Abstract
Using both confocal immunofluorescence microscopy and biochemical approaches, we have examined the role of β-arrestins in the activation and targeting of extracellular signal-regulated kinase 2 (ERK2) following stimulation of angiotensin II type 1a receptors (AT1aR). In HEK-293 cells expressing hemagglutinin-tagged AT1aR, angiotensin stimulation triggered β-arrestin-2 binding to the receptor and internalization of AT1aR-β-arrestin complexes. Using red fluorescent protein-tagged ERK2 to track the subcellular distribution of ERK2, we found that angiotensin treatment caused the redistribution of activated ERK2 into endosomal vesicles that also contained AT1aR-β-arrestin complexes. This targeting of ERK2 reflects the formation of multiprotein complexes containing AT1aR, β-arrestin-2, and the component kinases of the ERK cascade, cRaf-1, MEK1, and ERK2. Myc-tagged cRaf-1, MEK1, and green fluorescent protein-tagged ERK2 coprecipitated with Flag-tagged β-arrestin-2 from transfected COS-7 cells. Coprecipitation of cRaf-1 with β-arrestin-2 was independent of MEK1 and ERK2, whereas the coprecipitation of MEK1 and ERK2 with β-arrestin-2 was significantly enhanced in the presence of overexpressed cRaf-1, suggesting that binding of cRaf-1 to β-arrestin facilitates the assembly of a cRaf-1, MEK1, ERK2 complex. The phosphorylation of ERK2 in β-arrestin complexes was markedly enhanced by coexpression of cRaf-1, and this effect is blocked by expression of a catalytically inactive dominant inhibitory mutant of MEK1. Stimulation with angiotensin increased the binding of both cRaf-1 and ERK2 to β-arrestin-2, and the association of β-arrestin-2, cRaf-1, and ERK2 with AT1aR. These data suggest that β-arrestins function both as scaffolds to enhance cRaf-1 and MEK-dependent activation of ERK2, and as targeting proteins that direct activated ERK to specific subcellular locations.
The traditional model of G protein-coupled receptor function is that signals emanating from these heptahelical receptors arise from their ability to catalyze the activation of heterotrimeric G proteins, whose dissociated subunits interact with enzymatic effectors, such as adenylyl cyclases, phospholipases, and ion channels, to regulate intermediary metabolism (1). Over the past decade, it has become clear that these receptors additionally function as important regulators of cell proliferation and differentiation in a variety of physiologic and pathophysiologic settings (2). In many cases, these effects correlate with the ability of heptahelical receptors to stimulate mitogen-activated protein kinase (MAPK) cascades.
Efforts to understand how these receptors regulate MAPK activity have led to the discovery of several previously unappreciated mechanisms of heptahelical receptor signaling (3, 4). Among these is the finding that, in addition to their originally described role of uncoupling agonist-occupied, G protein-coupled receptor kinase (GRK) phosphorylated receptors from their cognate G proteins (5), β-arrestins can function as adapter proteins that promote the stable association of signaling proteins with the heptahelical receptor. By binding to both the nonreceptor tyrosine kinase, c-Src, and to agonist-occupied β2 adrenergic receptors, β-arrestin-1 can confer tyrosine kinase activity upon the receptor (6, 7). Similarly, β-arrestins are involved in recruiting c-Src to the Neurokinin-1 receptor in KNRK kidney epithelial cells (8) and recruiting the Src family kinases, Hck and c-Fgr, to the CXCR-1 receptor in neutrophils (9). In addition, recent reports have indicated that β-arrestins can interact directly with component kinases of the extracellular signal-regulated kinase (ERK) and c-Jun N-terminal kinase (JNK) MAPK cascades. β-Arrestins form complexes with internalized protease-activated receptor 2 (PAR-2), Raf-1 and ERK1/2 (10), and with internalized neurokinin-1 receptor, c-Src and ERK1/2 (8). β-Arrestin-2 also acts as a scaffold for the component kinases of the JNK3 cascade, facilitating JNK3 activation by binding directly to JNK3 and the MAPK kinase kinase, Ask1 (11). The formation of stable complexes between a heptahelical receptor, β-arrestin, and a MAPK may also affect substrate specificity by constraining the spatial distribution of the activated MAPK.
We have studied the role of β-arrestins in the activation and targeting of ERK2 by angiotensin II type 1a receptors (AT1aR). Confocal immunofluorescence microscopy was used to track the spatial redistribution of AT1aR, β-arrestin-2, and ERK2 following angiotensin stimulation in HEK-293 cells. Transiently transfected COS-7 cells were used to assay the interaction of AT1aR, β-arrestin-2, and the component kinases of the ERK2 cascade, and to determine the role of β-arrestin-2 in ERK2 activation. We find that the formation of complexes between ATa1R, β-arrestin-2, and the component kinases of the ERK MAPK cascade leads to the activation of β-arrestin-bound ERK2 and the targeting of a pool of activated ERK2 into endosomal vesicles containing the internalized AT1aR.
Materials and Methods
Materials.
The pDsRed1-N1 plasmid was obtained from CLONTECH. FuGene 6 was from Roche Diagnostics, and LipofectAMINE was from GIBCO/BRL. Dithiobis[succinimidylpropionate] (DSP) was from Pierce. Monoclonal M2 anti-Flag affinity agarose and FITC-conjugated monoclonal anti-phospho-ERK1/2 were obtained from Sigma. Monoclonal HA.11 anti-hemagglutinin (HA) affinity agarose was obtained from Covance (Princeton, NJ). Rhodamine-conjugated monoclonal anti-HA antiserum was obtained from Molecular Probes. Polyclonal anti-ERK1/2 and anti-phospho-ERK1/2 antisera were from New England Biolabs. Rabbit polyclonal anti-Flag antiserum and anti-cRaf-1 were from Santa Cruz Biotechnology, and monoclonal anti-MEK1 was from Transduction Laboratories (Lexington, KY). Horseradish peroxidase-conjugated polyclonal donkey anti-rabbit and anti-mouse IgG were obtained from Jackson ImmunoResearch.
DNA Constructs.
pcDNA3.1 expression plasmids encoding HA-tagged AT1aR (12), and green fluorescent protein (GFP)-tagged β-arrestin-2 (13) were provided by L. Barak, R. Oakley, and M. G. Caron (Duke University). The pEGFP-N1 expression plasmid encoding GFP-tagged ERK2 (10) was provided by K. DeFea and N. Bunnett (University of California at San Francisco). The expression plasmid for red fluorescent protein (RFP)-tagged ERK2 was created by subcloning a BamHI restriction fragment encoding ERK2 from the pEGFP-N1 ERK2 plasmid into the polylinker region of the pDsRed1-N1 plasmid. Expression plasmids encoding cRaf-1 (14) and MEK-1 (15) were provided by C. Der (University of North Carolina, Chapel Hill), E. Krebs (University of Washington), and R. Seger (The Weitzmann Institute of Science). All other expression plasmids were created in our laboratory as previously described (7, 11).
Cell Culture and Transfection.
HEK-293 and COS-7 cells were from the American Type Culture Collection. HEK-293 cells were maintained in Minimum Essential Medium with Earle's Salts supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C in a humidified 5% CO2 atmosphere. COS-7 cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Transient transfection of HEK-293 cells was performed by using FuGene 6 according to the manufacturer's instructions. Transient transfection of COS-7 cells was performed by using LipofectAMINE as previously described (16). Monolayers of transfected cells were incubated in serum-free growth medium supplemented with 10 mM Hepes, pH 7.4, 0.1% BSA, and 1% penicillin/streptomycin for 16–20 h before stimulation. Agonist stimulations were performed at 37°C in serum-free media as described in the figure legends.
Confocal Microscopy.
HEK-293 cells were used for experiments performed using confocal microscopy because of their favorable morphology for simultaneously examining the cytoplasmic and nuclear distribution of proteins. For visualization of HA-AT1aR, transiently transfected HEK-293 cells in collagen-coated 35-mm glass-bottom dishes were stained with a 1:500 dilution of rhodamine-conjugated monoclonal anti-HA antiserum for 1 h at 37°C. Cells were then stimulated as described in the figure legends, washed with Dulbecco's PBS, fixed with 10% paraformaldehyde for 30 min at room temperature, and again washed with PBS prior to examination. For visualization of GFP-β-arrestin-2 and RFP-ERK2, cells were stimulated and fixed with paraformaldehyde as described. For visualization of phospho-ERK1/2, cells were stimulated and fixed as described, permeabilized with absolute methanol for 10 min at −20°C, and blocked with 1% BSA/5% FBS for 3 h at room temperature. Cells were then stained using a 1:100 dilution FITC-conjugated monoclonal anti-phospho-ERK1/2 in 1% BSA/1% FBS for 24 h at 4°C, and washed with PBS prior to examination. Confocal microscopy was performed on a Zeiss LSM510 laser scanning microscope using a Zeiss 63 × 1.4 numerical aperture water immersion lens with dual line switching excitation (488 nm for GFP, 568 nm for rhodamine) and emission (515–540 nm GFP, 590–610 nm rhodamine) filter sets.
Immunoprecipitation and Protein Immunoblotting.
Immunoprecipitation experiments were performed by using transiently transfected COS-7 cells in 100-mm dishes. Cells were stimulated as described in the figure legends and solubilized in 1 ml of immunoprecipitation buffer containing 250 mM NaCl, 50 mM Hepes, 0.5% Nonidet P-40, 10% glycerol, 2 mM EDTA, 1 mM sodium orthovanadate, 1 mM PMSF, 10 μg/ml leupeptin, and 10 μg/ml aprotinin. To detect AT1aR-associated proteins, stimulated cells were subjected to covalent protein crosslinking by using the membrane permeable hydrolyzable crosslinker, DSP, as previously described (6). A 50-μl aliquot of each clarified whole-cell lysate was mixed with an equal volume of 2× Laemmli sample buffer and resolved by SDS/PAGE for confirmation of plasmid expression by immunoblotting.
Immunoprecipitation of full-length and truncated Flag-β-arrestin-2 was performed using monoclonal M2 anti-Flag affinity agarose, as previously described (7). Immunoprecipitation of HA-AT1aR was performed similarly, using monoclonal HA.11 anti-HA affinity agarose. Immunoprecipitates were resolved by SDS-PAGE and transferred to poly(vinylidene difluoride) membrane for immunoblotting. Protein immunoblotting for full-length and truncated Flag-β-arrestin-2 was performed by using rabbit polyclonal anti-FLAG antiserum. Coprecipitated cRaf-1, MEK1, ERK2, and phospho-ERK2 were detected by using polyclonal anti-cRaf-1, monoclonal anti-MEK1, and polyclonal anti-ERK1/2 and anti-phospho-ERK1/2 antisera, respectively. Horseradish peroxidase-conjugated polyclonal donkey anti-rabbit and anti-mouse IgG were used as secondary antibody as appropriate. Immune complexes on poly(vinylidene difluoride) filters were visualized by enzyme-linked chemiluminescence and quantified by scanning laser densitometry.
Results
Activated ERK Colocalizes with AT1aR-β-Arrestin Complexes Following Angiotensin II Stimulation.
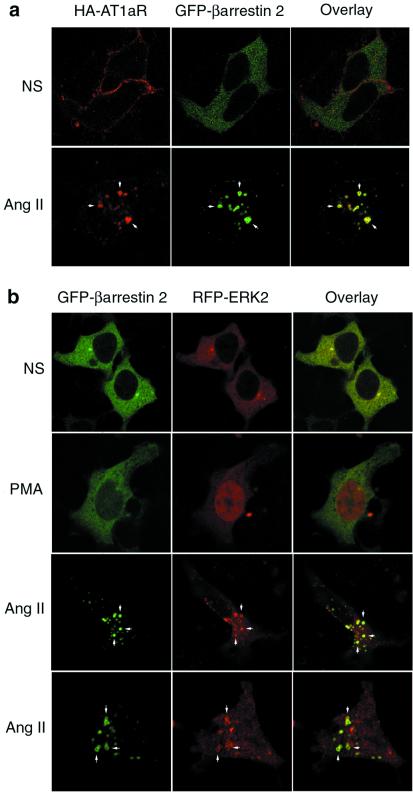
The stimulation of AT1aR results in the rapid recruitment of β-arrestins from the cytoplasm to the agonist-occupied receptor on the plasma membrane, followed by the trafficking of receptor-β-arrestin complexes into endosomal vesicles (12). This formation of stable internalized AT1aR-β-arrestin complexes is shown by confocal immunofluorescence microscopy in Fig. 1A. In the absence of ligand, HA-AT1aR were distributed along the plasma membrane (Top, red), whereas GFP-β-arrestin-2 (Top, green) was diffusely cytosolic in distribution. After 15 min of exposure to angiotensin II, receptor and β-arrestin each underwent a dramatic redistribution, with both proteins translocating to a common endosomal vesicle compartment (Bottom, yellow; arrows).
Figure 1.
Effect of angiotensin II on the cellular distribution of HA-AT1aR, GFP-β-arrestin-2, and RFP-ERK2. (a) HEK-293 cells were transiently transfected with plasmid DNA encoding HA-AT1aR and GFP-β-arrestin-2. Serum-starved cells were prestained with anti-HA rhodamine to label cell surface HA-AT1aR, treated with vehicle (NS) or angiotensin II (Ang II, 1 μM) for 15 min, fixed with paraformaldehyde, and examined by confocal microscopy. The distribution of rhodamine-labeled HA-AT1aR (red) and GFP-β-arrestin-2 (green) are shown in the single channel images. Colocalization of HA-AT1aR and GFP-β-arrestin-2 is shown in the overlay images (yellow; arrows). (b) Cells were transiently transfected with plasmid DNA encoding HA-AT1aR, GFP-β-arrestin-2, and RFP-ERK2. Serum-starved cells were treated with vehicle (NS) or PMA (100 nM) or angiotensin II (Ang II) for 15 min, and fixed cells were examined by confocal microscopy. The distribution of GFP-β-arrestin-2 (green) and RFP-ERK2 (red) are shown in the single channel images. Colocalization of GFP-β-arrestin-2 and RFP-ERK2 is shown in the overlay images (yellow; arrows). Each image depicts a representative confocal microscopic image from one of three separate experiments.
To determine the effect of angiotensin stimulation on the cellular distribution of ERKs, we used transiently expressed RFP-ERK2. Expression of this chimera permitted simultaneous imaging of RFP-ERK2 and GFP-β-arrestin-2. As shown in Fig. 1B, both GFP-β-arrestin-2 (Top, green) and RFP-ERK2 (Top, red) are diffusely cytosolic in nonstimulated cells. Treatment for 15 min with phorbol myristate acetate (PMA), which causes rapid protein kinase C-dependent activation of ERK1/2 (17), resulted in translocation of the RFP-ERK2 to the nucleus (Second row, red), without affecting the cytosolic distribution of GFP-β-arrestin-2 (Second row, green). In contrast, stimulation of AT1aR caused a distinctive redistribution of both proteins. After 15 min of agonist treatment, the GFP-β-arrestin-2, as expected, was found predominantly in endosomal vesicles (Third and Bottom rows, green; arrows). Although some of the RFP-ERK2 redistributed to the nucleus, a significant amount of the kinase colocalized with GFP-β-arrestin-2 in vesicles (Third and Bottom rows, red; arrows). Because these endosomal vesicles also contain the internalized AT1aR (Fig. 1A), these data suggest that internalized β-arrestin-bound AT1aRs can exist in complex with ERK2.
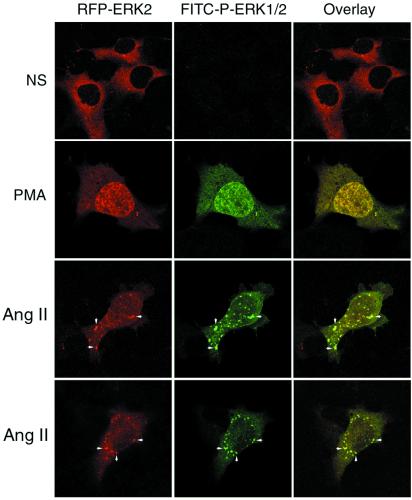
To determine the activation state of the RFP-ERK2 that colocalized with β-arrestin-2, cells coexpressing Flag-β-arrestin-2 and RFP-ERK2 were stained with FITC-conjugated anti-phospho ERK1/2 antiserum after stimulation. This antibody, which recognizes Thr202/Tyr204-phosphorylated ERK1/2, is specific for activated ERK1/2 (18). As shown in Fig. 2, there was no significant staining of cytosolic phospho-RFP-ERK2 in quiescent cells (Top). Stimulation for 15 min with PMA resulted in a redistribution of RFP-ERK2 from the cytoplasm to the nucleus (Second row, red) and an increase in FITC-stained phospho-ERK in both the cytoplasm and nucleus (Second row, green). Following angiotensin stimulation, FITC-stained phospho-ERK appeared both in the nucleus and in endosomal vesicles that colocalized with RFP-ERK2 (Third and Bottom rows; arrows). Thus, angiotensin stimulation resulted in the targeting of a significant fraction of the activated RFP-ERK2 to endosomal vesicles, resulting in a spatially constrained pool of activated ERK.
Figure 2.
Effect of angiotensin II on the cellular distribution of RFP-ERK2 and phospho-ERK1/2. HEK-293 cells were transiently transfected with plasmid DNA encoding HA-AT1aR, Flag-β-arrestin-2, and RFP-ERK2. Serum-starved cells were treated with vehicle (NS), PMA, or angiotensin II (Ang II) for 15 min, fixed with paraformaldehyde, permeabilized, and stained with FITC-conjugated monoclonal anti-phospho-ERK1/2 before examination by confocal microscopy. The distribution RFP-ERK2 (red) and FITC-stained phospho-ERK1/2 (green) are shown in the single channel images. Colocalization of RFP-ERK2 and phospho-ERK1/2 is shown in the overlay images (yellow; arrows). Each image depicts a representative confocal microscopic image from one of three separate experiments.
β-Arrestins Act as Scaffolds That Facilitate the Targeting and Raf-Dependent Activation of ERKs.
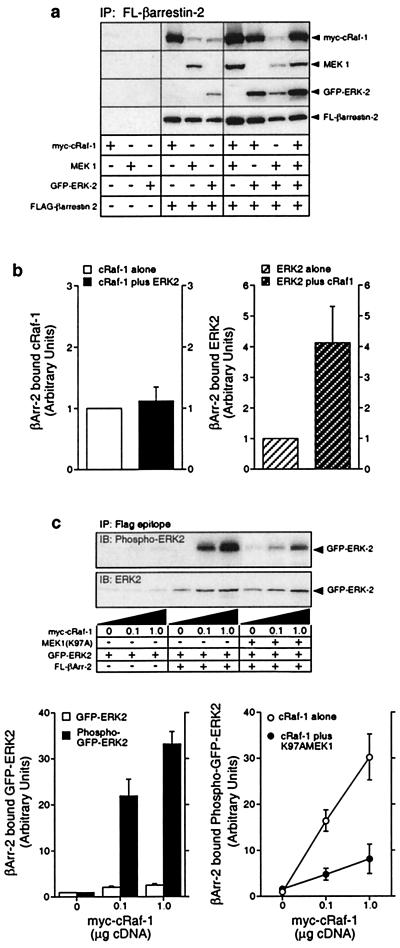
The data obtained by confocal microscopy suggest that AT1aR, β-arrestin-2, and activated ERK2 are targeted to endosomal vesicles following activation of the receptor. To examine the structural basis for this phenomenon, we used COS-7 cells transiently overexpressing Flag-β-arrestin-2 and each of the component kinases of the ERK cascade singly and in combination. As shown in Fig. 3A, myc-cRaf-1, MEK-1, and GFP-ERK2 were each detected in anti-Flag immunoprecipitates when coexpressed with Flag-β-arrestin-2. Simultaneous expression of either MEK1 or GFP-ERK2 had no effect on the amount of myc-cRaf-1 that precipitated with Flag-β-arrestin-2. Similarly, simultaneous expression of MEK1 and GFP-ERK2 did not affect the precipitation of either kinase with Flag-β-arrestin-2. In contrast, coexpression of myc-cRaf-1 enhanced the precipitation of both MEK1 and GFP-tagged ERK2 with Flag-β-arrestin-2. As shown in Fig. 3B, coexpression of myc-cRaf-1 increased ERK2 binding to Flag-β-arrestin-2 approximately 3- to 4-fold, suggesting that the binding of ERK2 to β-arrestin-2 may be at least partially indirect. These data suggest that the binding of cRaf-1 to β-arrestin-2 facilitates the assembly of a multiprotein complex on the β-arrestin that contains each of the component kinases of the ERK cascade.
Figure 3.
Binding of myc-cRaf-1, MEK1 and activated GFP-ERK2 to wild-type and mutant Flag-β-arrestin-2. (a) COS-7 cells were transiently transfected with plasmid DNA encoding myc-cRaf-1, MEK1, GFP-ERK2, and Flag-β-arrestin-2 in various combinations as indicated. GFP-ERK2, which undergoes agonist-stimulated phosphorylation and nuclear translocation like the wild-type kinase (10; not shown), was used in these studies to allow each overexpressed protein to have a unique epitope tag. Anti-Flag immunoprecipitates were probed for coprecipitated cRaf-1, MEK1, ERK2, and β-arrestin-2. Equivalent expression of each construct was confirmed by immunoblotting an aliquot of each whole cell lysate in parallel (not shown). Shown are representative immunoblots from one of six separate experiments. (b) Bar graphs depict the amount of myc-cRaf-1 (Left) and GFP-ERK2 (Right) present in Flag-β-arrestin-2 immunoprecipitates from cells coexpressing myc-cRaf-1 only, GFP-ERK2 only, or both myc-cRaf-1 and GFP-ERK2. Data are presented in arbitrary units where the amount of myc-cRaf-1 or GFP-ERK2 binding to Flag-β-arrestin-2 when singly expressed is defined as 1. Data shown represent the mean ± SD from four separate experiments. (c) Cells were transiently transfected with plasmid DNA encoding GFP-ERK2 and increasing amounts of myc-cRaf-1, in the presence and absence of Flag-β-arrestin-2 and MEK(K97A) as indicated. The amount of GFP-ERK2 and phospho-GFP-ERK2 present in Flag-β-arrestin-2 immunoprecipitates was determined by immunoblotting (Upper). Graphs depict the amount of GFP-ERK2 and phospho-GFP-ERK2 present in Flag-β-arrestin-2 immunoprecipitates (Left) and the effect of MEK(K97A) expression on the amount of phospho-GFP-ERK2 present in Flag-β-arrestin-2 immunoprecipitates (Right). Data are presented in arbitrary units where the amount of GFP-ERK2 or phospho-GFP-ERK2 present in Flag-β-arrestin-2 immunoprecipitates in the absence of overexpressed myc-cRaf-1 is defined as 1. Data shown represent the mean ± SD from three separate experiments.
To determine the effect of cRaf-1 on the phosphorylation of β-arrestin-bound ERK2, COS-7 cells expressing Flag-β-arrestin-2 and GFP-ERK2 were cotransfected with increasing amounts of the plasmid encoding myc-cRaf-1. As shown in Fig. 3C, increasing cRaf-1 expression resulted in a 2- to 3-fold increase in the amount of GFP-ERK2 that coprecipitated with Flag-β-arrestin-2. At the same time, the phosphorylation of β-arrestin-associated GFP-ERK2 increased 30- to 35-fold, consistent with cRaf-1 dependent phosphorylation of ERK2 in the β-arrestin complex (upper immunoblot and lower left bar graph). Because the activation of the ERK MAPK by the MAPK kinase kinase cRaf-1 should require a MAPK kinase, such as MEK1 (19), we used a dominant negative mutant of MEK1, MEK1(K97A), to determine whether the effect of cRaf-1 expression on β-arrestin-bound GFP-ERK2 depended on endogenous MEK activity. As shown, the myc-cRaf-1-dependent phosphorylation of GFP-ERK2 was significantly reduced in the presence of coexpressed MEK1(K97A) (upper immunoblot and lower right bar graph). These data suggest that β-arrestins, in addition to complexing with each of the component kinases of the ERK cascade, may function as scaffolds that facilitate cRaf-1 and MEK-dependent activation of β-arrestin-bound ERK.
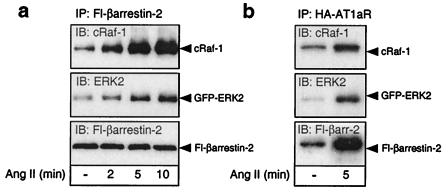
Fig. 4 depicts the effects of AT1aR stimulation on the binding of myc-cRaf-1 and GFP-ERK2 to Flag-β-arrestin-2, and on the recruitment of β-arrestin-2/cRaf-1/ERK2 complexes to the agonist-occupied AT1aR. As shown in Fig. 4A, angiotensin stimulation of COS-7 cells transiently expressing HA-AT1aR, Flag-β-arrestin-2, myc-cRaf-1, and GFP-ERK2 resulted in increased association of Flag-β-arrestin-2 with both myc-cRaf-1 and GFP-ERK2, suggesting that the conformational changes induced in β-arrestin-2 upon binding the receptor facilitates complex assembly. The agonist effect was detectable within 2 min of stimulation and maximal by 5 min, consistent with the time course of recruitment of GFP-β-arrestin-2 to agonist-occupied AT1aR (12). As shown in Fig. 4B, angiotensin stimulation increased the amount of Flag-β-arrestin-2, myc-cRaf-1, and GFP-ERK2 present in HA-AT1aR immunoprecipitates after covalent crosslinking with DSP. These data are consistent with the findings obtained by confocal microscopy in HEK-293 cells, that angiotensin stimulation caused the assembly of AT1aR/β-arrestin/ERK complexes that were subsequently targeted to endosomal vesicles.
Figure 4.
Effect of angiotensin II stimulation on the assembly of complexes containing HA-AT1aR, Flag-β-arrestin-2, myc-cRaf-1, and GFP-ERK2. (a) COS-7 cells were transiently transfected with plasmid cDNA encoding HA-AT1aR, Flag-β-arrestin-2, myc-cRaf-1, and GFP-ERK2. Serum-starved cells were stimulated with angiotensin II (Ang II) for the time indicated. The amount of myc-cRaf-1 and GFP-ERK2 present in Flag-β-arrestin-2 immunoprecipitates was determined by immunoblotting. (b) Serum-starved cells were stimulated with angiotensin II (Ang II) for 5 min before covalent crosslinking of receptor-associated proteins with DSP. The amount of myc-cRaf-1, GFP-ERK2, and Flag-β-arrestin-2 present in HA-AT1aR immunoprecipitates was determined by immunoblotting. Immunoblots shown are representative of three to five separate experiments.
Discussion
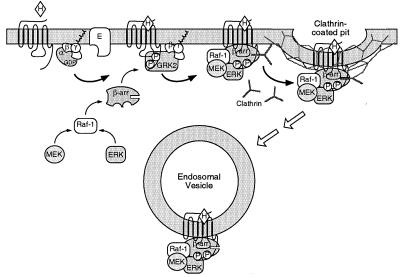
Fig. 5 depicts a model of β-arrestin-dependent ERK activation that is consistent with our data. The binding of agonist to heptahelical receptors results in the dissociation of heterotrimeric G proteins into Gα and Gβγ subunits, which go on to regulate various effectors. One consequence of Gβγ subunit release is the recruitment of G protein-coupled receptor kinases (e.g., GRK2 and GRK3) that phosphorylate the agonist-occupied receptor and increase its affinity for binding to β-arrestin (5). The component kinases of the ERK cascade, Raf-1, MEK, and ERK, subsequently assemble using β-arrestin as a scaffold and leading to activation of ERK. Endocytosis of these receptor/β-arrestin complexes by clathrin-coated pits results in the targeting of activated ERK to endosomal vesicles. The complexes, once formed, appear to be relatively stable, because they can be detected both by confocal microscopy and by immunoprecipitation techniques.
Figure 5.
Model of ERK activation and targeting by β-arrestin scaffolds. Agonist (H) binding to heptahelical receptors results in dissociation of heterotrimeric G proteins into Gα-GTP and Gβγ subunits, which activate G protein effectors (E). One consequence of Gβγ subunit release is enhanced GRK-mediated phosphorylation of the agonist-occupied receptor. β-Arrestins 1 (β-arr) bind to both GRK-phosphorylated receptor and to the component kinases of the ERK cascade, resulting in assembly of an ERK activation complex that is targeted into endosomal vesicles.
Multiple mechanisms are used by heptahelical receptors to regulate the activity of MAPK cascades (3, 4). These include second messenger-dependent pathways, such as the protein kinase A-dependent phosphorylation of the small G protein Rap1 in hematopoetic cells (20, 21), the protein kinase C-dependent activation of Raf isoforms (17), and the calcium- and cell adhesion-dependent activation of the focal adhesion kinase Pyk2 in neuronal cells (22, 23). In addition, many heptahelical receptors stimulate the transactivation of classical receptor tyrosine kinases, such as the epidermal growth factor (EGF) (24, 25) and platelet-derived growth factor (PDGF) receptors (26, 27). EGF receptor transactivation is the consequence of heptahelical receptor-mediated release of membrane-bound EGF-like ligands, such as heparin-binding EGF, through the action of as yet unidentified matrix metalloproteases (28). Signaling by means of β-arrestin scaffolds represents another, mechanistically distinct, means of MAPK regulation by heptahelical receptors.
The predominant mechanism used by any given heptahelical receptor to activate the ERK cascade varies between cell types and is apparently determined by the cellular context in which the receptor is expressed. In some cases, endogenous heptahelical receptors employ multiple mechanisms simultaneously (29). Indeed, AT1aR have been shown to trigger transactivation of both EGF (30) and PDGF receptors (31). Why should this seeming redundancy exist? One exciting possibility is that the consequences of ERK activation are determined to a significant degree by the mechanism by which they are activated. ERK substrates have been identified in the plasma membrane, cytoplasm, cytoskeleton, and nucleus. In addition to their role in regulating the phosphorylation of nuclear transcription factors (19), ERK1/2 have been reported to phosphorylate several proteins involved in heptahelical receptor signaling, such as β-arrestin-1 (32) and GRK2 (33, 34). Activation of the ERK cascade by transactivated EGF receptors or β-arrestin scaffolds, for example, may provide mechanisms for controlling both the time course and spatial distribution of ERK activity, leading to distinctly different consequences for the cell. Early evidence for this is provided by the finding that wild-type PAR-2 receptors, which cause β-arrestin-dependent activation of a cytosolic pool of ERK1/2, do not mediate mitogenic responses in KNRK cells. In contrast, mutant PAR-2 receptors that cannot bind β-arrestin, but still activate ERK1/2 through a calcium-dependent pathway, stimulate nuclear translocation of the ERK and provoke a proliferative response (10).
Our appreciation of β-arrestins as multifunctional adapter proteins is still evolving. In addition to their originally described roles in uncoupling heptahelical receptors from their cognate G proteins and in targeting receptors for endocytosis, β-arrestins increasingly appear to be directly involved in heptahelical receptor signaling. Through their ability to recruit Src kinases (6–8), and to act as scaffolds for the ERK (8, 10) and JNK (11) MAPK cascades, the binding of β-arrestins serves to trigger a second wave of signals emanating from the receptor. Thus, β-arrestins, like heterotrimeric G proteins, link heptahelical receptors to a defined subset of effector enzymes.
Acknowledgments
We thank D. Addison, M. Holben, and J. Turnbough for excellent secretarial assistance. R.J.L. is an investigator with the Howard Hughes Medical Institute. This work was supported by National Institutes of Health Grants DK55524 (to L.M.L.) and HL16037 (to R.J.L.).
Abbreviations
- MAPK
mitogen-activated protein kinase
- AT1aR
angiotensin II type 1a receptor
- DSP
dithiobis[succinimidylpropionate]
- EGF
epidermal growth factor
- ERK
extracellular signal-regulated kinase
- GFP
green fluorescent protein
- GRK
G protein receptor kinase
- HA
hemagglutinin
- JNK
c-Jun N-terminal kinase
- PMA
phorbol myristate acetate
- PAR-2
protease-activated receptor 2
- RFP
red fluorescent protein
References
- 1.Hepler J R, Gilman A G. Trends Biochem Sci. 1992;17:383–387. doi: 10.1016/0968-0004(92)90005-t. [DOI] [PubMed] [Google Scholar]
- 2.Dhanasekaran N, Heasley L E, Johnson G L. Endocrine Rev. 1995;16:259–270. doi: 10.1210/edrv-16-3-259. [DOI] [PubMed] [Google Scholar]
- 3.Gutkind J S. J Biol Chem. 1998;273:1839–1842. doi: 10.1074/jbc.273.4.1839. [DOI] [PubMed] [Google Scholar]
- 4.Pierce, K. L., Luttrell, L. M. & Lefkowitz, R. J. (2000) Oncogene, in press. [DOI] [PubMed]
- 5.Freedman N J, Lefkowitz R J. Recent Prog Horm Res. 1996;51:319. [PubMed] [Google Scholar]
- 6.Luttrell L M, Ferguson S S G, Daaka Y, Miller W E, Maudsley S, Della Rocca G J, Lin F-T, Kawakatsu H, Owada K, Luttrell D K, et al. Science. 1999;283:655–661. doi: 10.1126/science.283.5402.655. [DOI] [PubMed] [Google Scholar]
- 7.Miller W E, Maudsley S, Ahn S, Kahn K D, Luttrell L M, Lefkowitz R J. J Biol Chem. 2000;275:11312–11319. doi: 10.1074/jbc.275.15.11312. [DOI] [PubMed] [Google Scholar]
- 8.DeFea K A, Vaughn Z D, O'Bryan E M, Nishijima D, Dery O, Bunnett N W. Proc Nat Acad Sci USA. 2000;97:11086–11091. doi: 10.1073/pnas.190276697. . (First Published September 19, 2000; 10.1073/pnas.190276697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barlic J, Andrews J D, Kelvin A A, Bosinger S E, DeVries M E, Xu L, Dobransky T, Feldman R D, Ferguson S S G, Kelvin D J. Nat Immunol. 2000;1:227–233. doi: 10.1038/79767. [DOI] [PubMed] [Google Scholar]
- 10.DeFea K A, Zalevsky J, Thoma M S, Dery O, Mullins R D, Bunnett N W. J Cell Biol. 2000;148:1276–1281. doi: 10.1083/jcb.148.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonald P H, Chow C-W, Miller W E, LaPorte S A, Field M E, Lin F-T, Davis R J, Lefkowitz R J. Science. 2000;290:1574–1577. doi: 10.1126/science.290.5496.1574. [DOI] [PubMed] [Google Scholar]
- 12.Oakley R H, LaPorte S A, Holt J A, Caron M G, Barak L S. J Biol Chem. 2000;275:17201–17210. doi: 10.1074/jbc.M910348199. [DOI] [PubMed] [Google Scholar]
- 13.Barak L S, Ferguson S S G, Zhang J, Caron M G. J Biol Chem. 1997;272:27497–27500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]
- 14.Brtva T R, Drugan J K, Ghosh S, Terrell R S, Campbell-Burk S, Bell R M, Der C. J Biol Chem. 1995;270:9809–9812. doi: 10.1074/jbc.270.17.9809. [DOI] [PubMed] [Google Scholar]
- 15.Seger R, Seger D, Reszka A A, Munar E S, Eldar-Finkelman H, Dobrowolska G, Jensen A M, Campbell J S, Fischer E H, Krebs E G. J Biol Chem. 1994;269:25699–25709. [PubMed] [Google Scholar]
- 16.Luttrell L M, Della Rocca G J, van Biesen T, Luttrell D K, Lefkowitz R J. J Biol Chem. 1997;272:4637–4644. doi: 10.1074/jbc.272.7.4637. [DOI] [PubMed] [Google Scholar]
- 17.Hawes B E, van Biesen T, Koch W J, Luttrell L M, Lefkowitz R J. J Biol Chem. 1995;270:17148–17153. doi: 10.1074/jbc.270.29.17148. [DOI] [PubMed] [Google Scholar]
- 18.Her J H, Lakhani S, Zu K, Vila J, Dent P, Sturgill T W, Weber M J. Biochem J. 1993;296:25–31. doi: 10.1042/bj2960025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson M J, Cobb M H. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 20.Wan Y, Huang X-Y. J Biol Chem. 1998;273:14533–14537. doi: 10.1074/jbc.273.23.14533. [DOI] [PubMed] [Google Scholar]
- 21.Schmitt J M, Stork P J S. J Biol Chem. 2000;275:25342–25350. doi: 10.1074/jbc.M003213200. [DOI] [PubMed] [Google Scholar]
- 22.Lev S, Moreno H, Martinez R, Canoll P, Peles E, Musacchio J M, Plowman G D, Rudy B, Schlessinger J. Nature (London) 1995;376:737–745. doi: 10.1038/376737a0. [DOI] [PubMed] [Google Scholar]
- 23.Dikic I, Tokiwa G, Lev S, Courtneidge S A, Schlessinger J. Nature (London) 1996;383:547–550. doi: 10.1038/383547a0. [DOI] [PubMed] [Google Scholar]
- 24.Daub H, Weiss F U, Wallasch C, Ullrich A. Nature (London) 1996;379:557–560. doi: 10.1038/379557a0. [DOI] [PubMed] [Google Scholar]
- 25.Daub H, Wallash C, Lankenau A, Herrlich A, Ullrich A. EMBO J. 1997;16:7032–7044. doi: 10.1093/emboj/16.23.7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrlich A, Daub H, Knebel A, Herrlich P, Ullrich A, Schultz G, Gudermann T. Proc Natl Acad Sci USA. 1998;95:8985–8990. doi: 10.1073/pnas.95.15.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maudsley S, Zamah A M, Rahman N, Blitzer J T, Luttrell L M, Lefkowitz R J, Hall R A. Mol Cell Biol. 2000;20:8352–8363. doi: 10.1128/mcb.20.22.8352-8363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prenzel N, Zwick E, Daub H, Leserer M, Abraham R, Wallasch C, Ullrich A. Nature (London) 2000;402:884–888. doi: 10.1038/47260. [DOI] [PubMed] [Google Scholar]
- 29.Della Rocca G J, Maudsley S, Daaka Y, Lefkowitz R J, Luttrell L M. J Biol Chem. 1999;274:13978–13984. doi: 10.1074/jbc.274.20.13978. [DOI] [PubMed] [Google Scholar]
- 30.Eguchi S, Numaguchi K, Iwasaki H, Matsumoto T, Yamakawa T, Utsunomiya H, Motley E D, Owada K M, Hirata Y, Marumo F, Inagami T. J Biol Chem. 1998;273:8890–8896. doi: 10.1074/jbc.273.15.8890. [DOI] [PubMed] [Google Scholar]
- 31.Heeneman S, Haendeler J, Saito Y, Ishida M, Berk B C. J Biol Chem. 2000;275:15926–15932. doi: 10.1074/jbc.M909616199. [DOI] [PubMed] [Google Scholar]
- 32.Lin F-T, Miller W E, Luttrell L M, Lefkowitz R J. J Biol Chem. 1999;274:15971–15974. doi: 10.1074/jbc.274.23.15971. [DOI] [PubMed] [Google Scholar]
- 33.Pitcher J A, Tesmer J J, Freeman J L, Capel W D, Stone W C, Lefkowitz R J. J Biol Chem. 1999;274:34531–34534. doi: 10.1074/jbc.274.49.34531. [DOI] [PubMed] [Google Scholar]
- 34.Elorza A, Sarnago S, Mayor F., Jr Mol Pharmacol. 2000;57:778–783. doi: 10.1124/mol.57.4.778. [DOI] [PubMed] [Google Scholar]