, washed, and cell-wall extracts were prepared. All strains bound serum fH, as determined via Western blotting; a representative Western blot is shown in Fig. 1A. Reynolds and the CA-MRSA clinical isolate R7 bound significant fH by 30 minutes by Western-blot analysis (Fig. 1B). An alternate method of examining the binding of serum fH to S. aureus was performed by incubating R7 with 10% NHS followed by acid elution of bound proteins. Western blotting revealed a band for fH as well as bands that likely represent FHL-1/FHR-1β and FHR-1α (Fig 1C). To further assess the binding of serum fH to S. aureus, various concentrations of NHS were incubated with S. aureus Reynolds and R7 and fH binding was measured by ELISA (Fig. 1D). A positive correlation of increasing amounts of bound fH was found for increasing NHS concentrations.
Fig. 1.
Serum factor H binding to mid-logarithmic S. aureus. S. aureus were incubated with 10% normal human serum in GVBS++ for the time indicated at 37°C. Cell-wall proteins were solubilized from washed bacteria and analyzed via anti-fH Western blot: (A) Representative Western blot showing strain Newman (296), Reynolds and the clinical isolates S10 and S6; (B) Strain Reynolds and a clinical strain (R7). (C) R7 was incubated with 10% NHS in GVBS for 30 mins at 37°C, washed, then bound proteins eluted with 0.1M glycine, pH 2.5. (D) Bacteria were incubated with various concentrations of NHS in GVBS++ for 30 mins at 37°C. Bound fH was stripped from washed bacteria with 2% SDS at 95°C; fH binding was measured via ELISA. Data represent the mean of three independent experiments ± SEM. Abbreviations: factor H (fH), Factor H-like protein 1 (FHL-1), Factor H related protein 1 (FHR 1).
3.2. S. aureus binds fH independent of complement activation
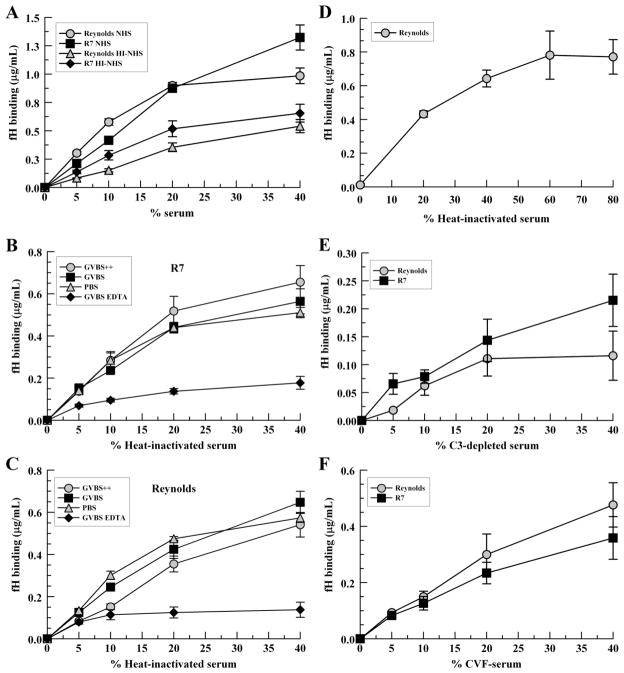
The secreted staphylococcal protein Sbi binds fH as determined using immobilized recombinant Sbi constructs; however, this interaction requires the presence of either C3b or C3d (Haupt et al., 2008). Therefore, we sought to determine the extent to which complement activation affects the binding of fH to intact S. aureus. In heat-inactivated serum, fH bound S. aureus strains in a positive dose-response manner demonstrating that complement activation is not necessary for fH binding. Overall, binding of fH was reduced by 45% (P <0.0001) and 50% (P = 0.0016) for Reynolds and R7, respectively, in heat-inactivated serum compared with NHS suggesting that complement activation, while not required, increases fH binding to S. aureus (Fig. 2A). Binding of fH in heat-inactivated serum was similar for all buffers except GVBS-EDTA, suggesting that the presence of EDTA inhibits fH binding independent of complement activation (Fig. 2B and 2C). To test for saturation of fH binding, strain Reynolds was incubated with various concentrations of heat-inactivated serum in PBS, as described, and bound fH was measured by ELISA. Saturation was achieved at 60% heat-inactivated serum (Fig 2D).
Fig. 2.
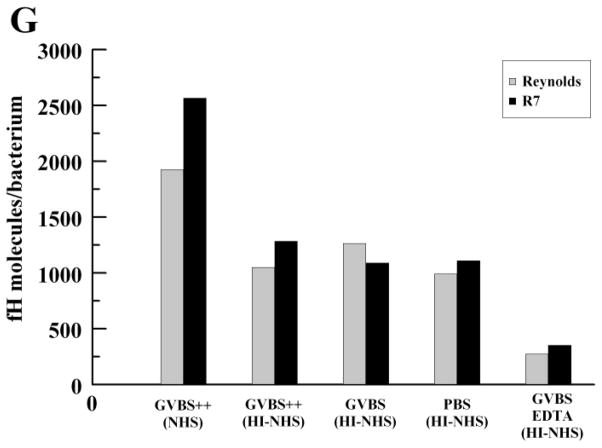
Serum factor H binding to S. aureus in the presence or absence of complement activation. S. aureus were incubated with various concentrations of NHS, heat-inactivated serum (HI-NHS), C3-depleted serum or cobra-venom factor treated serum (CVF-serum) for 30 mins at 37°C. Bound proteins were extracted with 2% SDS at 95°C for 5 mins and fH binding was measured by ELISA. (A) NHS vs. heat-inactivated serum; heat-inactivated serum in various buffers, (B) R7, (C) Reynolds; (D) heat-inactivated serum in PBS, Reynolds; (E) C3-depleted serum; (F) cobra-venom factor treated human serum; (G) fH molecules/bacterium at 40% serum per buffer type. GVBS++ buffer was used for all fH-binding assays, except where indicated. Data points represent the mean of three independent experiments ± SEM.
To further explore fH binding in conditions where C3 is depleted, we assayed the binding of fH to strain Reynolds and R7 with cobra-venom-factor treated serum and C3-depleted serum. CVF-treated serum is depleted of C3, but may have some C3b/iC3b present, whereas C3-depleted serum lacks C3, C3b, iC3b, and C3d. As shown in Fig. 2E and 2F, Reynolds and R7 can both bind fH from either type of sera, with an increase in binding correlating with an increase in serum concentration. To quantitate the amount of fH bound per bacterial cell, we converted the mean values of bound fH in 40% serum to molecules of fH per bacterium (Fig. 2G). In these conditions, NHS deposits ≥ 2,000 molecules of fH on the S. aureus surface. As a whole, these data show that neither complement activation nor C3/C3b/iC3b/C3d is required for S. aureus binding of fH.
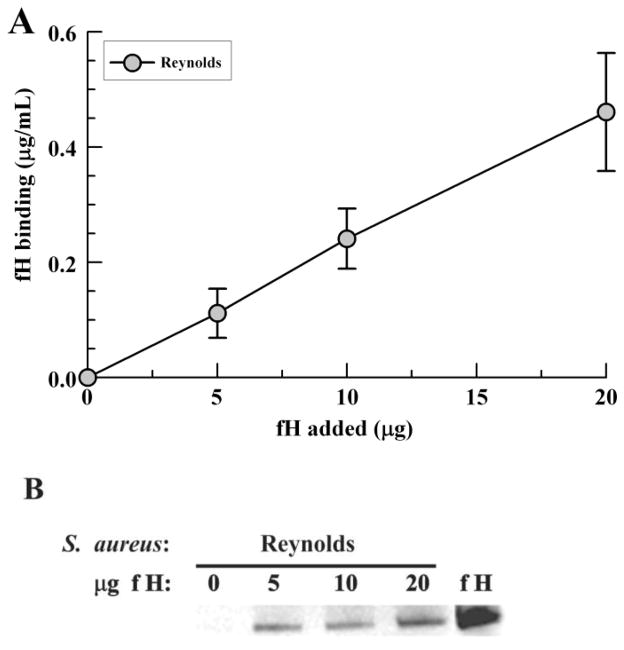
In order to evaluate whether any serum proteins are necessary for S. aureus to bind fH, various concentrations of purified fH were incubated with S. aureus strain Reynolds. Bound proteins were stripped in 2% SDS and examined for binding via Western blotting and ELISA. Similar to incubation with serum fH, higher concentrations of purified fH were associated with increased fH binding by ELISA (Fig. 3A). Binding of intact fH was confirmed by Western blot analysis (Fig. 3B).
Fig. 3.
Purified fH binding to S. aureus. Bacteria were incubated with various concentrations of purified fH for 1 hour at 37°C. Bound fH was stripped from washed bacteria with 2% SDS at 95°C and measured by (A) ELISA or (B) Western blot. Data points represent the mean of three independent experiments ± SEM.
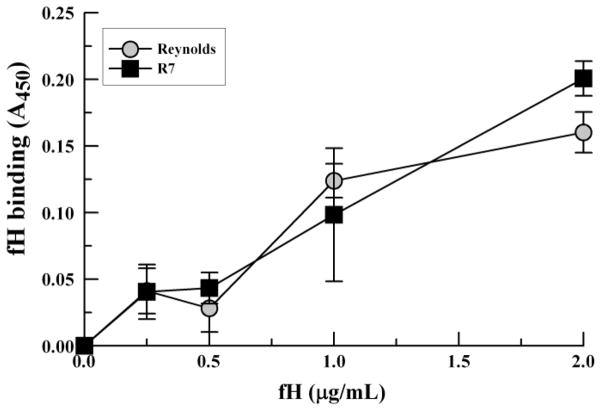
To further substantiate that S. aureus binds fH without additional serum components, we employed a whole-cell assay. S. aureus strains Reynolds and R7 were adsorbed to microtiter plates and incubated with various amounts of purified fH. The results depicted in Fig. 4 show increased fH binding correlating with increased fH concentration. These results further support that a dose-response relationship exists for fH binding to the S. aureus surface and that S. aureus binds fH efficiently in the absence of serum intermediary molecules.
Fig. 4.
Factor H binding via whole-cell ELISA. S. aureus were adsorbed to wells of a microtitre plate, incubated with various amounts of purified fH, and probed with chicken anti-fH antibody. Data points represent the mean of three independent experiments ± SEM.
3.3 fH retention and release by S. aureus
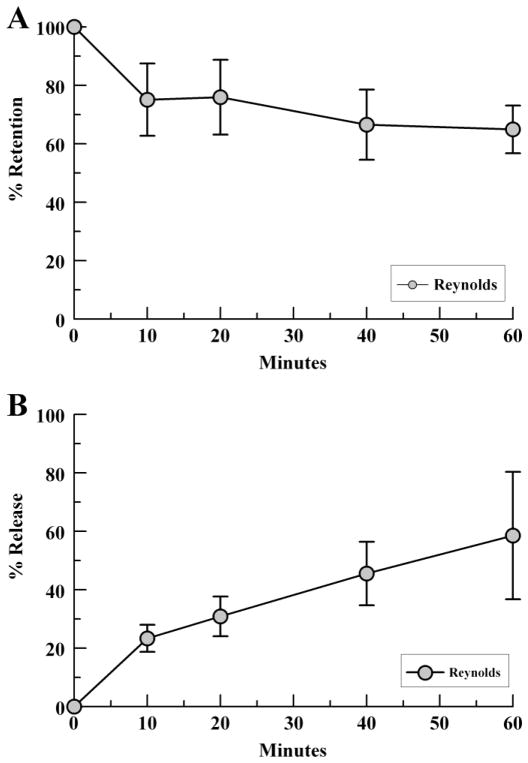
To further characterize the binding relationship of fH to intact S. aureus, we examined the retention and release rates of S. aureus-bound fH by ELISA. Strain Reynolds was incubated with 20% NHS, washed, and then incubated in 3% BSA/PBS. As shown in Fig. 5, retention of fH decreased over time, with a mean retention of 75% fH at 10 mins which dropped to 65% retention at 60 mins. These data correlated with increasing amount of fH released into the supernatant over time.
Fig. 5.
Factor H retention and release by S. aureus. Bacteria were incubated with 20% NHS to bind serum fH then washed and resuspended in 3%BSA/PBS. Samples were assessed for percent fH retention (pellet, A) and release (supernatant, B) at various time points via fH ELISA. Data points represent the mean of three independent experiments ± SEM.
3.4. S. aureus-bound fH increases the release of Bb and decreases C3 fragment deposition on the S. aureus surface
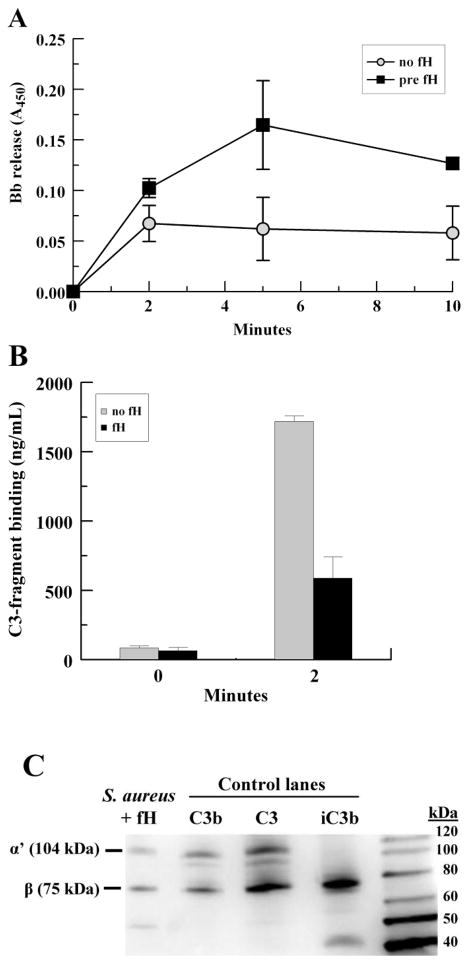
To date, the functional importance of fH recruitment to the S. aureus surface has not been elucidated. Therefore, we sought to determine the extent to which S. aureus-bound fH affects the alternative pathway C3-convertase by building the convertase on the bacterial surface using purified components. Mid-log strain Reynolds were incubated with the alternative pathway enzyme, C3bBb, and purified C3 to deposit C3b on the bacterial surface. Washed bacteria were then incubated with C3, properdin, fB and fD for various time points to provide conditions permissive for the formation of the alternative pathway C3-convertase and complement amplification. Shed Bb and bound C3 fragments were assessed via ELISA. To ascertain whether fH had an effect on released Bb or bound C3 fragments, one group of bacteria was pre-incubated with purified fH to allow fH binding prior to the amplification step.
For the group pre-incubated with fH, Bb release increased over the first 5 minutes. Overall, Bb release was 2-fold greater (P = 0.032) for S. aureus pre-incubated with fH compared with control (Fig. 6A). We also analyzed the deposition of C3-fragments on the S. aureus surface via ELISA. As shown in Fig. 6B, total C3 deposition was decreased 3-fold at 2 minutes in samples pre-incubated with fH, compared to control (P = 0.026). These data strongly suggest that S. aureus-bound fH disrupts the alternative pathway C3 convertase, as shown by an increase in released Bb and a marked reduction in total C3 deposition. The form of C3 deposited on the S. aureus surface was confirmed to be C3b by Western blot (Fig. 6C).
Fig. 6.
S. aureus- bound fH increases the release of Bb and decreases C3 fragment deposition on the S. aureus surface. Washed mid-logarithmic S. aureus were incubated with fluid phase C3bBb(Ni) enzyme and purified C3 at 20°C for 30 min to deposit C3b on the bacterial surface. After washing, organisms were incubated ± fH for 30 min, and then incubated with C3, fB, properdin and fD at 4°C for various time points. Released Bb was recovered in the supernatant and bound C3-fragments were stripped with 25mM methylamine. (A) Bb ELISA; (B) C3 ELISA; (C) Representative Western blot showing forms of C3 stripped from S. aureus surface compared to purified C3, C3b and iC3b control lanes; the type of C3 chain is indicated (α’, β). Data points represent the mean of three independent experiments ± SEM.
3.5. Cleavage of C3b to iC3b occurs in the presence of fH and factor I (fI)
To investigate whether S. aureus-bound fH maintains its cofactor activity for fI-mediated cleavage of C3b to iC3b, we tested three S. aureus strains: Reynolds, Newman and R7. Bacteria were incubated with 10% heat-inactivated serum for 30 minutes at 37°C to bind serum fH. Control samples were not incubated with serum. Bacteria were washed and either purified C3 or purified C3b, with or without fI, was added. Purified C3 may undergo spontaneous hydrolysis resulting in C3(H20), a C3b-like molecule that can be cleaved to iC3b by fI in the presence of fH (Oran and Isenman, 1999). Western blotting revealed that the generation of iC3b occurred only when bacteria were pre-incubated with serum fH and in the presence of fI (Fig. 7). This indicates that fH bound by S. aureus retains cofactor activity for fI cleavage of C3b. It should be noted, however, that the binding of FHL-1 and/or FHR 1 by S. aureus, as suggested in Fig. 1C, may also contribute to the generation of iC3b shown here, due to their presence in serum.
Fig. 7.
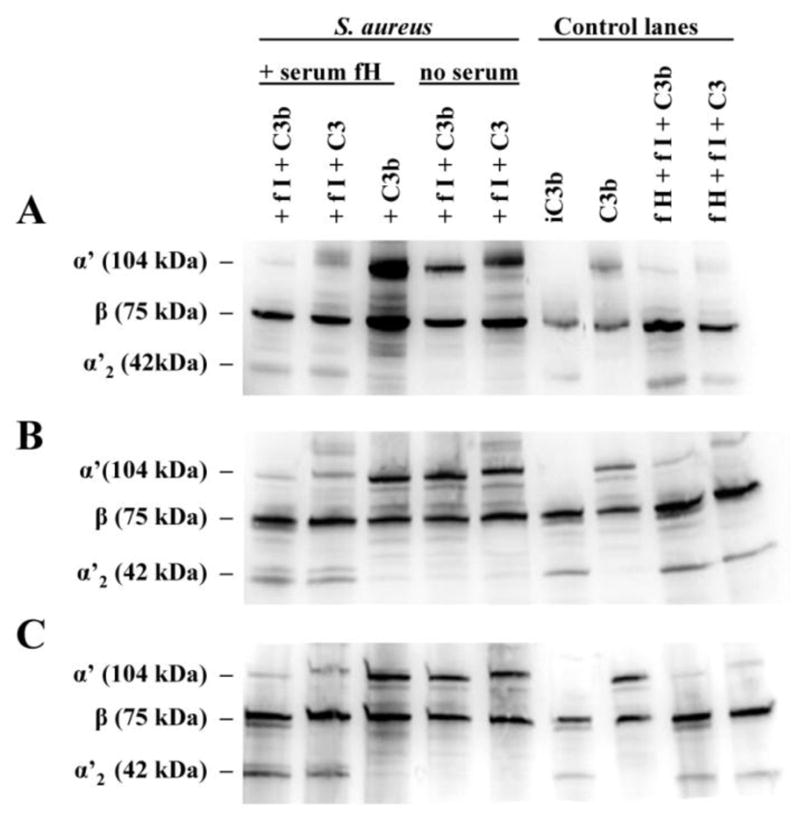
Cleavage of C3b to iC3b occurs in the presence of fH and fI. S. aureus were incubated with 10% heat-inactivated serum for 30 mins at 37°C to bind serum fH. After washing, various combinations of fI, C3b or C3 (1 ug each) were added and allowed to incubate for 1 hr at 37°C. Controls included bacteria not incubated with serum, and samples of purified fH and fI with purified C3 or C3b. The bacteria were sedimented and the supernatant was analyzed for the presence of C3b and iC3b via Western blotting, under reducing conditions; the types of C3 chains are indicated (α’, β, α’2). (A) R7; (B) Reynolds; (C) Newman. Representative Western blots of three independent experiments.
4. Discussion
A diverse group of pathogenic bacteria have developed strategies to circumvent host immune defenses, with a commonality for complement evasion (Blom et al., 2009; Jarva et al., 2002; Jarva et al., 2003; Kenedy et al., 2009; Kraiczy et al., 2001; Pandiripally et al., 2002; Perez-Caballero et al., 2000); however, the interaction between S. aureus and complement regulatory proteins is only beginning to be explored. We hypothesized that S. aureus binds fH in a manner that preserves its inherent regulatory functionality thereby providing an immune evasion mechanism for disrupting the alternative pathway C3 convertase via displacement of Bb from the bacterial surface. Resultant decreases in C3 activation and C3b binding to S. aureus inhibit opsonization along with downstream effector functions. Although Sbi has been shown to bind fH, Sbi-bound fH occurs via the formation of a tripartite complex with C3b or C3d, as demonstrated using immobilized recombinant Sbi III/IV constructs (Haupt et al., 2008). Moreover, Sbi is a secreted protein and not found in the cell-wall fraction (Burman et al., 2008). To elucidate the binding of fH to the surface of S. aureus, we employed fH-binding assays that permit fH binding to intact S. aureus followed by a functional analysis of S. aureus-bound fH. These assays provide a novel insight into the physiological activities of fH on the S. aureus surface.
In the present study, we tested sixteen strains of S. aureus, six laboratory and 10 clinical isolates, for the ability to bind fH; all of which bound serum fH suggesting that this is a common property for S. aureus strains. Interestingly, fH bound efficiently to S. aureus in the absence of C3/C3b/C3d or other serum proteins suggesting that S. aureus can bind fH via an as yet undescribed mechanism. However, it remains possible that Sbi could contribute to fH binding in the presence of complement activation, if secreted Sbi binds back to the bacterial surface.
Curiously, fH binding in GVBS EDTA buffer with heat-inactivated serum resulted in a marked decrease in fH binding compared to heat-inactivated serum in GVBS++. This suggests that EDTA inhibits fH binding to S. aureus by a mechanism unrelated to complement activation. As divalent cations are important components of bacterial cell membranes/walls (Weidenmaier and Peschel, 2008), we speculate that the chelating agent EDTA may affect the S. aureus surface molecule(s) to which fH binds. Although fH is not known to have an interaction with divalent cations, it is also possible that EDTA could affect fH.
Our functional analyses reveal that surface-bound fH inhibits C3 activation on S. aureus by displacing Bb, resulting in decreased binding of the complement effector C3b on the bacterial surface. C3b has been shown to be a vital opsonin for S. aureus phagocytosis (Cunnion et al., 2005; Cunnion et al., 2003) as well as important in the control of S. aureus bacteremia in animal models (Cunnion et al., 2004a). To our knowledge, this is the first report demonstrating functional activities for fH recruited to the S. aureus surface. Additionally, S. aureus-bound fH retains its cofactor function for fI, enhancing the cleavage of C3b.
S. aureus is known to express many immune evasion proteins, some of which inhibit the complement system at the C3 level leading to reduced opsonization. The staphylococcal proteins Ecb, SCIN, and Efb interfere with C3b deposition on the S. aureus surface via their interaction with C3b (Chen et al., 2010; Jongerius et al., 2010; Ricklin et al., 2009). However, disruption of the alternative pathway convertase by the release of Bb, shown here, cannot be explained by the action of any of these secreted proteins. Moreover, our assays used live bacteria such that secreted S. aureus factors could participate, thus controlling for the action of these molecules in all experimental groups. The potent effect we report mediated by the acquisition of fH to the staphylococcal surface is independent of previously described host-defense-evasion strategies. Thus, the mechanism shown here where fH is recruited to the bacterial surface leading to Bb displacement and decreased C3b deposition is novel for S. aureus.
Our Western-blot analysis suggests that S. aureus also binds FHL-1 and/or factor H-related proteins as seen with other pathogenic bacteria like S. pyogenes and B. burgdorferi (Johnsson et al., 1998; Kraiczy et al., 2001). S. aureus-bound serum proteins extracted under acidic conditions revealed a 42 kDa band, as assessed via anti-fH western blotting, which is likely FHL-1 and/or FHR-1β. Additionally, a 37 kDa band may represent FHR-1α. Although likely, it is unknown whether fH, FHL-1 and/or FHR-1 bind the same or different structures on the S. aureus surface.
In conclusion, our data clearly demonstrate that S. aureus binds fH in a dose-dependent manner requiring neither complement activation, nor C3/C3b/C3d, nor other serum components. Moreover, S. aureus-bound fH down-regulates the alternative pathway of complement by disrupting the alternative pathway C3-convertase by displacing Bb and decreasing C3b deposition. Since C3b is a critical opsonin for S. aureus, this mechanism likely contributes to S. aureus pathogenicity. Future studies will investigate the role of fH binding to intact S. aureus in the organism’s survival and killing.
Acknowledgments
This work was supported by the National Institutes of Health: R21AI082398. PFGE support was kindly provided by Dr. E. Stephen Buescher, Department of Pediatrics, Children’s Hospital of the King’s Daughters, Norfolk, VA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blom AM, Hallstrom T, Riesbeck K. Complement evasion strategies of pathogens-acquisition of inhibitors and beyond. Mol Immunol. 2009;46:2808–17. doi: 10.1016/j.molimm.2009.04.025. [DOI] [PubMed] [Google Scholar]
- Burman JD, Leung E, Atkins KL, O'Seaghdha MN, Lango L, Bernado P, Bagby S, Svergun DI, Foster TJ, Isenman DE, van den Elsen JM. Interaction of human complement with Sbi, a staphylococcal immunoglobulin-binding protein: indications of a novel mechanism of complement evasion by Staphylococcus aureus. J Biol Chem. 2008;283:17579–93. doi: 10.1074/jbc.M800265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004;32:470–85. doi: 10.1016/S0196655304005425. [DOI] [PubMed] [Google Scholar]
- Chang FY, MacDonald BB, Peacock JE, Jr, Musher DM, Triplett P, Mylotte JM, O'Donnell A, Wagener MM, Yu VL. A prospective multicenter study of Staphylococcus aureus bacteremia: incidence of endocarditis, risk factors for mortality, and clinical impact of methicillin resistance. Medicine (Baltimore) 2003;82:322–32. doi: 10.1097/01.md.0000091185.93122.40. [DOI] [PubMed] [Google Scholar]
- Chen H, Ricklin D, Hammel M, Garcia BL, McWhorter WJ, Sfyroera G, Wu YQ, Tzekou A, Li S, Geisbrecht BV, Woods VL, Jr, Lambris JD. Allosteric inhibition of complement function by a staphylococcal immune evasion protein. Proc Natl Acad Sci U S A. 2010;107:17621–6. doi: 10.1073/pnas.1003750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AL, Fischetti VA. Variation in the expression of cell wall proteins of Staphylococcus aureus grown on solid and liquid media. Infect Immun. 1988;56:1061–5. doi: 10.1128/iai.56.5.1061-1065.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnion KM, Benjamin DK, Jr, Hester CG, Frank MM. Role of complement receptors 1 and 2 (CD35 and CD21), C3, C4, and C5 in survival by mice of Staphylococcus aureus bacteremia. J Lab Clin Med. 2004a;143:358–65. doi: 10.1016/j.lab.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Cunnion KM, Buescher ES, Hair PS. Serum complement factor I decreases Staphylococcus aureus phagocytosis. J Lab Clin Med. 2005;146:279–86. doi: 10.1016/j.lab.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Cunnion KM, Hair PS, Buescher ES. Cleavage of complement C3b to iC3b on the surface of Staphylococcus aureus is mediated by serum complement factor I. Infect Immun. 2004b;72:2858–63. doi: 10.1128/IAI.72.5.2858-2863.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnion KM, Lee JC, Frank MM. Capsule production and growth phase influence binding of complement to Staphylococcus aureus. Infect Immun. 2001;69:6796–803. doi: 10.1128/IAI.69.11.6796-6803.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnion KM, Zhang HM, Frank MM. Availability of complement bound to Staphylococcus aureus to interact with membrane complement receptors influences efficiency of phagocytosis. Infect Immun. 2003;71:656–62. doi: 10.1128/IAI.71.2.656-662.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3:948–58. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- Friberg N, Carlson P, Kentala E, Mattila PS, Kuusela P, Meri S, Jarva H. Factor H binding as a complement evasion mechanism for an anaerobic pathogen, Fusobacterium necrophorum. J Immunol. 2008;181:8624–32. doi: 10.4049/jimmunol.181.12.8624. [DOI] [PubMed] [Google Scholar]
- Hair PS, Echague CG, Sholl AM, Watkins JA, Geoghegan JA, Foster TJ, Cunnion KM. Clumping factor A interaction with complement factor I increases C3b cleavage on the bacterial surface of Staphylococcus aureus and decreases complement-mediated phagocytosis. Infect Immun. 2010;78:1717–27. doi: 10.1128/IAI.01065-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hair PS, Ward MD, Semmes OJ, Foster TJ, Cunnion KM. Staphylococcus aureus clumping factor A binds to complement regulator factor I and increases factor I cleavage of C3b. J Infect Dis. 2008;198:125–33. doi: 10.1086/588825. [DOI] [PubMed] [Google Scholar]
- Hakim H, Mylotte JM, Faden H. Morbidity and mortality of Staphylococcal bacteremia in children. Am J Infect Control. 2007;35:102–5. doi: 10.1016/j.ajic.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Haupt K, Reuter M, van den Elsen J, Burman J, Halbich S, Richter J, Skerka C, Zipfel PF. The Staphylococcus aureus protein Sbi acts as a complement inhibitor and forms a tripartite complex with host complement Factor H and C3b. PLoS Pathog. 2008;4:e1000250. doi: 10.1371/journal.ppat.1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstmann RD, Pangburn MK, Muller-Eberhard HJ. Species specificity of recognition by the alternative pathway of complement. J Immunol. 1985;134:1101–4. [PubMed] [Google Scholar]
- Jarva H, Janulczyk R, Hellwage J, Zipfel PF, Bjorck L, Meri S. Streptococcus pneumoniae evades complement attack and opsonophagocytosis by expressing the pspC locus-encoded Hic protein that binds to short consensus repeats 8–11 of factor H. J Immunol. 2002;168:1886–94. doi: 10.4049/jimmunol.168.4.1886. [DOI] [PubMed] [Google Scholar]
- Jarva H, Jokiranta TS, Wurzner R, Meri S. Complement resistance mechanisms of streptococci. Mol Immunol. 2003;40:95–107. doi: 10.1016/s0161-5890(03)00108-1. [DOI] [PubMed] [Google Scholar]
- Johnsson E, Berggard K, Kotarsky H, Hellwage J, Zipfel PF, Sjobring U, Lindahl G. Role of the hypervariable region in streptococcal M proteins: binding of a human complement inhibitor. J Immunol. 1998;161:4894–901. [PubMed] [Google Scholar]
- Jongerius I, Garcia BL, Geisbrecht BV, van Strijp JA, Rooijakkers SH. Convertase inhibitory properties of Staphylococcal extracellular complement-binding protein. J Biol Chem. 2010;285:14973–9. doi: 10.1074/jbc.M109.091975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozsi M, Zipfel PF. Factor H family proteins and human diseases. Trends Immunol. 2008;29:380–7. doi: 10.1016/j.it.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Kenedy MR, Vuppala SR, Siegel C, Kraiczy P, Akins DR. CspA-mediated binding of human factor H inhibits complement deposition and confers serum resistance in Borrelia burgdorferi. Infect Immun. 2009;77:2773–82. doi: 10.1128/IAI.00318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraiczy P, Skerka C, Kirschfink M, Brade V, Zipfel PF. Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL-1/reconectin and Factor H. Eur J Immunol. 2001;31:1674–84. doi: 10.1002/1521-4141(200106)31:6<1674::aid-immu1674>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Liszewski MK, Fang CJ, Atkinson JP. Inhibiting complement activation on cells at the step of C3 cleavage. Vaccine. 2008;26(Suppl 8):I22–7. doi: 10.1016/j.vaccine.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V. Understanding how leading bacterial pathogens subvert innate immunity to reveal novel therapeutic targets. J Allergy Clin Immunol. 2007;120:13–22. doi: 10.1016/j.jaci.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Oran AE, Isenman DE. Identification of residues within the 727–767 segment of human complement component C3 important for its interaction with factor H and with complement receptor 1 (CR1, CD35) J Biol Chem. 1999;274:5120–30. doi: 10.1074/jbc.274.8.5120. [DOI] [PubMed] [Google Scholar]
- Pandiripally V, Gregory E, Cue D. Acquisition of regulators of complement activation by Streptococcus pyogenes serotype M1. Infect Immun. 2002;70:6206–14. doi: 10.1128/IAI.70.11.6206-6214.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D, Alberti S, Vivanco F, Sanchez-Corral P, Rodriguez de Cordoba S. Assessment of the interaction of human complement regulatory proteins with group A Streptococcus. Identification of a high-affinity group A Streptococcus binding site in FHL-1. Eur J Immunol. 2000;30:1243–53. doi: 10.1002/(SICI)1521-4141(200004)30:4<1243::AID-IMMU1243>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Ricklin D, Tzekou A, Garcia BL, Hammel M, McWhorter WJ, Sfyroera G, Wu YQ, Holers VM, Herbert AP, Barlow PN, Geisbrecht BV, Lambris JD. A molecular insight into complement evasion by the staphylococcal complement inhibitor protein family. J Immunol. 2009;183:2565–74. doi: 10.4049/jimmunol.0901443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal VD, Maki DG, Jamulitrat S, Medeiros EA, Todi SK, Gomez DY, Leblebicioglu H, Abu Khader I, Miranda Novales MG, Berba R, Ramirez Wong FM, Barkat A, Pino OR, Duenas L, Mitrev Z, Bijie H, Gurskis V, Kanj SS, Mapp T, Hidalgo RF, Ben Jaballah N, Raka L, Gikas A, Ahmed A, Thu le TA, Guzman Siritt ME. International Nosocomial Infection Control Consortium (INICC) report, data summary for 2003–2008, issued June 2009. Am J Infect Control. 2010;38:95–104. e2. doi: 10.1016/j.ajic.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Weidenmaier C, Peschel A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol. 2008;6:276–87. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- Zipfel PF, Skerka C, Hellwage J, Jokiranta ST, Meri S, Brade V, Kraiczy P, Noris M, Remuzzi G. Factor H family proteins: on complement, microbes and human diseases. Biochem Soc Trans. 2002;30:971–8. doi: 10.1042/bst0300971. [DOI] [PubMed] [Google Scholar]