Abstract
Objective
To connect a new family with early-onset Alzheimer disease (EOAD) in Germany to the American Volga German pedigrees.
Design
Pedigree molecular genetic analysis.
Setting
University Medical Centers in Fulda and Giessen, Germany, and in Seattle, Washington.
Results
The families from Fulda, Germany, and the American Volga German families with EOAD share the same N141I PSEN2 mutation on an identical haplotypic background. This establishes that the N141I mutation occurred prior to emigration of the families from the Hesse region to Russia in the 1760s, and documents that relatives of the original immigrant families are presently living in Germany with the mutation and the disease.
Conclusion
A family with the N141I mutation in PSEN2 that presently lives in Germany has been connected to the haplotype that carries the same mutation in pedigrees descended from the Volga Germans. This raises the possibility that the original patient with Alzheimer disease (Auguste D.), who had EOAD and lived in this same region of Germany, may also have had the PSEN2 N141I mutation.
In 1988, Bird and colleagues described several American pedigrees with familial early-onset Alzheimer disease (EOAD) of Volga German (VG) ancestry whose ancestors immigrated from 2 adjacent villages in Russia.1 These families share a single mutation, N141I, in the gene that encodes presenilin-2 (PSEN2; OMIM 600759).2,3 Although more than 10 additional mutations have been described in PSEN2, no families other than those of VG ancestry have been found with the N141I mutation. Recently, Nikisch et al4 described a 3-generation family in Germany with the N141I mutation. Because this family lives in the same region of Germany (Hesse) from which the VG individuals emigrated, we have performed a study attempting to link these families through shared genetic markers.
METHODS
Blood samples were obtained and DNA extracted from the American and German families with informed consent approved by the relevant institutions. Haplotype analysis of the PSEN2 region, based on multiple genetic markers, was performed on the DNA samples.
HAPLOTYPE ANALYSIS
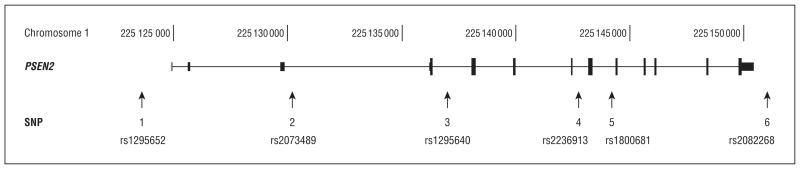
HapMap genotype data (http://www.hapmap.org, release 22) and Haploview 4.1 software were used to assess the PSEN2 haplotype structure and to select tagging single-nucleotide polymorphisms (SNPs). Genotypes of 20 SNPs from 60 CEU singletons were downloaded and analyzed using aggressive tagging, with the linkage disequilibrium (r2) threshold set at r2 > 0.8. Six tagging SNPs (Figure) were selected, capturing all information from the original 20 SNPs. TaqMan allele discrimination assays were performed to genotype the 6 SNPs in 130 subjects from the VG pedigrees as well as in the proband with EOAD from Fulda, Germany. To extend these haplotypes to the regions flanking PSEN2, including to the more informative multiallelic short tandem repeats loci flanking PSEN2, we also genotyped D1S479, which isapproximately4 cM (1.2 megabases [Mb]) 5′ of PSEN2, and D1S225, which is approximately1.5 cM (4.1 Mb) 3′ of PSEN2.
Figure.
Tagging single-nucleotide polymorphisms (SNPs) for PSEN2 haplotype analysis.
We used 2 strategies to estimate haplotype frequencies to accommodate data sets containing either unrelated or related individuals. We estimated SNP haplotype frequencies among 60 unrelated Centre d’Etude du Polymorphisme Humain (CEPH) Europeans using a maximum-likelihood expectation-maximization algorithm5 executed by the software package Arlequin 3.11 (http://cmpg.unibe.ch/software/arlequin3/). A single individual with missing SNP data was excluded from the haplotype estimation. Haplotype frequencies among the VGs were estimated by haplotype counting, using the SNP genotypes, pedigree information, and flanking marker data (D1S479, D1S225, N141I). Allele and haplotype counting provide unbiased estimates of allele frequencies, even in the presence of related individuals. Forced haplotypes were identified, where forced haplotypes are defined as haplotypes observed among individuals heterozygous for no more than a single SNP locus. When faced with ambiguous genotypes, we favored combinations of forced haplotypes over combinations of novel haplotypes. Haplotypes are identified by the allelic state at each of the 6 loci, in the order presented in the Figure: rs1295652, rs2073489, rs1295640, rs2236913, rs1800681, and rs2802268.
RESULTS
The Fulda (Hesse) patient had onset of memory loss at 47 years of age. Her father had onset of dementia at 64 years of age and died at 76 years. The paternal grandmother had onset at 48 years of age and died at 52 years. The American VG families have a mean age at onset of 55.5 years and mean age at death of 64 years. Not only are the onset ages similar between the VG and German families, the surname of the German family also occurs in one of the American VG families.
The genotype data for the 6 PSEN2 tagging SNPs was obtained for the CEPH and VG subjects. A total of 11 forced haplotypes were identified among the CEPH European and VG samples. The GTTACG haplotype (H1) is predominant (42%) among subjects from the VG families; this haplotype is only the fourth most frequent (13%) among the HapMap CEU sample (Table). All affected persons in the VG families share an identical PSEN2 haplotype based on these SNPs. The same haplotype was also found in the affected individual from the German family (patient III-24).
Table.
Frequencies of Haplotypes and Alleles of the Markers Segregating With the N141I Mutation
| Marker | Haplotype or Allele | CEPH Europeans | VG N141I Noncarriers | VG N141I Carriers |
|---|---|---|---|---|
| PSEN2 haplotype | GTTACG | 0.13 | 0.26 | 0.64 |
| D1S479 | 112 | 0.04 | 0.03 | 0.50 |
| D1S225 | 113 | 0.21 | 0.24 | 0.52 |
Abbreviations: CEPH, Centre d’Etude du Polymorphisme Humain; VG, Volga German.
The N141I mutation resides on the H1 haplotype (H1mut) shared by the affected subjects from the VG families; however, a wild-type version of the H1 haplotype (H1wt) is also present in the families in high frequency (Table). Thus, the H1mut/H1wt diplotype is common among the affected subjects in the VG families. The proband from Fulda also carries this diplotype but, because of the high frequency of the H1 haplotype, this alone is not sufficient to demonstrate shared ancestry of the VG pedigrees and the proband from Fulda. Attempts to identify additional genetic variants within this H1 haplotype group by genomic resequencing were unsuccessful, as no additional polymorphisms were identified.
To demonstrate that the proband from Fulda has the identical PSEN2 mutation haplotype as the VG families, we tested and defined subhaplotypes for the H1 haplotype group by genotyping short tandem repeats markers that flank the PSEN2 gene. Among the VG subjects, single alleles at D1S479 and D1S225 segregate with affectation status. The Table summarizes the frequencies of these alleles and the H1 SNP haplotype among genotyped VG and CEPH European subjects.
If we assume that the 2 short tandem repeats loci and haplotypes in PSEN2 are in linkage equilibrium, which is reasonable given their physical distance,6 then using the CEPH European allele frequencies, we can estimate the probability that the Fulda proband would share both risk alleles with the VG families as 0.0084. Under the assumption of linkage equilibrium among all 3 loci, the probability of the Fulda proband sharing the 2 risk alleles by chance with the VG families is between 0.1% and 0.2%, depending on whether one uses the CEPH European or the VG N141I noncarrier PSEN2 H1 haplotype frequency for estimation.
COMMENT
There are 3 interesting implications of this study. First, the N141I mutation in PSEN2 must have occurred prior to the emigration of the VGs from the Hesse region to Russia in the 1760s during the reign of Catherine the Great. Many of the immigrants assembled and then departed for Russia from the German village of Budingen, only a few kilometers from Fulda. Second, the finding of a family with this mutation living in modern Ger-many suggests that there are probably additional cases in Germany who share this common ancestral heritage and mutation. Third, it is of interest to note that Alois Alzheimer’s original patient, Auguste D. (the quintessential case of AD), lived in the same Hesse region as the modern family, and that Auguste D. had early-onset dementia (age, 51 years).7,8 Alzheimer was working in Frankfurt (Hesse) at the time (1901). This raises the interesting possibility that Auguste D. also had the N141I mutation in PSEN2. Little is known about her family history, but her father died at the relatively young age of 45 years. Neuropathology slides from her brain have been identified, and the tissue has been tested to determine her APOE genotype (ε3/ε3), which does not contain the high-risk ε4 allele.9 It would therefore be of historical and genetic interest to determine if she carried the N141I mutation in PSEN2 because this study provides strong circumstantial evidence to support this hypothesis.
Acknowledgments
Funding/Support: This study was supported by Veterans Affairs Research funds and National Institutes of Health/National Institute on Aging grant AG005136-24.
Footnotes
Financial Disclosure: Dr Bird reports receiving licensing fees and being on the Speaker’s Bureau of Athena Diagnostics, Inc.
Author Contributions: Study concept and design: Yu, Marchani, Müller, Wijsman, and Bird. Acquisition of data: Yu, Nolte, Hertel, and Bird. Analysis and interpretation of data: Yu, Marchani, Nikisch, Hertel, and Wijsman. Drafting of the manuscript: Yu, Marchani, Wijsman, and Bird. Critical revision of the manuscript for important intellectual content: Yu, Marchani, Nikisch, Müller, Nolte, Hertel, Wijsman, and Bird. Statistical analysis: Marchani and Wijsman. Obtained funding: Wijsman and Bird. Administrative, technical, and material support: Yu, Nolte, Hertel, Wijsman, and Bird. Study supervision: Yu, Nikisch, Müller, and Wijsman.
References
- 1.Bird TD, Lampe TH, Nemens EJ, Miner GD, Sumi SM, Schellenberg GD. Familial Alzheimer’s disease in American descendents of the Volga Germans: probable genetic founder effect. Ann Neurol. 1988;23(1):25–31. doi: 10.1002/ana.410230106. [DOI] [PubMed] [Google Scholar]
- 2.Levy-Lahad E, Wijsman EM, Nemens E, et al. A familial Alzheimer’s disease locus on chromosome 1. Science. 1995;269(5226):970–973. doi: 10.1126/science.7638621. [DOI] [PubMed] [Google Scholar]
- 3.Levy-Lahad E, Wasco W, Poorkaj P, et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science. 1995;269(5226):973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 4.Nikisch G, Hertel A, Kiessling B, et al. Three-year follow-up of a patient with early-onset Alzheimer’s disease with presenilin-2 N141I mutation case report and review of the literature. Eur J Med Res. 2008;13(12):579–584. [PubMed] [Google Scholar]
- 5.Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12(5):921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 6.Huttley GA, Smith MW, Carrington M, O’Brien SJ. A scan for linkage disequilibrium across the human genome. Genetics. 1999;152(4):1711–1722. doi: 10.1093/genetics/152.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurer K. The history of Alois Alzheimer’s first case Auguste D.: how did the eponym “Alzheimer’s Disease” come into being? In: Jucker M, Beyreuther K, Haass C, Nitsch R, Christen Y, editors. Alzheimer: 100 Years and Beyond. Berlin, Germany: Springer-Verlag; 2006. pp. 13–34. [Google Scholar]
- 8.Maurer K, Volk S, Gerbaldo H. Auguste D and Alzheimer’s disease. Lancet. 1997;349(9064):1546–1549. doi: 10.1016/S0140-6736(96)10203-8. [DOI] [PubMed] [Google Scholar]
- 9.Graeber MB, Kosel S, Grasbon-Frodi E, Moller HJ, Mehraein P. Histopathology and ApoE genotype of the first Alzheimer disease patient, Auguste D. Neurogenetics. 1998;1(3):223–228. doi: 10.1007/s100480050033. [DOI] [PubMed] [Google Scholar]