Abstract
The impact of water deficit on stilbene biosynthesis in wine grape (Vitis vinifera) berries was investigated. Water deficit increased the accumulation of trans-piceid (the glycosylated form of resveratrol) by 5-fold in Cabernet Sauvignon berries but not in Chardonnay. Similarly, water deficit significantly increased the transcript abundance of genes involved in the biosynthesis of stilbene precursors in Cabernet Sauvignon. Increased expression of stilbene synthase, but not that of resveratrol-O-glycosyltransferase, resulted in increased trans-piceid concentrations. In contrast, the transcript abundance of the same genes declined in Chardonnay in response to water deficit. Twelve single nucleotide polymorphisms (SNPs) were identified in the promoters of stilbene synthase genes of Cabernet Sauvignon, Chardonnay, and Pinot Noir. These polymorphisms resulted in eight changes within the predicted cis regulatory elements in Cabernet Sauvignon and Chardonnay. These results suggest that cultivar-specific molecular mechanisms might exist that control resveratrol biosynthesis in grapes.
Keywords: Grape berry, stilbene synthase, stilbenes, Vitis vinifera, water deficit
Introduction
Resveratrol, a polyphenolic compound, exhibits anti-inflammatory, antioxidative, and antiproliferative properties (1−4) in humans. Resveratrol in wines is thought to contribute to the well-known health benefits of red wine, including protective effects against cardiovascular diseases (5) and extension of life span in animals (6−8). Its average concentration in red wines is 1.9 mg L−1trans-resveratrol, ranging from nondetectable levels to 14.3 mg L−1 and is recognized to account, in part, for the famous “French Paradox” (9−12). Low concentrations of stilbenes, especially resveratrol and piceid, were found to protect against Alzheimer’s disease (13−16) and skin cancer (17). In the plant kingdom, resveratrol (3,4′,5-trihydroxystilbene) biosynthesis occurs in various plants including grapes, berries, and peanuts (18) and is found in leaves, skin, and seed coats of the fruit (10,19). Multiple stilbene-derived compounds, including isomers, polymers, and glycosylated forms, have also been characterized in grapes (11,18,19).
The resveratrol biosynthetic pathway consists of four enzymes: phenylalanine ammonia lyase (PAL), cinnamic acid 4-hydroxylase (C4H), 4-coumarate: CoA ligase, (4CL), and resveratrol synthase, also known as stilbene synthase (STS) (18). Only plants with STS, the last enzyme in the resveratrol biosynthetic pathway, are capable of synthesizing resveratrol (18). Downstream of these reactions, a resveratrol glucosyltransferase transfers a glucose moiety onto the resveratrol backbone to produce piceid-derived compounds. This last enzyme was identified recently in Vitis labrusca grape berries (20). Likewise, another step in this pathway was recently characterized in grapes involving a resveratrol O-methyl transferase (ROMT) cDNA associated with the biosynthesis of pterostilbene, which has attracted much attention recently because of its promising pharmacological properties (21,22).
Grape resveratrol biosynthesis appears to be dependent upon multiple factors including environmental and fungal stresses on the vine (23). Stilbene synthesis in Vitis spp. leaves can be influenced by fertilizer application, with less resveratrol accumulating with increasing nitrogen supply (24). UV light irradiation greatly induces STS steady state transcript abundance in unripe berries (25). In addition, ectopic expression of STS genes improves pathogen resistance in several plant species (26−30).
In the recently sequenced Vitis genome, 43 members of the STS gene family were identified (31). However, only 20 of them appear to be expressed in grapes, based upon transcript evidence. Previously, we reported that water deficit increases the specific steady state transcript abundance of a STS gene and phenylpropanoid metabolism in general in Cabernet Sauvignon berries (32). Here, we provide evidence in support of the hypothesis that stilbene concentrations are increased in drought stressed grapes.
Materials and Methods
Field Experiments and Physiological Data
Grape berries harvested at seven different developmental stages from Cabernet Sauvignon and Chardonnay (Vitis vinifera) vines, respectively, were collected during the summer of 2004 from the Shenandoah Vineyard in Plymouth, CA, and the Valley Road experimental vineyard belonging to the University of Nevada, Reno, NV, USA. Additional details about the training system, plant density, ripeness parameter (total soluble solids, titratable acidity) and stem water potentials were published previously (33). Berry cluster samples were collected on a weekly basis, and seven time points encompassing the growing season were selected to perform a global transcript profiling (33). To avoid any row effects, plants used for this experiment were located in the middle of the vineyard. All vines were equipped with drip irrigation. Irrigation was withheld until the desired range of stem water potentials was reached. Stem water potentials were measured with a pressure chamber as described previously (34). Two grape clusters were harvested weekly on the south (sunny) and the north (shady) side of each plant. The clusters were pooled together in order to avoid any light and temperature effects. No visible symptoms of disease were observed on the grapevines or grape clusters. Clusters from three plants under differential water regime supply in Cabernet Sauvignon and Chardonnay were compared for their global transcript profiles. The effect of water deficit on the resveratrol biosynthetic pathway in Cabernet Sauvignon and Chardonnay grape varieties was investigated during berry development. Global transcript profiling throughout berry development was performed on Cabernet Sauvignon (CS) and Chardonnay (CH) cultivars of wine grape with two irrigation levels: well-watered (sufficient water) and water deficit (see Figure 1 in ref (33)). Throughout berry development, well-watered grapevines were irrigated to maintain stem water potentials between −0.8 and −0.6 MPa for control treatment conditions, while stem water potentials for water deficit stressed grapevines were maintained between −1.25 and −0.8 MPa for the water deficit treatment (33).
RNA Extraction, Microarray Hybridization, and Microarray Data Processing
Global transcript profiling throughout the berry development was performed on Cabernet Sauvignon (CS) and Chardonnay (CH) cultivars with two irrigation levels: well-watered (sufficient water) and water deficit (see Figure 1 in ref (33)). Total RNA was extracted from whole berries, which had not been deseeded, finely ground in liquid nitrogen using Qiagen RNeasy Plant MidiKit columns (Qiagen Inc., CA) as described previously (35). The total RNA was further purified using a Qiagen RNeasy Plant Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RNA integrity was confirmed by electrophoresis on 1.5% agarose gels containing formaldehyde, and quality was confirmed by analysis on an Agilent 2100 Bioanalyzer using RNA LabChip assays according to the manufacturer’s instructions. Biotinylated complementary RNAs (cRNAs) were purified, fragmented, and hybridized in the GeneChip Vitis vinifera Genome Array cartridge (Affymetrix, Santa Clara, CA). Microarrays were scanned using a Hewlett-Packard GeneArray scanner, and image data were collected and processed on a GeneChip workstation using Affymetrix GCOS software. Three biological replicates per experimental treatment (well-watered (WW) and water deficit (WD) treatments of Chardonnay (CH) and Cabernet Sauvignon (CS)) were processed to evaluate intravarietal variability. Expression data were processed by Robust Multi-Array Average (RMA) (36) using the R package AFFY previously described (37). Genes that were differentially expressed throughout berry development were detected by ANOVA of the RMA expression values. A simple, three-way fixed effects analysis of variance (ANOVA) was performed on the RMA-normalized and processed data to examine probesets with significant treatment effects, treatment and cultivar interaction effects, and treatment, cultivar, and time interaction effects. A multiple testing correction was applied to the p-values of the F-statistics to control the false discovery rate associated with multiple comparisons. Genes with adjusted F-statistic p-values of less than 0.05 were extracted for further analysis. The raw data have been deposited in PlexDB (www.plexdb.org; experiment: VV5). The microarray data was validated by qPCR experiments as previously shown by Deluc and Grimplet (32,33,37).
Quantification of Stilbene Compounds
Freeze-dried powder of grape berries (50 mg) was extracted with methanol (4 mL) overnight at 4 °C. After centrifugation (750g, 5 min), 3 mL of the supernatant was evaporated to dryness under a vacuum with a Speedvac SVC200 centrifugal evaporator (Farmingdale NY, USA) and recovered with 100 μL of methanol and 1 mL of water. The extract was chromatographically separated on a cation exchange resin column (6 mm × 40 mm) and eluted with 75% (v/v) aqueous methanol to obtain polyphenols. Analysis of polyphenols was performed with an Agilent HPLC 1100 system on a Prontosil Eurobound C18 (5 μm) reverse-phase column (4 mm i.d. × 250 mm) (Bischoff, Stuttgart, Germany). Solvents used for the separation were A, water with 0.1% trifluoroacetic acid; and B, acetonitrile with 0.1% trifluoroacetic acid. The elution program at a rate of 1 mL min−1 was as follows: 0 min, 10% B in A; 50 min, 35% B in A; 51 min, 100% B in A; 60 min 100% B in A; 61 min 10% B in A. The chromatogram was monitored at 286 and 306 nm. Stilbene contents were estimated according to calibration curves prepared with standards of trans-resveratrol (Sigma-Aldrich, Saint Quentin-Fallavier, France) and trans-piceid (Sequoia Research Products, Pangbourne, UK). For mass spectrometry identification, the same solvents were used, but with less trifluoroacetic acid, which provokes signal suppression. The solvent concentrations were 0.025% TFA in water and acetonitrile. The analytical HPLC separations were also conducted in the same column and the HPLC effluent was introduced into the electrospray source in a postcolumn splitting ratio of 9:1. The mass spectrometry was acquired on a Thermo Finnigan LCQ Advantage ion trap spectrometer, equipped with an electrospray source. The software used for data acquisition and retreatment was Xcalibur (http://www.xcalibur.com/). Data were obtained both in positive and negative ionization modes; most of the compounds were assigned by their positive [ionization] mass spectra (higher sensitivity).
DNA Extraction and Amplification of the STS Promoter Region
DNA was extracted from the rachis of Chardonnay and Cabernet Sauvignon grape berry clusters according to Lodhi et al. (38). The transcript sequence used for the amplification of the STS promoter was related to the probe set (1608009_s_at). The annotated identifier from Genoscope (39) for this transcript was GSVIVT00008253001. Primers were designed upstream using Primer3 software, based on the sequence of the STS gene deposited on the Genoscope Web site, which contained the 8X assembly of the partially inbred derived Pinot noir genome (http://www.genoscope.cns.fr). PCR products were amplified using an AccuPrimeTaq DNA polymerase (Invitrogen, Carlsbad, CA), and PCR products were cloned into a pGEM-T vector (PROMEGA, Madison, WI). Clones of interest were sequenced in both strands by an ABI prism 3730 DNA analyzer using the Sanger method (40). Alignment of the sequence of the promoters from the two cultivars was performed using MEGA4 software (http://www.megasoftware.net/) (41). Aligned sequences were edited using MacVector 10 (Cary, NC; http://www.macvector.com/).
Results and Discussion
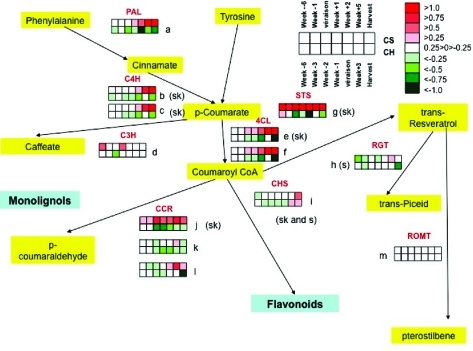
Transcript abundance of genes related to the resveratrol biosynthetic pathway has been investigated in two cultivars with two irrigation levels: sufficient water and water deficit. These transcripts were among the most differentially expressed genes that exhibited significant cultivar, treatment, and time interaction effects (Table 1). One probeset associated with stilbene synthase gene was found significantly differentially expressed among 13 probesets related to stilbene synthase genes present in the Affymetrix microarrays, which represent a total of 8 genes out of the 42 genes identified in the Pinot Noir genome (42).
Table 1. The Set of Transcripts Differentially Expressed by Water Deficit and Associated with the Resveratrol Biosynthetic Pathwaya.
| affy probe set ID | putative function | treatment effect | p-value | adjusted p-valueb | cultivarbtreatment effects | p-value | adjusted p-valueb | cultivarbtreatmentbdevelopment effects | p-value | adjusted p-valueb |
|---|---|---|---|---|---|---|---|---|---|---|
| 1607732_at | chalcone synthase | X | 9.54 × 10−3 | 2.87 × 10−2 | X | 9.54 × 10−3 | 2.87 × 10−2 | − | − | − |
| 1610415_at | cinnamate 3′hydroxylase | X | 1.73 × 10−2 | 4.64 × 10−2 | X | 2.75 × 10−6 | 1.34 × 10−4 | X | 2.25 × 10−6 | 1.08 × 10−3 |
| 1610821_at | cinnamate 4 hydroxylase | − | − | − | X | 2.97 × 10−7 | 2.83 × 10−5 | X | 5.85 × 10−4 | 2.60 × 10−2 |
| 1616191_s_at | cinnamate 4 hydroxylase | − | − | − | X | 4.23 × 10−8 | 6.02 × 10−6 | X | 4.97 × 10−4 | 2.40 × 10−2 |
| 1609307_at | 4 coumaroyl-CoA ligase | − | − | − | X | 1.17 × 10−10 | 4.99 × 10−8 | X | 4.21 × 10−6 | 1.58 × 10−3 |
| 1608094_at | cinnamoyl-CoA reductase | X | 1.42 × 10−3 | 6.13 × 10−3 | X | 1.42 × 10−3 | 6.13 × 10−3 | − | − | − |
| 1619513_at | cinnamoyl-CoA reductase | X | 6.09 × 10−3 | 2.00 × 10−2 | − | − | − | − | − | − |
| 1621163_at | cinnamoyl-CoA reductase | X | 1.47 × 10−4 | 9.30 × 10−4 | − | − | − | − | − | − |
| 1613113_at | phenylalanine ammonia lyase | − | − | − | X | 2.56 × 10−11 | 1.52 × 10−8 | X | 1.42 × 10−5 | 2.98 × 10−3 |
| 1611265_at | 4 Coumarate CoA Ligase | X | 3.48 × 10−4 | 1.90 × 10−3 | − | − | − | − | − | − |
| 1617078_at | resveratrol glucosyl transferase | X | 7.96 × 10−4 | 3.78 × 10−3 | − | − | − | − | − | − |
| 1608009_s_at | stilbene synthase | X | 1.92 × 10−4 | 1.15 × 10−3 | X | 1.55 × 10−10 | 6.27 × 10−8 | − | − | − |
X, significantly differentially expressed. −, Not significantly differentially expressed for the treatment effect, cultivar X treatment effects, cultivar X treatment X development effects.
Multiple testing correction applied to p-value.
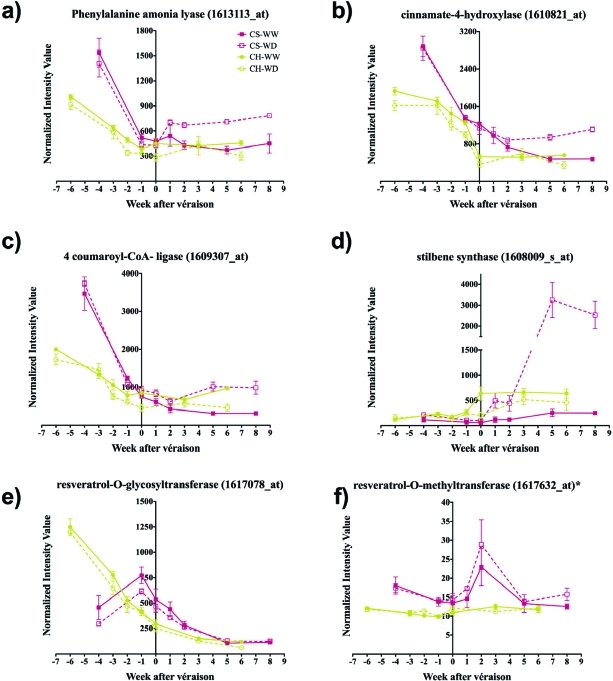
The first committed step associated with the resveratrol biosynthetic pathway is controlled by phenylalanine ammonia-lyase. One gene (1613113_at; GSVIVT00018175001) was found to have a statistically different mRNA expression pattern (Figures 1a and 2). This unigene encodes a protein that shares strong homology with a protein sequence of Arabidopsis thaliana AtPAL2, which has functional specialization in abiotically, environmentally triggered flavonoid synthesis (43). In our study, there was a coordinated and gradual decline in transcript abundance of this gene over berry development in both cultivars under well-watered conditions. In Cabernet Sauvignon, both genes had coordinately increased transcript abundance in water-deficit-treated compared to well-watered berries from the véraison stage through harvest (Figures 1a and 2). In contrast, transcript abundance in Chardonnay was reduced in water-deficit-treated berries. This indicates cultivar specificity with respect to the relative transcriptional regulation of these two genes.
Figure 1.
Expression of potential candidate unigenes associated with the resveratrol pathway. (a) Phenylalanine ammonia lyase (1613113_at; GSVIVT00018175001); (b) cinnamate-4-hydroxylase (1610821_at; GSVIVT00023932001); (c) 4-coumarate:CoA ligase (1609307_at; GSVIVT00031383001); (d) stilbene synthase (1608009_s_at; GSVIVT00008253001); (e) resveratrol-O-glycosyltransferase (1617078_at; GSVIVT00036670001); (f) resveratrol-O-methyltransferase (1617632_at, GSVIVT00003032001). The symbol “*” indicates a nonsignificant difference observed for the treatment, cultivar-treatment interaction effects, and the cultivar-treatment-development interaction effects. Symbols represent means ± standard errors (SE); n = 3.
Figure 2.
Heatmap profiles of transcripts involved in the resveratrol biosynthetic pathway. Each box represents a different developmental stage of berry development for each probe set and corresponds to the log2 of the ratio between the intensity value obtained in WD conditions and the intensity value in WW conditions. Cabernet Sauvignon is on the top row of boxes and Chardonnay is on the bottom row (see the key of white boxes in the upper right-hand corner). The gene expression ratio levels are indicated by the color legend in the upper right-hand corner of the figure. (a) (1613113_at, GSVIVT00018175001- phenylalanine ammonia lyase), (b) (1610821_at, GSVIVT00023932001- cinnamate-4-hydroxylase), (c) (1616191_s_at, GSVIVT00023932001-cinnamate-4-hydroxylase), (d) (1610415_at, GSVIVT00026288001-cinnamate-3′-hydroxylase), (e) (1611265_at; GSVIVT00009148001-4-coumarate:CoA ligase), (f) (1609307_at, GSVIVG00031383001), (g) (1608009_s_at; GSVIVT00008253001, stilbene synthase), (h) (1617078_at, GSVIVT00036670001-resveratrol/hydroxycinnamic acid-O-glucosyltransferase), (i) (1607732_at, GSVIVT00037969001-chalcone synthase), (j) (1621163_at, GSVIVT00024039001-cinnamoyl-CoA reductase-like), (k) (1619513_at, GSVIVT00002938001-cinnamoyl-CoA reductase), (l) (1608094_at, GSVIVT00038153001-cinnamoyl-CoA reductase), (m) (1617632_at, GSVIVT00003032001, resveratrol-O-methyltransferase). Sk and s: preferentially expressed in skin and in seed according to the data from the Vv3 experiment in PLEXdb (http://www.plexdb.org/).
The second step in the resveratrol biosynthetic pathway is catalyzed by cinnamate 4-hydroxylase (C4H). In Arabidopsis, the C4H gene responds to various biotic stresses and abiotic stresses, such as light and wounding, indicating that it might have diverse functions in phenylpropanoid metabolism (44,45). In Cabernet Sauvignon, one C4H transcript (1610821_at; GSVIVT00023932001) showed increased abundance under water deficit from véraison through harvest, whereas the expression of this same gene was not affected in Chardonnay (Figures 1b and 2).
The third step of resveratrol biosynthesis is related to the expression of the 4-coumarate coenzyme A: ligase (4CL) gene (1611265_at; GSVIVT00009148001). There was a significant increase in transcript abundance due to water deficit in Cabernet Sauvignon following véraison through harvest, but a decrease in transcript abundance in water-deficit-treated Chardonnay berries during the same period (Figures 1c and 2). The closest ortholog of this gene is the tobacco 4-coumarate coenzyme A: ligase 2 gene, which was found to utilize 4-coumarate as a substrate, supporting its direct involvement in the biosynthesis of coumaroyl-CoA, precursor of trans-resveratrol (46). Constitutive expression in Saccharomyces cerevesiae and in Escherichia coli of 4-coumarate:coenzyme A ligase 4CL from tobacco enhances resveratrol biosynthesis indicating a potential regulatory role of this enzyme in the accumulation of resveratrol in grapes (47).
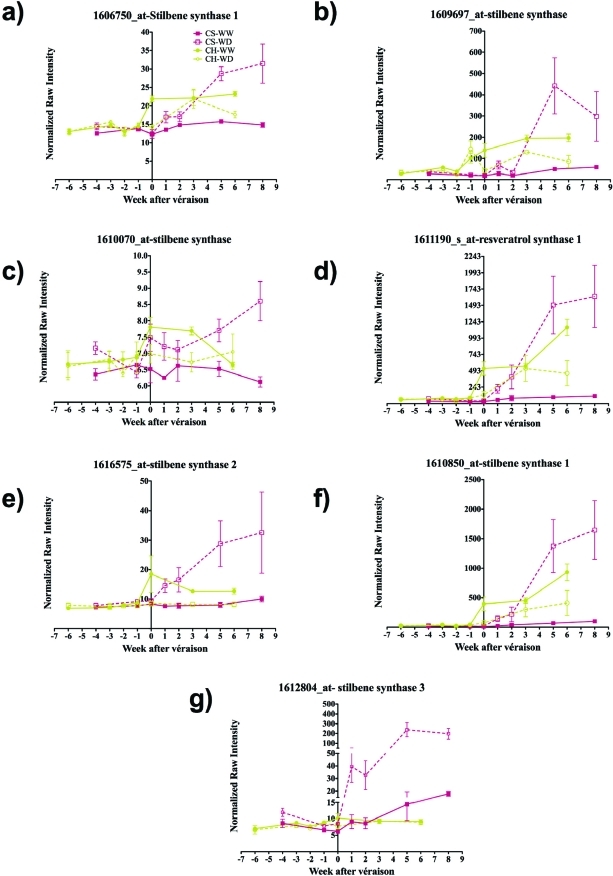
The next step in the resveratrol biosynthetic pathway is catalyzed by stilbene synthase (STS) or resveratrol synthase (RS). The most remarkable and statistically significant difference was seen with a probe set (1608009_s_at; GSVIVT00008253001) that underwent a 10-fold increase upon water deficit one week following véraison in Cabernet Sauvignon, reaching maximal expression five weeks after véraison (Figures 1d and 2). In contrast, the transcript abundance of this gene in Chardonnay was always higher in well-watered vines than in vines exposed to water deficit. Interestingly, the expression of seven other, but less strongly expressed, STS isogenes showed very similar transcript expression patterns to GSVIVT00008253001 (Figure 3). However, these results should be interpreted with caution because the expression of these isogenes was not statistically different between well-watered and water-deficit-stressed plants in both cultivars.
Figure 3.
Gene expression profiles of other stilbene synthases on the microarray. (a) (1606750_at, GSVIVG00009225001, stilbene synthase 1), (b) (1609697_at, GSVIVT00005194001, stilbene synthase), (c) (1610070_at, GSVIVT00009232001, stilbene synthase), (d) (1611190_s_at, GSVIVT00010117001, resveratrol synthase 1), (e) (1616575_at, GSVIVT00009225001, stilbene synthase 2), (f) (1610850_at, GSVIVT00009216001, stilbene synthase 1), (g) (1612804_at, GSVIVT00004047001, stilbene synthase 3). The lines and symbols are the same as in Figure 1. Symbols represent means ± SE; n = 3.
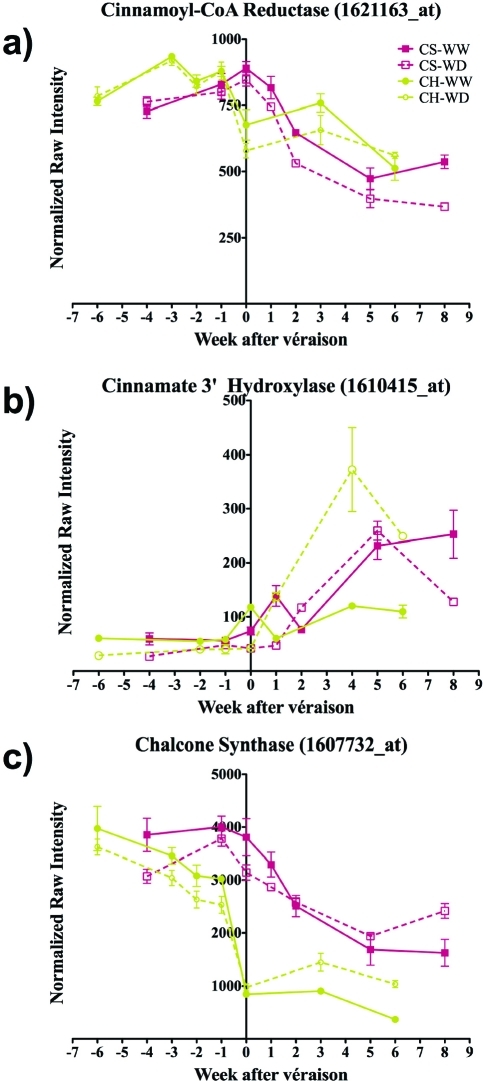
Trans-resveratrol is converted to trans-piceid by resveratrol-O-glucosyltransferase. One gene was recently characterized in Vitis labrusca; however, other substrates such as p-hydroxybenzoic and hydroxycinnamic acids might be acted up by this enzyme (48). The steady state abundance of the transcript of resveratrol-O-glucosyltransferase (1617078_at; GSVIVT00036670001) declined steadily over the course of development and was unaffected by water deficit in either cultivar. Two other genes (GSVIVT00036668001; GSVIVT00036670001) potentially associated with this enzyme are present in the Pinot Noir genome according to the 8X assembly. However, none of them had a related probeset (Figures 1e and 2). The transcript abundance of resveratrol-O-methyltransferase (1617632_at; GSVIVG00003032001) was unaffected by water deficit even though the pattern of expression was different between the two cultivars (Figure 1f). Because of the crosstalk between monolignol and the phenylpropanoid biosynthetic pathways, the expression of genes encoding other enzymes including chalcone synthase (CHS), which leads to flavonoid production, and cinnamate 3′-hydroxylase (C3H) and cinnamoyl CoA reductase (CCR), which lead to monolignol production, was surveyed. The transcript abundance of four out of six genes analyzed showed small increases upon water deficit in Cabernet Sauvignon after véraison (Figure 4), and three of these transcripts showed similar increases in water-deficit-stressed Chardonnay.
Figure 4.
Expression profiles of unigenes related to flavonoid and monolignol biosynthesis. (a) 1621163_at, GSVIVT00024039001, cinnamoyl CoA reductase, (b) 1617740_at, GSVIVT00001045001, cinnamate 3′ hydroxylase, (c) 1607732_at, GSVIVT00037969001, chalcone synthase. The lines and symbols are the same as in Figure 1. Symbols represent means ± SE; n = 3.
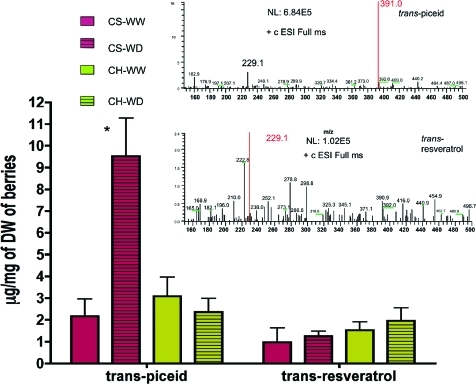
Upon the basis of the above transcript abundance changes, stilbene concentrations were hypothesized to increase following water deficit treatment. Stilbene accumulation was determined by HPLC-coupled mass spectrometry (MS) with the same berry samples used for the transcriptomic analyses. Cabernet Sauvignon and Chardonnay were analyzed from whole berries harvested at six and eight weeks, after véraison, respectively. The abundance of two stilbene-derived compounds, trans-piceid and trans-resveratrol, was not significantly different between the two cultivars when well-watered (Figure 5). The most abundant stilbene in both cultivars was trans-piceid, the glycosylated form of trans-resveratrol. Trans-piceid concentration significantly increased 5-fold in response to water deficit in berries of Cabernet Sauvignon, but not in Chardonnay. Given that resveratrol-O-glycosyltransferase gene expression was unaffected by water stress, STS gene expression appears to be a regulatory step for the accumulation of trans-piceid. This result is consistent with findings in Sorghum bicolor in which ectopic expression of STS enhanced the accumulation of cis-piceid (49).
Figure 5.
The concentrations of trans-piceid and trans-resveratrol in whole berries for both cultivars at EL-stage 38 (harvest stage) for the two irrigation treatments. CH = Chardonnay, CS = Cabernet Sauvignon, WW = well watered, WD = water deficit. Values are means ± SE; n = 6. The “*” indicates a significant difference from well-watered plants (P < 0.001; two-way ANOVA), (inset) mass spectrometric data with fragmentation patterns to confirm the presence of trans-piceid and trans-resveratrol in the samples.
In summary, the hypothesis that resveratrol biosynthesis would increase due to water deficit in Cabernet Sauvignon berries was supported by both transcript and metabolite data. Transcript data revealed that the majority of resveratrol biosynthetic genes exhibited increased steady state transcript abundance following water deficit in Cabernet Sauvignon, but not in Chardonnay. These results were confirmed by the metabolite profiling of resveratrol and piceid, indicating that under water deficit, the synthesis of stilbenes might be differentially regulated at the transcriptional level between the two cultivars.
Interestingly, the impact of water deficit on stilbene synthesis was previously investigated in V. vinifera cv. Barbera (50). In this study, the application of water deficit did not significantly influence the stilbene concentrations in Barbera berries, whereas cumulative applications of methyl jasmonate did induce trans-resveratrol synthesis at the ripening stage (50). Piceid concentrations were not measured, so it is uncertain whether this result was due to cultivar differences.
To provide a preliminary assessment of the genomic origin of the cultivar-specific interactions with water deficit in the transcriptional regulation of STS genes, a 1.2 kb proximal upstream region of the most differentially expressed stilbene synthase gene of this experiment (GSVIVT00008253001) was sequenced for Chardonnay and Cabernet Sauvignon. The sequences were compared to the corresponding promoter region from the partially inbred derived Pinot Noir genome (31). Interestingly, the nucleic acid alignment of this promoter region indicates a closer homology between Pinot Noir and Cabernet Sauvignon, even though Chardonnay is a progeny of Pinot Noir (51).
Computational analysis of this particular promoter region in both cultivars, using the PLACE algorithm (http://www.dna.affrc.go.jp/PLACE/) revealed 12 single nucleotide polymorphisms (SNP) resulting in the modification of eight canonical cis-elements within the promoter region (Table 2). Particularly interesting were two single nucleotide changes in regions that are related to abiotic stress regulation (Table 2). The first polymorphism, positioned at −407 from the transcription start site, lies within a regulatory element named the RAV1AAT-box. This polymorphism present in 50% of the cloned Cabernet Sauvignon sequences surveyed by PCR amplification indicates that only one allele is affected by this mutation. This particular cis-element is bound by the RAV transcription factor family, from which ectopic expression results in enhanced plant tolerance to biotic and abiotic stresses (52). Moreover, the transcript abundance of one member of this transcription factor family was found to be induced by abscisic acid (ABA) in soybean (53). Further investigations will be needed to correlate the potential role of ABA in the resveratrol biosynthetic pathway, and a more detailed functional analysis of the resveratrol promoter region will be critical to clearly identify potential ABA regulatory elements of this promoter.
Table 2. Putative cis-Acting Elements Harboring Polymorphisms within the Proximal Region of the STS2 Promoter in Cabernet Sauvignon and Chardonnaya.
| regulatory elements |
||||
|---|---|---|---|---|
| Chardonnay |
Cabernet Sauvignon |
|||
| nature of mutation | sequences | metabolism or response | sequences | metabolism or response |
| -823 − substitution in CH | GAATGG | − | GAATGA | − |
| -553 − substitution in CH | CAATGCGT | − | CAATGTGT | − |
| -407 − substitution in CH | GAACA | − | C/GAACA *(RAV1-Box) | drought stress response elements |
| -273 − substitution in CH | AGTCTTTA | − | AGTCC/TTTA | light regulated response elements |
| -190 − deletion in CH | CACATA | MYC consensus elements | CACAAATA | − |
| -181 − substitution in CH | AACACAT | − | AACGCAT | − |
| + 20 - substitution in CH | TAGGCCTTA | embryo development | TAGGCTTTA | − |
| carbon metabolism | ||||
| guard cell control | ||||
The symbol * means that the DNA sequencing of 50% of the PCR products result in the presence of a cytosine residue.
The second nucleotide change located at −273, from the transcription start site, leads to the formation of an I-Box core in 50% of the clones sequenced from Cabernet Sauvignon, but not in Chardonnay, indicating a heterozygous state for this cis-element in Cabernet Sauvignon. This I-Box core is commonly found upstream of light-regulated genes (54,55). In grapevine, water deficit alters canopy architecture by reducing shoot growth and basal shoot foliage, leading to higher levels of sun exposure within the cluster zone (56). Therefore, transcriptional regulation of some target genes might be due to light or heat effects, as already proposed for the flavonoid pathway in vines exposed to water deficit (57,58). Recently, comparative analysis between gene expression and protein accumulation of resveratrol synthase indicated that both are strongly influenced by UV−C irradiation (59). Whether this environmental cue is directly involved in a differential cis-regulatory activity of the STS gene between cultivars should be tested experimentally. Polymorphisms in promoter regions have been identified previously in Vitis. For instance, some of them are associated with traits selected during domestication that yield greater color (60,61).
To the best of our knowledge, this is the first report showing an increase in trans-piceid concentration in grape berry under water deficit conditions. Water deficit does not induce the transcript abundance of the gene most directly related to the piceid accumulation (resveratrol-O-glycosyltransferase) but rather affects the expression of the STS gene. Even though this glycosylated derivative is not the most active form of the stilbene compounds in wine, hydrolysis of trans-piceid releases trans-resveratrol in the small intestine and liver of humans (62). The elucidation of differences in the promoter of STS gene might allow both cultural and genetic manipulation of this important class of nutraceuticals, leading to increased concentrations of stilbene-derived compounds in grape berry. In addition to the potential human health benefits, increased stilbene concentrations in berries may contribute to improved vine resistance to pathogens and reduce the use of pesticides in vineyards.
Acknowledgments
This work was supported by funding from the National Science Foundation NSF (DBI-0217653) and the University of Nevada Agricultural Experiment Station (to G.R.C. and J.C.C.). The authors thank Craig Osborn of the Nevada Genomic Center for performing microarray analyses, and Kitty Spreeman for invaluable technical support. We would also like to thank Mary Ann Cushman for her critical reading of the manuscript. This publication was also made possible by NIH Grant Number P20 RR-016464 from the INBRE Program of the National Center for Research Resources through its support of the Nevada Genomics, Proteomics and Bioinformatics Centers.
This work was supported by funding from the National Science Foundation NSF (DBI-0217653) and the University of Nevada Agricultural Experiment Station (to G.R.C. and J.C.C.). This publication was also made possible by NIH Grant Number P20 RR-016464 from the INBRE Program of the National Center for Research Resources through its support of the Nevada Genomics, Proteomics and Bioinformatics Centers.
Funding Statement
National Institutes of Health, United States
References
- Kasdallah-Grissa A.; Mornagui B.; Aouani E.; Hammami M.; Gharbi N.; Kamoun A.; El-Fazaa S. Protective effect of resveratrol on ethanol-induced lipid peroxidation in rats. Alcohol Alcoholism 2006, 41 (3), 236–239. [DOI] [PubMed] [Google Scholar]
- Cheng J. C.; Fang J. G.; Chen W. F.; Zhou B.; Yang L.; Liu Z. L. Structure-activity relationship studies of resveratrol and its analogues by the reaction kinetics of low density lipoprotein peroxidation. Bioorg. Chem. 2006, 34 (3), 142–157. [DOI] [PubMed] [Google Scholar]
- Olas B.; Wachowicz B.; Majsterek I.; Blasiak J. Resveratrol may reduce oxidative stress induced by platinum compounds in human plasma, blood platelets and lymphocytes. Anticancer Drugs 2005, 16 (6), 659–665. [DOI] [PubMed] [Google Scholar]
- Chen G.; Shan W.; Wu Y.; Ren L.; Dong J.; Ji Z. Synthesis and anti-inflammatory activity of resveratrol analogs. Chem. Pharm. Bull (Tokyo) 2005, 53 (12), 1587–1590. [DOI] [PubMed] [Google Scholar]
- Vidavalur R.; Otani H.; Singal P. K.; Maulik N. Significance of wine and resveratrol in cardiovascular disease: French paradox revisited. Exp. Clin. Cardiol. 2006, 11 (3), 217–225. [PMC free article] [PubMed] [Google Scholar]
- Lagouge M.; Argmann C.; Gerhart-Hines Z.; Meziane H.; Lerin C.; Daussin F.; Messadeq N.; Milne J.; Lambert P.; Elliott P.; Geny B.; Laakso M.; Puigserver P.; Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 2006, 127 (6), 1109–1122. [DOI] [PubMed] [Google Scholar]
- Molnar A.; Nicak A.; Skvarla J.; Navesnakova Y.; Skvarlova A.; Beno P.; Krcmery V. Effect of Tokaj wine in combination with water and 10% alcohol solution on long-life and development of alcohol progeria in Wistar SPF rats. Neuro Endocrinol. Lett. 2007, 28 (Suppl 4), 4–6. [PubMed] [Google Scholar]
- Mader I.; Wabitsch M.; Debatin K. M.; Fischer-Posovszky P.; Fulda S.. Identification of a novel proapoptotic function of resveratrol in fat cells: SIRT1-independent sensitization to TRAIL-induced apoptosis. FASEB J. 201024, 1997−2009. [DOI] [PubMed] [Google Scholar]
- Liu B. L.; Zhang X.; Zhang W.; Zhen H. N. New enlightenment of French Paradox: resveratrol’s potential for cancer chemoprevention and anti-cancer therapy. Cancer Biol. Ther. 2007, 6 (12), 1833–6. [DOI] [PubMed] [Google Scholar]
- Bavaresco L.; Fregoni C.; van Zeller de Macedo Basto Goncalves M.; Vezzulli S., Grapevine Molecular Physiology & Biotechnology; Springer Science: New York, 2009; pp 341−364. [Google Scholar]
- Bavaresco L.; Fregoni M.; Trevisan M.; Mattivi F.; Vrhovsek U.; Falchetti R. The occurrence of the stilbene piceatannol in grapes. Vitis 2002, 41 (3), 133–136. [Google Scholar]
- Bavaresco L.; Pezzutto S.; Ragga A.; Ferrarri F.; Terevisan M. Effect of nitrogen supply on trans-resveratrol concentration in berries of V. vinifera L cv Cabernet Sauvignon. Vitis 2001, 40, 229–230. [Google Scholar]
- Conte A.; Pellegrini S.; Tagliazucchi D. Effect of resveratrol and catechin on PC12 tyrosine kinase activities and their synergistic protection from beta-amyloid toxicity. Drugs Exp. Clin. Res. 2003, 29 (5−6), 243–255. [PubMed] [Google Scholar]
- Conte A.; Pellegrini S.; Tagliazucchi D. Synergistic protection of PC12 cells from beta-amyloid toxicity by resveratrol and catechin. Brain Res. Bull. 2003, 62 (1), 29–38. [DOI] [PubMed] [Google Scholar]
- Riviere C.; Delaunay J. C.; Immel F.; Cullin C.; Monti J. P. The polyphenol piceid destabilizes preformed amyloid fibrils and oligomers in vitro: hypothesis on possible molecular mechanisms. Neurochem. Res. 2009, 34 (6), 1120–1128. [DOI] [PubMed] [Google Scholar]
- Riviere C.; Richard T.; Quentin L.; Krisa S.; Merillon J. M.; Monti J. P. Inhibitory activity of stilbenes on Alzheimer’s beta-amyloid fibrils in vitro. Bioorg. Med. Chem. 2007, 15 (2), 1160–1167. [DOI] [PubMed] [Google Scholar]
- Kowalczyk M.; Kowalczyk P.; Tolstykh O.; Hanausek M.; Walaszek Z.; Slaga T. Synergistic effects of combined phytochemicals and skin cancer prevention in SENCAR mice. Cancer Prev. Res. (Phila) 2010, 3 (2), 170–178. [DOI] [PubMed] [Google Scholar]
- Halls C.; Yu O. Potential for metabolic engineering of resveratrol biosynthesis. Trends Biotechnol. 2008, 26 (2), 77–81. [DOI] [PubMed] [Google Scholar]
- Jeandet P.; Douillet-Breuil A. C.; Bessis R.; Debord S.; Sbaghi M.; Adrian M. Phytoalexins from the Vitaceae: biosynthesis, phytoalexin gene expression in transgenic plants, antifungal activity, and metabolism. J. Agric. Food Chem. 2002, 50 (10), 2731–2741. [DOI] [PubMed] [Google Scholar]
- Hall D.; De Luca V. Mesocarp localization of a bi-functional resveratrol/hydroxycinnamic acid glucosyltransferase of Concord grape (Vitis labrusca). Plant J 2007, 49 (4), 579–591. [DOI] [PubMed] [Google Scholar]
- Schmidlin L.; Poutaraud A.; Claudel P.; Mestre P.; Prado E.; Santos-Rosa M.; Wiedemann-Merdinoglu S.; Karst F.; Merdinoglu D.; Hugueney P.. A stress-inducible resveratrol o-methyltransferases involved in the biosynthesis of pterostilbene in grapevine. Plant Physiol. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J. G.; Alosi J. A.; McDonald D. E.; McFadden D. W. Pterostilbene inhibits lung cancer through induction of apoptosis. J. Surg. Res. 2010, 161 (1), 18–22. [DOI] [PubMed] [Google Scholar]
- Gatto P.; Vrhovsek U.; Muth J.; Segala C.; Romualdi C.; Fontana P.; Pruefer D.; Stefanini M.; Moser C.; Mattivi F.; Velasco R. Ripening and genotype control stilbene accumulation in healthy grapes. J. Agric. Food Chem. 2008, 56 (24), 11773–11785. [DOI] [PubMed] [Google Scholar]
- Bavaresco L.; Eibach R. Investigations on the influence of N fertilizer on resistance to powdery mildew (Oidium tuckeri), downy mildew (Plasmapora viticola) and on phytoalexin synthesis in different grapevine varieties. Vitis 1987, 26, 192–200. [Google Scholar]
- Versari A.; Parpinello G. P.; Tornielli G. B.; Ferrarini R.; Giulivo C. Stilbene compounds and stilbene synthase expression during ripening, wilting, and UV treatment in grape cv. Corvina. J. Agric. Food Chem. 2001, 49 (11), 5531–5536. [DOI] [PubMed] [Google Scholar]
- Hain R.; Reif H. J.; Krause E.; Langebartels R.; Kindl H.; Vornam B.; Wiese W.; Schmelzer E.; Schreier P. H.; Stocker R. H.; et al. Disease resistance results from foreign phytoalexin expression in a novel plant. Nature 1993, 361 (6408), 153–156. [DOI] [PubMed] [Google Scholar]
- Lim J.; Yun S.; Chung I.; Yu C. Resveratrol synthase transgene expression and accumulation of resveratrol glycoside in Rehmannia glutinosa. Mol. Breed. 2005, 16, 219–233. [Google Scholar]
- Schwekendiek A.; Spring O.; Heyerick A.; Pickel B.; Pitsch N. T.; Peschke F.; de Keukeleire D.; Weber G. Constitutive expression of a grapevine stilbene synthase gene in transgenic hop (Humulus lupulus L.) yields resveratrol and its derivatives in substantial quantities. J. Agric. Food Chem. 2007, 55 (17), 7002–7009. [DOI] [PubMed] [Google Scholar]
- Coutos-Thévenot P.; OPoinssot B.; Bonomelli A.; Yean H.; Breda C.; Buffard D.; Esnault R.; Hain R.; Boulay M. In vitro tolerance to Botrytis cinerea of grapevine 41B rootstock in transgenic plants expressing the stilbene synthase Vst1 gene under the control of a pathogen-inducible PR 10 promoter. J. Exp. Bot. 2001, 52, 901–910. [DOI] [PubMed] [Google Scholar]
- Zhu Y. J.; Agbayani R.; Jackson M. C.; Tang C. S.; Moore P. H. Expression of the grapevine stilbene synthase gene VST1 in papaya provides increased resistance against diseases caused by Phytophthora palmivora. Planta 2004, 220 (2), 241–250. [DOI] [PubMed] [Google Scholar]
- Jaillon O.; Aury J. M.; Noel B.; Policriti A.; Clepet C.; Casagrande A.; Choisne N.; Aubourg S.; Vitulo N.; Jubin C.; Vezzi A.; Legeai F.; Hugueney P.; Dasilva C.; Horner D.; Mica E.; Jublot D.; Poulain J.; Bruyere C.; Billault A.; Segurens B.; Gouyvenoux M.; Ugarte E.; Cattonaro F.; Anthouard V.; Vico V.; Del Fabbro C.; Alaux M.; Di Gaspero G.; Dumas V.; Felice N.; Paillard S.; Juman I.; Moroldo M.; Scalabrin S.; Canaguier A.; Le Clainche I.; Malacrida G.; Durand E.; Pesole G.; Laucou V.; Chatelet P.; Merdinoglu D.; Delledonne M.; Pezzotti M.; Lecharny A.; Scarpelli C.; Artiguenave F.; Pe M. E.; Valle G.; Morgante M.; Caboche M.; Adam-Blondon A. F.; Weissenbach J.; Quetier F.; Wincker P. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449 (7161), 463–467. [DOI] [PubMed] [Google Scholar]
- Grimplet J.; Deluc L. G.; Tillett R. L.; Wheatley M. D.; Schlauch K. A.; Cramer G. R.; Cushman J. C. Tissue-specific mRNA expression profiling in grape berry tissues. BMC Genomics 2007, 8, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L. G.; Quilici D. R.; Decendit A.; Grimplet J.; Wheatley M. D.; Schlauch K. A.; Merillon J. M.; Cushman J. C.; Cramer G. R. Water deficit alters differentially metabolic pathways affecting important flavor and quality traits in grape berries of Cabernet Sauvignon and Chardonnay. BMC Genomics 2009, 10, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer G. R.; Ergul A.; Grimplet J.; Tillett R. L.; Tattersall E. A.; Bohlman M. C.; Vincent D.; Sonderegger J.; Evans J.; Osborne C.; Quilici D.; Schlauch K. A.; Schooley D. A.; Cushman J. C. Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Funct. Integr. Genomics 2007, 7 (2), 111–134. [DOI] [PubMed] [Google Scholar]
- Tattersall E. A. R.; Ergul A.; Alkayal F.; Deluc L.; Cushman J. C.; Cramer G., R. Comparison of methods for isolating high-quality RNA from leaves of grapevine. Am. J. Enol. Vitic. 2005, 56, 400–406. [Google Scholar]
- Irizarry R. A.; Hobbs B.; Collin F.; Beazer-Barclay Y. D.; Antonellis K. J.; Scherf U.; Speed T. P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4 (2), 249–264. [DOI] [PubMed] [Google Scholar]
- Deluc L. G.; Grimplet J.; Wheatley M. D.; Tillett R. L.; Quilici D. R.; Osborne C.; Schooley D. A.; Schlauch K. A.; Cushman J. C.; Cramer G. R. Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics 2007, 8, 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodhi M.; Guang-Ning Y.; Weeden N.; Reisch B. A simple and efficient method for DNA extraction from grapevine cultivars, Vitis species and Ampelopsis. Plant Mol. Biol. Reporter 1994, 12 (1), 6–13. [Google Scholar]
- Howe K. L.; Chothia T.; Durbin R. GAZE: a generic framework for the integration of gene-prediction data by dynamic programming. Genome Res. 2002, 12 (9), 1418–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F.; Nicklen S.; Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 1977, 74 (12), 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K.; Dudley J.; Nei M.; Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24 (8), 1596–1599. [DOI] [PubMed] [Google Scholar]
- Jaillon O.; Aury J.-M.; Noel B.; Policriti A.; Clepet C.; Casagrande A.; Choisne N.; Aubourg S.; Vitulo N.; Jubin C.; Vezzi A.; Legeai F.; Hugueney P.; Dasilva C.; Horner D.; Mica E.; Jublot D.; Poulain J.; Bruyère C.; Billault A.; Segurens B.; Gouyvenoux M.; Ugarte E.; Cattonaro F.; Anthouard V.; Vico V.; Del Fabbro C.; Alaux M.; Di Gaspero G.; Dumas V.; Felice N.; Paillard S.; Juman I.; Moroldo M.; Scalabrin S.; Canaguier A.; Le Clainche I.; Malacrida G.; Durand E.; Pesole G.; Laucou V.; Chatelet P.; Merdinoglu D.; Delledonne M.; Pezzotti M.; Lecharny A.; Scarpelli C.; Artiguenave F.; Pè M. E.; Valle G.; Morgante M.; Caboche M.; Adam-Blondon A.-F.; Weissenbach J.; Quétier F.; Wincker P. French-Italian Public Consortium for Grapevine Genome Characterization. . The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449 (7161), 463−467. [DOI] [PubMed] [Google Scholar]
- Olsen K. M.; Lea U. S.; Slimestad R.; Verheul M.; Lillo C. Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J. Plant Physiol. 2008, 165 (14), 1491–9. [DOI] [PubMed] [Google Scholar]
- Raes J.; Rohde A.; Christensen J. H.; Van de Peer Y.; Boerjan W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003, 133 (3), 1051–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M.; Ohta D.; Sato R. Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol. 1997, 113 (3), 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.; Douglas C. J. Two divergent members of a tobacco 4-coumarate:coenzyme A ligase (4CL) gene family. cDNA structure, gene inheritance and expression, and properties of recombinant proteins. Plant Physiol. 1996, 112 (1), 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekwilder J.; Wolswinkel R.; Jonker H.; Hall R.; de Vos C. H. R.; Bovy A. Production of resveratrol in recombinant microorganisms. Appl. Environ. Microbiol. 2006, 72 (8), 5670–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D.; De Luca V.. Mesocarp localization of a bi-functional resveratrol/hydroxycinnamic acid glucosyltransferase of Concord grape (Vitis labrusca). Plant J. 2007, 49, 579−591. [DOI] [PubMed] [Google Scholar]
- Yu C. K.; Lam C. N.; Springob K.; Schmidt J.; Chu I. K.; Lo C. Constitutive accumulation of cis-piceid in transgenic Arabidopsis overexpressing a sorghum stilbene synthase gene. Plant Cell Physiol. 2006, 47 (7), 1017–1021. [DOI] [PubMed] [Google Scholar]
- Vezzulli S.; Civardi S.; Ferrari F.; Bavaresco L. Methyl jasmonate treatment as a trigger of resveratrol synthesis in cultivated grapevine. Am. J. Enol. Vitic. 2007, 58 (4), 530–533. [Google Scholar]
- Bowers J.; Boursiquot J. M.; This P.; Chu K.; Johansson H.; Meredith C. Historical genetics: the parentage of Chardonnay, Gamay, and other wine grapes of Northeastern France. Science 1999, 285 (5433), 1562–1565. [DOI] [PubMed] [Google Scholar]
- Sohn K. H.; Lee S. C.; Jung H. W.; Hong J. K.; Hwang B. K. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol. Biol. 2006, 61 (6), 897–915. [DOI] [PubMed] [Google Scholar]
- Zhao L.; Luo Q.; Yang C.; Han Y.; Li W. A RAV-like transcription factor controls photosynthesis and senescence in soybean. Planta 2008, 227 (6), 1389–1399. [DOI] [PubMed] [Google Scholar]
- Martinez-Hernandez A.; Lopez-Ochoa L.; Arguello-Astorga G.; Herrera-Estrella L. Functional properties and regulatory complexity of a minimal RBCS light-responsive unit activated by phytochrome, cryptochrome, and plastid signals. Plant Physiol. 2002, 128 (4), 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G.; Pichersky E.; Malik V.; Timko M.; Scolnik P.; Cashmore A. An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. U. S. A. 1988, 85, 7089–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves M.; Santos T.; Souza C.. Deficit irrigation in grapevine improves water−use efficiency while controlling vigour and production quality. Ann. Appl. 2007, 150, 237−252. [Google Scholar]
- Castellarin S. D.; Pfeiffer A.; Sivilotti P.; Degan M.; Peterlunger E.; G D. I. G. Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ. 2007, 30 (11), 1381–1399. [DOI] [PubMed] [Google Scholar]
- Fujita A.; Goto-Yamamoto N.; Aramaki I.; Hashizume K. Organ-specific transcription of putative flavonol synthase genes of grapevine and effects of plant hormones and shading on flavonol biosynthesis in grape berry skins. Biosci. Biotechnol. Biochem. 2006, 70 (3), 632–638. [DOI] [PubMed] [Google Scholar]
- Wang W.; Tang K.; Yang H. R.; Wen P. F.; Zhang P.; Wang H. L.; Huang W. D. Distribution of resveratrol and stilbene synthase in young grape plants (Vitis vinifera L. cv. Cabernet Sauvignon) and the effect of UV-C on its accumulation. Plant Physiol. Biochem. 2010, 48 (2−3), 142–152. [DOI] [PubMed] [Google Scholar]
- Kobayashi S.; Goto-Yamamoto N.; Hirochika H. Retrotransposon-induced mutations in grape skin color. Science 2004, 304 (5673), 982. [DOI] [PubMed] [Google Scholar]
- Walker A. R.; Lee E.; Bogs J.; McDavid D. A. J.; Thomas M. R.; Robinson S. P. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007, 49 (5), 772–785. [DOI] [PubMed] [Google Scholar]
- Henry-Vitrac C.; Desmouliere A.; Girard D.; Merillon J. M.; Krisa S. Transport, deglycosylation, and metabolism of trans-piceid by small intestinal epithelial cells. Eur. J. Nutr. 2006, 45 (7), 376–382. [DOI] [PubMed] [Google Scholar]