Abstract
We report here that butyrate, a naturally occurring fatty acid commonly used as a nutritional supplement and differentiation agent, greatly enhances the efficiency of induced pluripotent stem (iPS) cell derivation from human adult or fetal fibroblasts. After transient butyrate treatment, the iPS cell derivation efficiency is enhanced by 15- to 51-fold using either retroviral or piggyBac transposon vectors expressing 4 to 5 reprogramming genes. Butyrate stimulation is more remarkable (>100- to 200-fold) on reprogramming in the absence of either KLF4 or MYC transgene. Butyrate treatment did not negatively affect properties of iPS cell lines established by either 3 or 4 retroviral vectors or a single piggyBac DNA transposon vector. These characterized iPS cell lines, including those derived from an adult patient with sickle cell disease by either the piggyBac or retroviral vectors, show normal karyotypes and pluripotency. To gain insights into the underlying mechanisms of butyrate stimulation, we conducted genome-wide gene expression and promoter DNA methylation microarrays and other epigenetic analyses on established iPS cells and cells from intermediate stages of the reprogramming process. By days 6 to 12 during reprogramming, butyrate treatment enhanced histone H3 acetylation, promoter DNA demethylation, and the expression of endogenous pluripotency-associated genes, including DPPA2, whose overexpression partially substitutes for butyrate stimulation. Thus, butyrate as a cell permeable small molecule provides a simple tool to further investigate molecular mechanisms of cellular reprogramming. Moreover, butyrate stimulation provides an efficient method for reprogramming various human adult somatic cells, including cells from patients that are more refractory to reprogramming.
Keywords: Reprogramming, Induced pluripotent stem cells, Sodium butyrate, piggyBac DNA transposition, Sickle cell disease
Introduction
The efficiency of generating induced pluripotent stem (iPS) cells by reprogramming from human adult fibroblasts and other easily accessible somatic cells remains low (up to 0.01% to 0.05%) even when the standard four factors are delivered by retroviral or lentiviral vectors [1]. Contributing factors include nature of the starting somatic cell type and genetic composition, a requirement for the appropriate (absolute and relative) expression levels of the various transgenes, and undefined epigenetic events that take place during the weeks-long reprogramming process [1]. Consequently, small molecules that are known to remodel chromatin and alter gene expression are actively being investigated [1–5]. These may act by reducing the epigenetic barrier for reprogramming from differentiated somatic cells, and potentially also improve the efficiency and quality of the derived iPS cells. However, details of the underlying mechanisms are poorly understood. Here we report our systematic evaluation of a group of commonly used small molecules for human cell reprogramming. These compounds were selected based on either known functionality in chromatin remodeling and reprogramming of mouse or human cells [2–5], or their ability to modulate endogenous gene expression of pluripotency-associated genes such as KLF4 [6]. In addition to small molecules that were tested in the previous reports [2–5], we also tested butyrate, a small-chain fatty acid, which exhibits pleiotropic effects on mammalian cells, including activating KLF4 gene expression in colon cells [6], and is a well known histone deacetylase (HDAC) inhibitor when used at mM levels [7]. We report that butyrate at sub-mM levels improved the iPS cell derivation efficiency by up to 51-fold when using either retroviral transduction or piggyBac transposition for reprogramming of either adult or fetal human fibroblasts. Transient butyrate treatment greatly enhances the reprogramming efficiency so that we can readily generate iPS cell lines from adult fibroblastic cells from healthy donors or patients, including cells that are more refractory to conventional reprogramming methods.
Materials and Methods
Cell Culture
Human embryonic stem (hES) or iPS cells were maintained in KnockOut D-MEM (Invitrogen, Carlsbad, California), 20% KnockOut Serum (Invitrogen), nonessential amino acids (NEAA), L-glutamine, 0.1 mM β-mercaptoethanol, and 10 ng/ml of basic fibroblast growth factor. Human fibroblasts were grown in Dulbecco's modified Eagle's medium (DMEM, low glucose) with Earle's Salts, 10% fetal bovine serum (FBS), NEAA, and L-glutamine. We obtained approval from the Johns Hopkins Internal Review Board for conducting laboratory research using anonymous human cells. See more details of materials and methods in the supporting information.
Human iPS Derivation
A detailed protocol for the derivation of iPS lines using either retroviral transduction or piggyBac transposition is provided in the supporting information materials and methods. Briefly, transduced or electroporated fibroblasts were maintained in FBS-containing media for 6 days, and subsequently either passaged onto mouse embryonic fibroblasts (MEFs) or maintained as is and cultured in hES media from day 7 on. Treatment with butyrate was typically carried out for the entire duration of the reprogramming process (usually day 7 on until the day TRA-1-60 positive colonies were picked). Colonies were picked between days 12 and 24 and subsequently maintained and expanded under standard hES culture conditions.
Genome-Wide Signature Analyses
Data of relative messenger ribonucleic acid (mRNA) expression were obtained using Agilent (Santa Clara, California) whole human genome 4 × 44K microarrays. The DNA methylation data was obtained using Infinium Human DNA Methylation27 chip (Illumina, San Diego, California). All analysis and data visualization were performed using MATLAB software (The Math-Works, Natick, Massachusetts). K-means clustering and classical multidimensional scaling was used to categorize the genes into various clusters and analyze their ensemble dynamics.
Results
Evaluation of Small-Molecule Epigenetic Modulators That Enhance Reprogramming by Four Factors
To screen a panel of cell-permeable small molecules for enhancing human cell reprogramming, we first used the established human IMR90 fetal fibroblast line that has been extensively studied for epigenetic signatures and previously used for reprogramming [8, 9]. We followed the standard Yama-naka reprogramming protocol [10], with the four reprogramming factors Oct4, Sox2, Klf4, and c-Myc (collectively called OSKM) delivered by the classic retroviral vector pMXs. Cell reprogramming was monitored daily by morphologic changes and acquired expression of markers associated with human embryonic stem (hES) and iPS cells on emerging colonies. We found that TRA-1-60 surface expression 2 to 3 weeks after gene transduction is a more reliable and specific marker for predicting successful reprogramming and iPS cell derivation than other markers such as alkaline phosphatase (supporting information Fig. S1). These results were also recently reported by another group [11]. We developed a method for identifying pre-iPS colonies by live staining for TRA-1-60 (supporting information Fig. S1C), which is related to another cell-surface epitope TRA-1-81 that is expressed in hES/iPS cells and previously used for live staining [12]. To further validate the functionality of these TRA-1-60 positive (+) pre-iPS colonies, we followed and characterized 22 such clones that were derived from various starting cell types, picked after live staining, and subsequently expanded. All 22 proved to be bona fide iPS cell lines (supporting information Table S1).
After optimizing the amount of viruses and other parameters for reprogramming (supporting information Fig. S1), we consistently achieved the efficiency of ∼32 TRA-1-60+ (or pre-iPS) colonies at day 16 after transduction of 104 IMR90 fibroblasts (Fig. 1A). Using this robust assay, we next tested the effects of five small-molecule chromatin modifiers added at various concentrations and stages of reprogramming (days 2 to 11 post transduction). Individual chemicals were added into the culture either at an early stage (condition #1, days 2 to 6), a late stage (condition #2, days 7 to 11), or throughout (days 2 to 11, condition #3). We tested DNA methyltransferase inhibitors: 5-Aza-deoxycytidine (5-Aza) and RG108; a selective inhibitor of G9a histone methyltransferase: BIX01294; and HDAC inhibitors: valproic acid (VPA), and sodium butyrate (NaB). A solution of 5-Aza at effective concentrations reported previously was found to be too toxic when added at either stage, and thus was excluded from further studies. In comparison, other chemicals added at one or both stages showed stimulatory effects at optimal concentrations (Fig. 1A). Specifically, RG108, BIX01294, and VPA showed significant stimulation (2- to 15-fold) of reprogramming by four factors in our system. Surprisingly, NaB was significantly more stimulatory (by an additional threefold to sixfold). If present throughout (days 2 to 11, condition #3), butyrate elevated the reprogramming efficiency by 51-fold, resulting in the formation of hES-like colonies at the level of 15% to 20% of the input cells (Fig. 1A). Notably, the level of butyrate stimulation under condition #2 is significantly greater than condition #1, although both treatments last for 4 days. Butyrate, however, did not further increase IMR90 cell proliferation with or without OSKM transduction in the first 6 days. The butyrate stimulation was also evident by the TRA-1-60 expression at a single cell level when the whole cell populations were analyzed by fluorescence-activated cell sorting (FACS) on day 12 (Fig. 1B).
Figure 1.
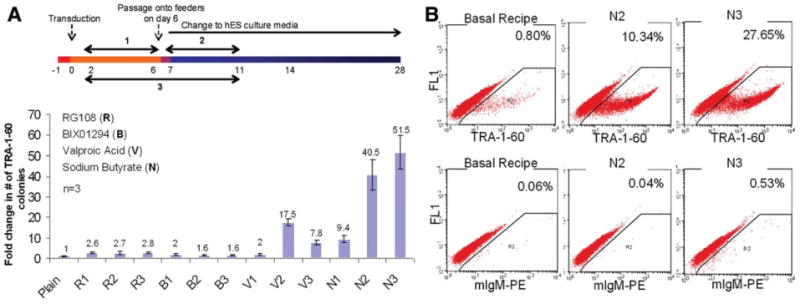
Butyrate stimulation of reprogramming of human fibroblasts. (A): IMR90 fibroblasts transduced by the standard four reprogramming factors, Oct4, Sox2, Klf4, and c-Myc (collectively called OSKM), were treated by individual small-molecule chromatin modifiers which were added in the cell culture in three different ways as shown: at an early stage (#1), a late stage (#2), or throughout (#3). The relative fold stimulation of TRA-1-60+ hES-like colonies compared with plain OSKM reprogramming (defined as one) was enumerated on day 16 after live staining (mean ± SEM, n = 3). Concentrations of the small molecules used in these experiments were: RG108: 5 μM; BIX01294: 0.5 μM; valproic acid: 1 mM; and sodium butyrate: 0.5 mM. (B): Flow cytometric analysis of TRA-1-60 expression in cells undergoing reprogramming. Cells at day 12 after OSKM transduction (basal recipe), or further treated with butyrate (N2 or N3), were harvested and stained. Abbreviations: B, BIX01294; hES, human embryonic stem; n, number; N, sodium butyrate; R, RG108; V, valproic acid.
Human iPS cell lines such as MR45 and MR46 that were established using butyrate stimulation and picked at day 18 (vs. day 24 to 30 without butyrate) show the expression of pluripotency markers, a normal karyotype, and pluripotency by both in vitro and in vivo assays (supporting information Figs. S2, S3). These iPS cell lines also show great similarity to hES cell lines in genome-wide molecular signature analyses (supporting information Fig. S4).
Efficient Derivation of Patient-Specific iPS Cells from Adult Mesenchymal Cells
We next tested the stimulatory effects of butyrate on reprogramming of adult human somatic cells, in particular adult marrow stromal cells (also called mesenchymal stem cells or MSCs) that can be obtained from a few milliliters of marrow aspirate of healthy donors or patients. We tested MSCs from four donors, and butyrate stimulation (3- to 27-fold) was observed in all of these. A similar degree of stimulation is also seen when adult MSCs are not passaged on day 6 onto mouse feeder cells, but instead the culture condition is switched to hES media (supporting information Fig. S5). The stimulation by butyrate is higher than other histone deacetylase (HDAC) inhibitors such as VPA and Trichostatin A (TSA) over a wide range of concentrations. Further, we were able to pick butyrate-stimulated colonies as early as day 12 that proceeded to give rise to bona fide iPS cell lines with full differentiation functionality (supporting information Table S1 and Fig. S6).
We next attempted to derive patient-specific iPS cell lines from adult MSCs. Three milliliters of marrow aspirate from a patient with sickle cell disease (SCD) with the homozygous HbS mutation was obtained, leading to the establishment of MSC culture (referred hence forth as BASC cells) by our standard method [13]. Butyrate treatment showed ∼24-fold stimulation over the four factors alone (Fig. 2A), and we derived five independent iPS lines following TRA-1-60+ colony picking at day 12 or 18 (supporting information Table S1). The homozygous HbS mutation status in derived iPS cell lines was verified. We have characterized these SCD iPS cell lines for the expression of pluripotency markers, retention of a normal karyotype, developmental potential by in vitro and in vivo differentiation assays, and genome-wide molecular signatures (supporting information Figs. S4, S7, S8).
Figure 2.
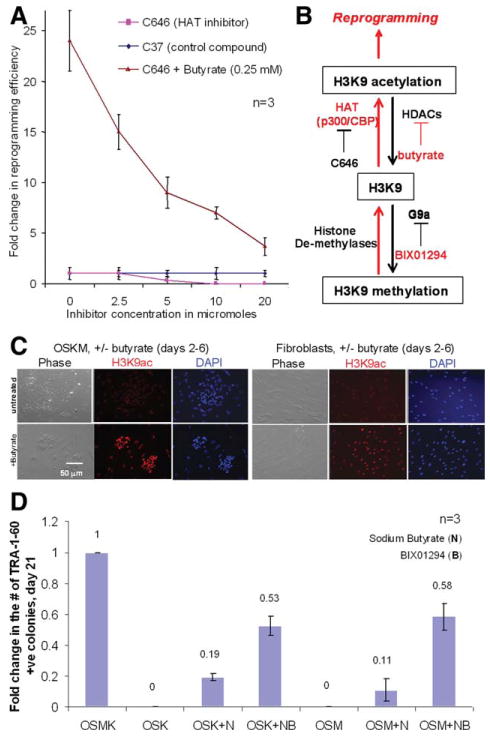
Probing the mechanisms of butyrate stimulation using various inhibitors affecting protein acetylation. (A): Protein acetylation is critical to successful reprogramming. BASC fibroblasts were reprogrammed in the absence or presence of butyrate (0.25 mM). A HAT inhibitor (C646) or a control compound (C37) was added at increasing concentrations. HAT inhibition at high concentrations prevented reprogramming and also significantly diminished reprogramming efficiency, even in the presence of butyrate (from 27-fold to 4-fold). (B): A diagram of protein acetylation and deacetylation mediated by HATs and HDACs (and their inhibitors) using histone H3 lysine nine (H3K9) as a substrate. The compounds that promote protein acetylation are marked in red. (C): Detection of H3K9 acetylation by antibody staining in BASC fibroblasts with or without OSKM transduction and in the presence or absence of butyrate (day 6). (D): Butyrate stimulated reprogramming in the presence of fewer reprogramming factors. IMR90 cells were transduced by OSK or OSM and cultured in the presence or absence of sodium butyrate (N) in combination with BIX01294 (B, 0.5 μM). The level of TRA-1-60+, human embryonic stem cell (hES)-like colonies at day 21 under each condition (mean ± SEM, n = 3) was compared with that by the optimal OSKM reprogramming. Abbreviations: HAT, histone acetyltransferase; N, sodium butyrate; OSKM, Oct4, Sox2, Klf4, and c-Myc collectively; OSK, Oct4, Sox2, Klf4; OSM, Oct4, Sox2, c-Myc.
Gaining Insights into Mechanisms of Butyrate Actions
To gain insights into the great stimulation of reprogramming efficiency by butyrate, we took four distinct approaches. In the first approach, we attempted to examine the importance of the HDAC inhibitory activity of butyrate. Opposing lysine deacetylation by multiple forms of HDACs, mammalian cells express a large family of histone acetyltransferases (HATs), including p300 and CREB Binding Protein (CBP) that acetylate lysine residues in various protein targets which thus gain new functions [14, 15]. A balance between HATs and HDACs, or experimentally by their respective inhibitors, will determine the level of acetylation of histones or nonhistone protein targets. We used a p300/CBP specific (HAT) inhibitor C646 and a structurally related but inactive compound C37 (E.M. Bowers et al., manuscript submitted for publication). We observed that with increased concentrations (2.5 to 20 μM) of the HAT inhibitor C646, the basal reprogramming is diminished (Fig. 2A). These data imply that the p300/CBP HAT activity blocked by C646 is critical to reprogramming. Interestingly, C646 also inhibited the butyrate-stimulated reprogramming; the relative enhancement reduced from 24-fold to 4-fold at the highest concentration tested. The remaining butyrate-stimulation could be caused by C646-insensitive HATs. Together we conclude that butyrate stimulation is largely a function of its HDAC activity (leading to increased protein acetylation), although other butyrate activities not associated with HDAC inhibition may contribute [7]. A diagram of protein acetylation and deacetylation mediated by HATs and HDACs, protein methylation and demethylation mediated by histone methylases and demethylases (and their inhibitors) is shown in Figure 2B using histone H3 lysine nine (H3K9) as a substrate. We further confirmed directly that butyrate promotes protein acetylation by staining OSKM-transduced fibroblasts using the specific antibody recognizing acetylated H3K9 with or without butyrate treatment for 4 days (Fig. 2C).
The second approach is to examine if butyrate stimulation is sustained or more critical when fewer reprogramming factors are used (Fig. 2D), similar to the previously studies describing stimulatory effects of small molecules [2, 4]. As previously reported, omitting c-Myc transgene (OSK only) resulted in significantly fewer TRA-1-60+ hES-like colonies, and appeared at a later stage (day 28 or later) as compared with the standard OSKM combination (supporting information Fig. S9). However, butyrate stimulation in these reduced factor scenarios is more remarkable when c-Myc or Klf4 transgene is absent (OSK or OSM in Fig. 2D). The iPS colonies picked at day 21 after OSK transduction and butyrate treatment (a total of 8 days) formed bona fide iPS cell lines such as MR31, as shown in supporting information Table S1 and Figure S10 BIX01294 could synergize with butyrate to further enhance the butyrate-stimulated reprogramming by three factors (Fig. 2D). Together our data show that small molecules that promote histone acetylation enhance reprogramming (Fig. 2B).
In the third approach, we analyzed the effects of butyrate on genome-wide gene expression during the reprogramming process (Fig. 3). IMR90 cells after OSKM transduction and treated by butyrate or VPA (for 4 days) were harvested at days 6 or 12 (conditions #1 or #2, respectively; Fig. 1A). We focused on the gained expression of 25 genes that are highly enriched in hES and iPS cells as compared with IMR90 cells [9] (Fig. 3A and supporting information Fig. S4). Two overall aspects are noticed: 1) both VPA and NaB stimulated the expression of these hES/iPS-enriched genes at day 6 and 12; and 2) VPA shows a similar pattern of stimulation as butyrate, but generally the stimulation is lesser. For example, DPPA2 expression at day 6 post OSKM transduction was elevated 23-fold and 47-fold by VPA and NaB, respectively (Fig. 3A). In addition to the genome-wide mRNA microarray analysis, we also performed conventional reverse transcription polymerase chain reaction (RT-PCR) using human-specific primers to confirm the butyrate stimulation on the endogenous expression of key pluripotency-associated genes (Fig. 3B). Butyrate significantly elevated gene expression of DPPA2, DPPA3, DPPA4, DPPA5, and NANOG at day 6. In contrast, butyrate did not stimulate the expression of the KLF4 endogenous gene or the four transgenes (all mouse origins) used for reprogramming (Fig. 3B). Similar to the previous data with VPA [2], it appears that butyrate and VPA directly act on human fibroblasts and stimulate endogenous genes as the enhancement is observed at day 6 in the absence of feeder cells and after 4 days of treatment (days 2 to 6).
Figure 3.
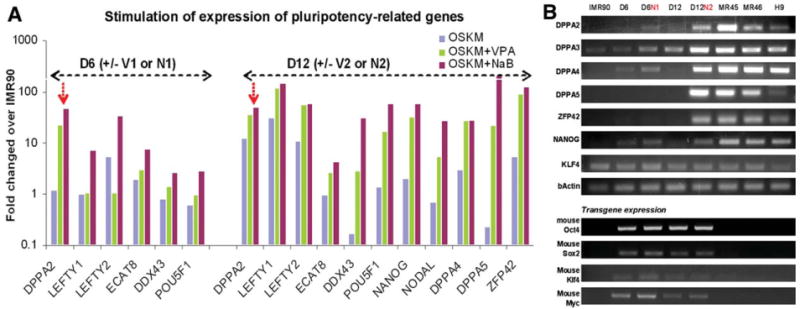
Butyrate stimulated the expression of a group of pluripotency-associated genes during reprogramming. (A): IMR90 cells after OSKM transduction and treated by NaB or VPA (for 4 days) were harvested at day 6 (D6) or day 12 (D12). The genome-wide gene expression at these intermediate stages of reprogramming was analyzed in comparison with the parental IMR90 cells. The relative expression levels of the top genes such as POU5F1/OCT4 with the greatest stimulation by NaB under condition #1 (days 2 to 6) are shown [9]. By day 12, elevation of pluripotency-associated genes upon small molecule stimulation was overt, generally more so by NaB than by VPA. (B): Reverse transcription polymerase chain reaction validated the stimulation of endogenous genes such as DPPA2, DPPA3, DPPA4, DPPA5, ZFP42/REX1, and NANOG (but not KLF4). However, the expression from the transgenes used for reprogramming was not significantly affected by either treatment. Abbreviations: NaB, sodium butyrate; OSKM, Oct4, Sox2, Klf4, and c-Myc collectively; VPA, valproic acid.
Finally, as the fourth approach, we examined the effects of butyrate during the reprogramming on dynamics of key epigenetic markers genome-wide. Because limited numbers of intermediate cells are available at day 6 and 12 during reprogramming, we used Illumina's Human DNA Methylation27 Analysis (Infinium) BeadChips that require ≤1 μg DNA but offer a genome-wide signature analysis (Fig. 4). This allows us to interrogate DNA methylation of 27,578 informative cytosine-phosphate-guanine (CpG) sites at single-nucleotide resolution and compare 12 samples per chip economically. The selected CpG sites are located close to the promoter region of 14,475 RefSeq genes. We analyzed two ES lines (H1 and H9), nine iPS lines, and their parental fibroblasts: IMR90, MSC1640, and BASC, using a K-means clustering algorithm to analyze the dynamics of loci/genes undergoing similar changes (gain, loss, or unchanged before or after reprogramming) (Fig. 4A and supporting information Fig. S4B). We found that iPS cells showed a pattern highly similar to hES cells in contrast to the parental fibroblasts. We observed that most of the methylome (>78%) stayed unchanged in either a methylated (cluster #1) or an unmethylated (cluster #4) state. However, DNA methylation is reduced after reprogramming in loci of cluster #2 (1,174 or 4.4%), which is enriched for pluripotency-associated genes such as POU5F1 and DPPA2. In cluster #3 (17.5%), there was a net gain of methylation after fibroblast reprogramming to iPS cells. As a result, iPS cells are more methylated than their fibroblast counterparts. These results are consistent with an earlier report about the murine system comparing ES and differentiated cells [16] and a recent report comparing DNA methylation in human somatic cells, iPS cells, and ES cells using a different platform [17].
Figure 4.
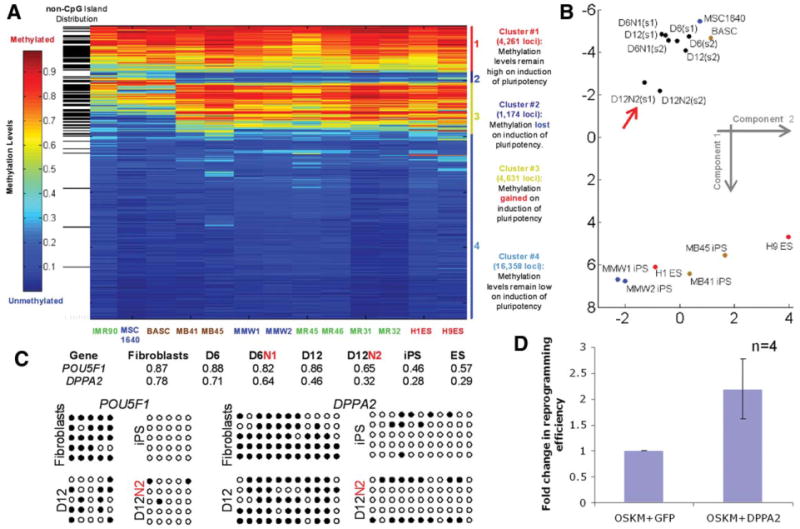
Butyrate enhanced promoter DNA demethylation of pluripotency-associated genes. (A): We used Infinium HumanMethylation27 Bead-Chip to analyze the DNA methylome before and after successfully reprogramming. The level of DNA methylation (from 0 to 1) at gene promoters containing a CpG island (white) or without (black) is illustrated by color from blue to red. Loci are grouped as four clusters based on loss (#2), gain (#3), or unchanged (#1 remaining high and #4 remaining low) of DNA methylation. We used the K-means clustering algorithm to analyze the methylation data from three fibroblast lines (IMR90, MSC1640, and BASC), eight successfully reprogrammed iPS cells, and two ES lines (H1 and H9). The eight validated iPS cells showed a pattern highly similar to hES cells versus their parental fibroblasts. (B): Multidimensional scaling analysis of the relationship among different cell types based on DNA methylation profiles of 1,174 loci in cluster #2 was performed. Data along the first two components are plotted here. Two types of mesenchymal stem cells (MSCs) (MSC1640 and BASC), derived iPS cell lines, and intermediate cells during MSC reprogramming at day 6 (D6, condition #1) or day 12 (D12, condition #2) with or without sodium butyrate (N) were used. Duplicate intermediate MSC samples are referred by s1 and s2. It is evident that the butyrate-treated samples at day 12 (D12N2, red arrow) cluster away from MSCs and move closer to the iPS/ES group. (C): Detection of CpG DNA methylation by the microarray (numbers for 0 to 1.00) and by direct sequencing after bisulfite conversion (filled circles) of the promoter regions of POU5F1/OCT4 and DPPA2 genes. Reduction of DNA methylation in multiple CpG sites at both promoters is evident from parental MSCs to reprogramming intermediate cells at 12 (especially after butyrate treatment) and ultimately in successfully reprogrammed iPS cells. (D): DPPA2 transgene expression partially substitutes butyrate and further enhances reprogramming by the four reprogramming factors OSKM. BASC or IMR90 cells were transduced by a retroviral vector expressing either DPPA2 or GFP (as a control) together with OSKM, and fold change in TRA-1-60+ colonies was enumerated (mean ± SEM, n = 4). Abbreviations: BASC, Bone marrow Aspirate derived marrow Stromal Cells from patient with Sickle Cell Disease; GFP, green fluorescent protein; ES, ES, embryonic stem; hES, human embryonic stem; iPS, induced pluripotent stem; n, number; N, sodium butyrate; OSKM, Oct4, Sox2, Klf4, and c-Myc collectively.
We used this metric to analyze epigenetic changes during the reprogramming process (day 6 or 12 with or without butyrate treatment). Using the whole cluster #2 methylation data, we analyzed these intermediate cells during reprogramming in relationship with their parental MSCs or iPS/ES cells by multidimensional scaling. A relationship plot based on scaling to two dimensions is shown (Fig. 4B). As expected, successfully reprogrammed iPS cell lines cluster together with ES cell lines at the bottom, whereas parental MSCs group at the top. Reprogramming cells at earlier stages show the proximity to the parental MSCs. Notably, the butyrate-treated samples at day 12 (D12N2) lie outside the latter group and move to the direction of the iPS/ES group (Fig. 4B). Consistent with the gene expression data, promoters of several pluripotency-associated genes such as POU5F1 and DPPA2 showed greater demethylation in the presence of butyrate stimulation, especially at day 12 (Fig. 4C).
To test the hypothesis that increased expression of DPPA2 gene may be a novel effector of the butyrate stimulation, we tested whether its enforced expression would further stimulate reprogramming after OSKM transduction. We observed that the DPPA2 cDNA expression vector increased the efficiency in both IMR90 and BASC fibroblasts by more than twofold as compared with the green fluorescent protein (GFP) control (Fig. 4D). Thus, DPPA2 is likely a butyrate target responsible in part for its stimulatory effect. Although the stimulation over the basal OSKM-mediated reprogramming by DPPA2 transgene expression is far less than by butyrate and practically not useful for stimulating reprogramming efficiency, our novel observations provide the first set of data regarding the functions of DPPA2, whose expression is associated with pluripotent and germline cells [18] and that were recently shown to be important to the self-renewal of mouse ES cells [19]. Together, our results in Figure 2 through Figure 4 provide strong evidence that butyrate greatly enhances human cell reprogramming, mainly by promoting epigenetic remodeling, resulting in histone acetylation, promoter DNA demethylation, and the expression of endogenous pluripotency-associated genes.
Butyrate Stimulation of Reprogramming Mediated by piggyBac DNA Transposition
To evaluate the generality of butyrate stimulation on human cell reprogramming, we also tested its applicability in techniques that enable generation of transgene-free and integration-free iPS cell lines. In particular, we chose piggyBac DNA transposition that has been shown to be effective in generations of iPS cell lines from mouse and human embryonic fibroblasts [20, 21]. Transient piggyBac transposase expression can catalyze both integration and scar-free excision of the transposon vector expressing the reprogramming genes. This unique feature of piggyBac DNA transposition has been used to generate mouse iPS cells in which the integrated transgenes were completely removed [20, 21].
We used the same piggyBac transposon vector containing the five transgenes (OSKM + Lin28) that showed the best result in deriving mouse iPS cells [21] to reprogram human BASCs, the MSCs established from the patient with sickle cell disease as described in Figure 2A. In the absence of butyrate (plain) and when early passage MSCs (passages 3 and 4) were used, the reprogramming efficiency by DNA transposition with an improved piggyBac transposase vector is ∼50-fold lower than that by the four retroviral vectors (Fig. 5A). No TRA-1-60+ colonies were formed from the serially-passaged MSCs at later stages (passages 5 through 7). Butyrate treatment under conditions #2 (days 2 to 6) and #3 (days 2 to 11) significantly stimulated the efficiency of reprogramming BASCs at various passages, and was essential for successful derivation of iPS cells from BASCs at later passages. Combined together, we observed a similar level of butyrate stimulation on reprogramming of BASCs by either piggyBac transposon (∼25-fold, Fig. 5A) or the four retroviral vectors (∼24-fold, Fig. 2A). Five iPS lines (MBP5) were expanded and characterized (Fig. 5B–5E) for the expression of pluripotency markers, retention of a normal karyotype, pluripotency by in vitro and in vivo differentiation assays, and genome-wide molecular signatures (supporting information Fig. S4B).
Figure 5.
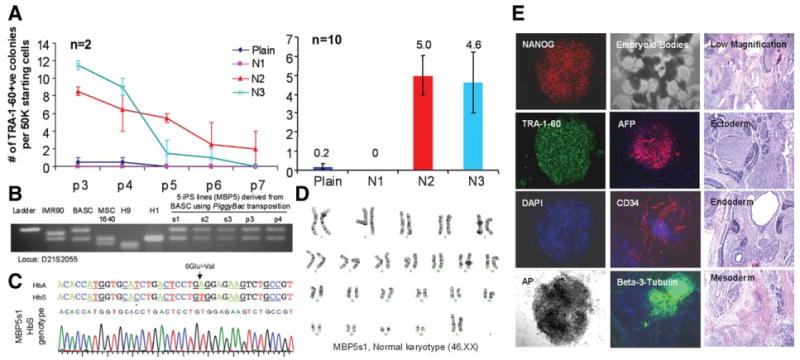
Butyrate similarly stimulates human iPS cell derivation when piggyBac transposition is used for reprogramming. (A): Serially-passaged BASCs (passage 3 through passage 7), the MSCs from an adult female patient with sickle cell disease, were reprogrammed using piggyBac transposition. Butyrate significantly stimulated the formation of TRA-1-60+ colonies under conditions N2 and N3 for MSCs at each passage (passage 3 through passage 7), although later passage MSCs generated less TRA-1-60+ colonies (5- to 10-fold lower). Data from MSCs of five different passages were combined (n = 10) and plotted. Under the conditions #2 and #3 (N2 and N3), butyrate increased the reprogramming efficiency by 23- to 25-fold. (B): Five iPS lines (s1, s2, and s3) or pools (passage 3 and passage 4) were derived by picking TRA-1-60+ colonies at day 24 after piggyBac transposition. DNA fingerprinting was used to confirm the origin of the BASC line. (C): One of the five established lines, s1, was confirmed to have the homozygous HbS mutation (A to T) and a polymorphic C (*) found in the patient with sickle cell disease compared with T in the general population. (D): The line shows a normal (female) karyotype. (E): It expresses the characteristic pluripotency markers (left panels), readily differentiates in suspension culture to form cystic embryoid bodies with elements of all three germ layers (middle), and forms teratomas in nude mice comprising of various cell types derived from all of the three embryonic germ layers (right). Abbreviations: iPS, induced pluripotent stem; MSC, mesenchymal stem cells; BASC, Bone marrow Aspirate derived marrow Stromal Cells from patient with Sickle Cell Disease.
Discussion
We report that butyrate, a naturally occurring fatty-acid commonly used as a nutritional supplement and differentiation agent, greatly enhances the efficiency of iPS cell derivation from human adult or fetal fibroblasts using either retroviral or piggyBac transposon vectors that express 4 to 5 reprogramming genes. Although a transient butyrate treatment (4 to 8 days) during the reprogramming suffices to achieve a 15- to 51-fold stimulation, adding butyrate continuously during the reprogramming did not negatively affect the derivation or expansion of iPS cells. We did, however, observe differences in the degree of stimulation between various cell lines, although no overt differences in toxicity were observed when butyrate was used at an optimal concentration. This could be caused by variations between different MSC preparations or other unknown intrinsic differences between different cell types. In spite of our incomplete understanding of the underlying mechanisms here, we found that butyrate consistently stimulated the formation of more (TRA-1-60+) pre-iPS cells from human fetal and adult mesenchymal cells by increasing the pace and percentage of TRA-1-60+, hES-like colony formation. Combined with live staining for the TRA-1-60 cell surface marker, we were able to select pre-iPS colonies as early as day 12 after OSKM transduction following butyrate treatment for 8 days. Some human adult somatic cells are relatively more refractory to reprogramming, likely because of the presence of genetic defects [22], cellular senescence, difficulties in transgene delivery, or other unknown factors. We envision that butyrate stimulation may be particularly useful for reprogramming in scenarios like these. In particular, butyrate stimulation by 23- to 25-fold enabled reliable generation of patient-specific iPS cell lines by piggyBac transposition of human adult mesenchymal cells, which is ∼50-fold less efficient than by retroviral vectors (Fig. 5). We have recently begun testing possible synergistic effects of butyrate stimulation with other small molecules that were recently shown to enhance human fibroblast reprogramming [5, 23]. Indeed, we also observed the stimulation by a transforming growth factor-β (TGFβ) signaling inhibitor (SB431532) on reprogramming of human adult MSCs by the same four retroviral vectors, although the stimulation (sixfold, unpublished) is less than by butyrate (∼30-fold). When the two were combined, however, we saw ∼70-fold synergistic stimulation (Mali, unpublished). It remains to be determined whether an HDAC inhibitor such as butyrate will also enhance reprogramming of other human cell types such as blood cells as we did recently by retroviral vectors [24]. We predict that butyrate with or without other small molecules will likely enhance the efficiency of iPS derivation when using transient gene or protein transduction techniques, which are currently much less efficient [25–27].
To gain insights into the mechanism underlying butyrate stimulation, we used four distinct approaches to probe molecular events during the reprogramming and in established human iPS cells after butyrate stimulation. We found that butyrate promotes protein acetylation at targets such as H3K9. Concurrently, it also accelerates promoter DNA demethylation and expression of pluripotency-associated genes such as POU5F1/OCT4 and DPPA2 (Figs. 2 through 4). It has been shown that DNA demethylation at certain loci is required for reprogramming by nuclear transfer and cell fusion [28, 29]. In our system using ecotropic gene expression to initiate nuclear reprogramming, we observed both DNA demethylation at the promoter regions of pluripotency associated genes (cluster #2) and gained promoter DNA methylation at many more loci (cluster #3) that are likely responsible for silencing fibroblast-specific genes (Fig. 4). How butyrate-stimulated histone acetylation during reprogramming affects both promoter DNA demethylation and methylation at different loci is unclear. Moreover, enhancing chromatin histone acetylation may not be the only mechanism of reprogramming stimulation, as butyrate exhibits diverse cellular effects in culture [7]. Whether or how butyrate also stimulates reprogramming by modulating activities of nonhistone key regulators, regardless it is HAT/HDAC dependent or not, remains to be investigated.
Ware et al. [30] recently reported that butyrate at 0.2 mM supported the maintenance of undifferentiated hES cells without otherwise required feeder cells or derived conditioned media, and upregulated a group of pluripotency/germline-associated genes that included DPPA2, ECAT8, DDX43, POU5F1/OCT4, and DPPA5. Butyrate is more potent than other HDAC inhibitors such as VPA and TSA in promoting ES and other pluripotent cells to a more primitive state [30]. Notably, many genes that are upregulated by butyrate in hES cells in the latter study overlapped with the most upregulated genes we identified upon butyrate stimulation during reprogramming of fibroblasts (Fig. 3). A convergent picture from both studies suggests that butyrate can facilitate cell fate changes by promoting epigenetic remodeling and the expression of pluripotency-associated genes, among other possible conserved mechanisms. It would be of great interest to test whether butyrate could reduce epigenetic barriers and enhance other types of epigenetic reprogramming such as transdifferentiation.
Summary
We report here that butyrate greatly enhances the efficiency of induced pluripotent stem (iPS) cell derivation from human adult or fetal fibroblasts. After transient butyrate treatment, the iPS cell derivation efficiency is enhanced by 15- to 51-fold using either retroviral or piggyBac transposon or based vectors expressing 4 to 5 reprogramming genes. Butyrate treatment is especially critical for successful reprogramming by the plasmid-based piggyBac delivery system, in which the baseline reprogramming efficiency is about 50-fold lower than when using retroviruses. Derived iPS cell lines, including those from an adult patient with sickle cell disease by multiple vector systems, show normal karyotypes and pluripotency. To gain insights into the mechanism of butyrate stimulation, we conducted genome-wide gene expression and promoter DNA methylation microarrays on established iPS cells and cells from intermediate stages of the reprogramming process. By days 6 to 12 during reprogramming, butyrate treatment enhanced histone H3 acetylation, promoter DNA demethylation, and the expression of endogenous pluripotency-associated genes. Using Illumina's HumanMethylation27 platform to analyze the genome-wide methylation status of cells at days 6 to 12 during the reprogramming process, we found that, consistent with gene expression data, butyrate treatment promotes the DNA demethylation dynamics of pluripotency-associated genes. Thus, butyrate as a cell permeable small molecule provides a simple tool to further investigate molecular mechanisms of cellular reprogramming. Moreover, butyrate stimulation provides an efficient method for reprogramming various human adult somatic cells using multiple transgene delivery systems and, including from cells that are more refractory to reprogramming.
Supplementary Material
Acknowledgments
We thank Dr. Sophie M. Lanzkron for SCD patient recruitment, and staff members in the laboratory of Dr. Nancy Craig (Johns Hopkins University) for stimulating discussions about using piggyBac DNA transposition. The work conducted at Johns Hopkins University was funded by the Institute for Cell Engineering and NIH (RC2 HL101582) to LC, and NIH GM62437 and Flight Attendant Medical Research Institute (FAMRI) Foundation support to PAC. The work at Sanger Institute was supported by the Wellcome Trust to AB. BK is supported by the Taiwan Merit Scholarship NSC-095-SAF-I-564-019-TMS. JZ was supported by a Maryland Stem Cell Research Postdoctoral Fellowship Grant. KY was supported by the Japan Society for Promotion of Science.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors indicate no potential conflicts of interest.
Author contributions: P.M.: Conception and design, provision of study material, collection and assembly of data, data analysis and interpretation, and manuscript writing; B.-K. C.: Provision of study material, collection and analysis of data; J.Y., Z.Y., J.Z., S.D.: Collection and analysis of data; J.E.O., W.Y.: Collection and assembly of data; S.B.B.: Data analysis and interpretation; R.A.B., K.Y., A.B, D.J.M., C.M., P.A.C.: Provision of study material or patients; L.C.: Conception and design, data analysis and interpretation, financial and administrative support, and manuscript writing.
References
- 1.Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huangfu D, Osafune K, Maehr R, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 3.Mikkelsen TS, Hanna J, Zhang X, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi Y, Desponts C, Do JT, et al. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Lin T, Ambasudhan R, Yuan X, et al. A chemical platform for improved induction of human iPSCs. Nat Methods. 2009;6:805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen ZY, Rex S, Tseng CC. Kruppel-like factor 4 is transactivated by butyrate in colon cancer cells. J Nutr. 2004;134:792–798. doi: 10.1093/jn/134.4.792. [DOI] [PubMed] [Google Scholar]
- 7.Miller SJ. Cellular and physiological effects of short-chain fatty acids. Mini Rev Med Chem. 2004;4:839–845. doi: 10.2174/1389557043403288. [DOI] [PubMed] [Google Scholar]
- 8.Mali P, Ye Z, Hommond HH, et al. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells. 2008;26:1998–2005. doi: 10.1634/stemcells.2008-0346. [DOI] [PubMed] [Google Scholar]
- 9.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Chan EM, Ratanasirintrawoot S, Park IH, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 12.Lowry WE, Richter L, Yachechko R, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng L, Hammond H, Ye Z, et al. Human adult marrow cells support prolonged expansion of human embryonic stem cells in culture. Stem Cells. 2003;21:131–142. doi: 10.1634/stemcells.21-2-131. [DOI] [PubMed] [Google Scholar]
- 14.Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 15.Cole PA. Chemical probes for histone-modifying enzymes. Nat Chem Biol. 2008;4:590–597. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doi A, Park IH, Wen B, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maldonado-Saldivia J, van den Bergen J, Krouskos M, et al. Dppa2 and Dppa4 are closely linked SAP motif genes restricted to pluripotent cells and the germ line. Stem Cells. 2007;25:19–28. doi: 10.1634/stemcells.2006-0269. [DOI] [PubMed] [Google Scholar]
- 19.Du J, Chen T, Zou X, et al. Dppa2 knockdown-induced differentiation and repressed proliferation of mouse embryonic stem cells. J Biochem. 2010;147:265–271. doi: 10.1093/jb/mvp161. [DOI] [PubMed] [Google Scholar]
- 20.Woltjen K, Michael IP, Mohseni P, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yusa K, Rad R, Takeda J, et al. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raya A, Rodriguez-Piza I, Guenechea G, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esteban MA, Wang T, Qin B, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Ye Z, Zhan H, Mali P, et al. Human induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009;114:5473–5480. doi: 10.1182/blood-2009-04-217406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, Kim CH, Moon JI, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J, Hu K, Smuga-Otto K, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou H, Wu S, Joo JY, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhutani N, Brady JJ, Damian M, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simonsson S, Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol. 2004;6:984–990. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


