A new crystal form of the R163K variant of human thymidylate synthase (hTS) with five subunits per asymmetric part of the unit cell, all with loop 181–197 in the active conformation, is reported.
Keywords: thymidylate synthase, asymmetry, glutathione, conformational switching, FdUMP, dUMP
Abstract
Thymidylate synthase (TS) is a well validated target in cancer chemotherapy. Here, a new crystal form of the R163K variant of human TS (hTS) with five subunits per asymmetric part of the unit cell, all with loop 181–197 in the active conformation, is reported. This form allows binding studies by soaking crystals in artificial mother liquors containing ligands that bind in the active site. Using this approach, crystal structures of hTS complexes with FdUMP and dUMP were obtained, indicating that this form should facilitate high-throughput analysis of hTS complexes with drug candidates. Crystal soaking experiments using oxidized glutathione revealed that hTS binds this ligand. Interestingly, the two types of binding observed are both asymmetric. In one subunit of the physiological dimer covalent modification of the catalytic nucleophile Cys195 takes place, while in another dimer a noncovalent adduct with reduced glutathione is formed in one of the active sites.
1. Introduction
Thymidylate synthase (TS) catalyzes the reductive methylation of the nucleotide deoxyuridylate (dUMP) to thymidylate (TMP) using 5,10-methylenetetrahydrofolate (mTHF) as a cosubstrate (Humphreys & Greenberg, 1958 ▶). Substrates bind to the active site in an ordered manner, with dUMP binding prior to mTHF. The active-site cysteine (Cys195 in human TS) attacks the 6-position of the pyrimidine base of the nucleotide, resulting in the formation of a covalent bond between the enzyme and the nucleotide, thus activating the 5-position of the dUMP for subsequent covalent-bond formation with the C-11 substituent of mTHF, forming a ternary complex (reviewed in Stroud & Finer-Moore, 2003 ▶).
TS is a dimer of identical subunits that generate asymmetry upon substrate/ligand binding (Danenberg & Danenberg, 1979 ▶; Dev et al., 1994 ▶). The enzyme is ubiquitous and highly conserved among species; it is the sole source of de novo thymidylate, which is required for DNA synthesis. Cessation of this reaction not only halts cell replication but also leads to apoptosis or necrosis of rapidly dividing cells, an effect named ‘thymineless death’ (Houghton, 1999 ▶). Inhibitors mimicking either the substrate or cosubstrate have been developed as chemotherapeutic agents. Their binding leads to strained conformations of TS (Matthews et al., 1990 ▶; Montfort & Weichsel, 1997 ▶).
Human TS (hTS) differs from bacterial TSs in three regions: the N-terminus is extended by 28–29 residues and there are two insertions of 12 and eight residues at positions 117 and 146, respectively (Carreras & Santi, 1995 ▶). hTS was originally crystallized in high ammonium sulfate conditions and when its structure was compared with those of other TS enzymes it was observed that the catalytic loop 181–197 was rotated by approximately 180° (Schiffer et al., 1995 ▶). This conformation, thus far seen only in hTS, has the catalytic cysteine (Cys195 in hTS) out of the active site and therefore must be inactive. In this inactive conformation of hTS, one of the eukaryotic inserts, loop 107–128, is disordered and four sulfates per subunit can be seen; they appear to stabilize the conformer. However, hTS in an inhibitory complex with dUMP and raltitrexed (Phan, Koli et al., 2001 ▶) has loop 107–128 ordered and loop 181–197 in the active conformation. The conformational equilibrium of hTS is also affected by the N-terminus: structural studies of hTS with residues 7–29 deleted showed the enzyme in the active conformation (Almog et al., 2001 ▶), while some Val3 mutants had an altered equilibrium (Huang et al., 2010 ▶). The position of Trp182 is quite different in the active and inactive conformations, which allowed Phan and coworkers to monitor the conformational equilibrium through the use of intrinsic fluorescence (Phan, Steadman et al., 2001 ▶). The addition of phosphate, which stabilizes the inactive conformer, leads to an increase in hTS fluorescence, while the subsequent addition of dUMP quenches this increase. In solution, the active and inactive conformations are in equilibrium (Phan, Steadman et al., 2001 ▶). The hTS variant R163K was engineered to shift the equilibrium towards the active conformation and it is apparent that the design was successful (Gibson et al., 2008 ▶).
In general, TS subunits show negative cooperativity for substrate, cosubstrate or inhibitor binding. The ratio of the binding constants, K b, for the two subunits depends strongly on the ligand studied and on the source of the enzyme. For hTS, measurements of dUMP binding in a binary complex by equilibrium dialysis yielded a dissociation constant K d of 7.5 µM, with n = 0.90 per dimer (Dev et al., 1994 ▶). This value corresponds to the tighter binding site, while the K d for the weaker site could not be measured using this technique. The binding of 5-fluoro-2′-deoxyuridine monophosphate (FdUMP) to hTS Q214E showed a biphasic pattern with a ratio of binding constants of about 6. Successful crystal binding studies typically use ligand concentrations that are much higher than the K d value, which therefore leads to binding in both subunits. The only reported structure of a TS dimer with half of the sites filled by ligands is that of Pneumocystis carinii (PcTS), for which the folate K b ratio is about 8000. In this structure (PDB code 1ci7) PcTS is in complex with dUMP, which is present in both active sites, and an antifolate cosubstrate inhibitory analogue CB3717, which is present in only one (Anderson et al., 1999 ▶). The subunit with bound inhibitor represents a ternary inhibitory complex with a covalent bond between TS and dUMP, while the other subunit represents only an adduct of dUMP. For hTS, asymmetric binding of Pi was observed in form 1 of R163K and the reactivity of hTS was also asymmetric; the mutant displayed an asymmetric dimer in which the catalytic cysteine was covalently modified in only one subunit (Gibson et al., 2008 ▶).
Here, we report a new crystal form of hTS mutant R163K (form 3) and crystal structures with bound FdUMP, dUMP and glutathione. These structures display asymmetric ligand binding in dimers.
2. Materials and methods
2.1. Chemicals
Salts, β-mercaptoethanol (BME), polyethylene glycols (PEGs), 2′-deoxyuridine-5′-monophosphate (dUMP) and 5-fluoro-2′-deoxyuridine monophosphate (FdUMP) were obtained from Sigma (St Louis, Missouri, USA). (6S)-5,6,7,8-Tetrahydrofolic acid (H4folate) was prepared from folic acid and converted to (6R)-CH2H4folate (mTHF) as described previously (Steadman et al., 1998 ▶). l-Oxidized glutathione was obtained from Acros Organics (Morris Plains, New Jersey, USA).
2.2. Protein expression and crystallization
A plasmid containing the hTS mutant R163K (Gibson et al., 2008 ▶) was transformed into the Escherichia coli TX61− (thyA−) bacterial strain, which does not produce its own thymidylate synthase enzyme (Dev et al., 1988 ▶). R163K hTS was expressed and purified following procedures previously developed for hTS (Phan, Steadman et al., 2001 ▶) with only minor modifications. Briefly, cell-free extract was separated on a Q-Sepharose anion-exchange column using a 0–100% saturated KCl gradient. The active fractions were pooled, dialyzed and separated on a Blue Sepharose CL-6B column using a similar gradient. The active fractions were again pooled, dialyzed and separated a second time on the Q-Sepharose anion-exchange column using a similar gradient. The active fractions were then pooled, concentrated to 12 mg ml−1 and analyzed for purity on an SDS–PAGE gel. R163K crystals were grown under low-salt conditions (100 mM Tris pH 9.0, 20 mM BME, 3 mM KH2PO4, 10–20% PEG 4K) by hanging-drop vapor diffusion at 277 K.
2.3. Data collection and processing
Crystals were transferred into soaking solution (100 mM Tris pH 9.0, 20 mM BME, 3 mM KH2PO4, 10–20% PEG 4K) plus ligand (3 mM FdUMP and 2 mM mTHF or 3 mM dUMP and 2 mM mTHF) or soaking solution without BME and with 1 mM l-oxidized glutathione and soaked for 2 min. Crystals were then transferred to a cryosolution containing an additional 20% ethylene glycol and flash-frozen in a liquid-N2 vapor stream.
X-ray diffraction data were collected on the SER-CAT 22ID or 22BM beamlines at the Advanced Photon Source, Argonne National Laboratory. The data were indexed and processed with the HKL-2000 software (Otwinowski & Minor, 1997 ▶); processing parameters and statistics are summarized in Table 1 ▶.
Table 1. Crystallographic data and refinement statistics for R163K form 3.
Values in parentheses are for the outer shell.
| FdUMP | dUMP | Glutathione | |
|---|---|---|---|
| X-ray source | APS SER-CAT 22ID | APS SER-CAT 22BM | APS SER-CAT 22BM |
| Wavelength (Å) | 1.0 | 1.0 | 0.92 |
| No. of frames (high pass/low pass) | 200/200 | 360 | 288 |
| Oscillation range (°) | 1.0 | 0.5 | 0.5 |
| Crystal-to-detector distance (mm) | 300/350 | 300 | 300 |
| Temperature (K) | 100 | 100 | 100 |
| Space group | C2 | C2 | C2 |
| Unit-cell parameters | |||
| a (Å) | 200.284 | 196.459 | 201.80 |
| b (Å) | 122.190 | 122.149 | 123.11 |
| c (Å) | 99.828 | 99.157 | 99.93 |
| β (°) | 115.18 | 114.551 | 115.8 |
| Volume (Å3) | 2210928 | 2164371 | 2236989 |
| Solvent content (%) | 60.53 | 59.68 | 60.97 |
| Matthews coefficient (Å3 Da−1) | 3.11 | 3.05 | 3.15 |
| Mosaicity (°) | 0.52 | 0.64 | 0.89 |
| Resolution range (Å) | 50.0–2.65 (2.74–2.65) | 50.0–2.74 (2.84–2.74) | 45.5–2.75 (2.85–2.75) |
| Average multiplicity | 4.9 | 3.0 | 2.7 |
| Average I/σ(I) | 11.4 | 18.3 | 13.7 |
| Total No. of reflections | 312575 | 157053 | 138780 |
| No. of unique reflections | 59470 | 50175 | 46360 |
| Completeness (%) | 99.7 (98.0) | 96.7 (82.6) | 89.1 (55.8) |
| Total linear Rmerge | 11.0 | 6.9 | 7.6 |
| R (CNS) (%) | 21.4 | 21.0 | 23.8 |
| Rfree (CNS) (%) | 26.0 | 26.2 | 27.5 |
| Ramachandran statistics, residues in | |||
| Most favored regions (%) | 84.3 | 85.5 | 77.2 |
| Additional allowed regions (%) | 15.3 | 14.2 | 21.1 |
| Generously allowed regions (%) | 0.4 | 0.3 | 1.7 |
| Disallowed regions (%) | 0.0 | 0.0 | 0.0 |
| Average B factors (Å2) | |||
| Subunit A | 47.1 | 49.1 | 65.4 |
| Subunit B | 56.5 | 57.5 | 77.3 |
| Subunit C | 52.9 | 50.1 | 79.4 |
| Subunit D | 71.0 | 62.3 | 92.1 |
| Subunit E | 60.8 | 54.8 | 92.0 |
2.4. Structure determination and refinement
The initial structure of R163K form 3 was solved by molecular replacement with the CNS software (Brünger et al., 1998 ▶) using the hTS–dUMP–raltitrexed structure (PDB code 1hvy; Phan, Koli et al., 2001 ▶) without ligands as the search model. Subsequent structures of R163K form 3 with ligands bound were solved by molecular replacement with the CNS software (Brünger et al., 1998 ▶) using the native R163K form 3 structure as the search model. Structure rebuilding and subsequent refinements were performed using the TURBO software (Roussel & Cambillau, 1991 ▶) and CNS (Brünger et al., 1998 ▶), respectively. Dictionary files for the cysteine residue covalently modified by glutathione were created starting from GSH and CME topology and parameter files from HIC-Up (Kleywegt & Jones, 1998 ▶). Superpositions were calculated using the LSQKAB program (Kabsch, 1976 ▶) from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). Ligand occupancies were calculated using SHELXL (Sheldrick, 2008 ▶). Figures were prepared using TURBO (Roussel & Cambillau, 1991 ▶), MolScript (Kraulis, 1991 ▶) and Raster3D (Merritt & Bacon, 1997 ▶).
3. Results and discussion
3.1. Crystallization
It has been shown previously that hTS mutant R163K has a propensity for polymorphism (Gibson et al., 2008 ▶). Crystals of R163K form 3 appeared after 3–4 d in the same crystallization conditions as the previously reported R163K crystal forms 1 and 2. Form 3 belongs to space group C2 and has five subunits per asymmetric part of the unit cell (two dimers, A/B and D/E, are in general positions, while subunit C forms a dimer with its symmetry equivalent via a twofold crystallographic axis). The previously reported structures of R163K crystal forms 1 and 2 belonged to space group P3221 and had six and two subunits per asymmetric part of the unit cell, respectively (Gibson et al., 2008 ▶).
3.2. Conformation of loop 181–197
Loop 181–197 of wild-type hTS (wt-hTS) populates two conformations in solution: one active and the other inactive. The number of conformers appears to be similar: kinetic data suggest that in the minimum energy state an asymmetric dimer exists with one subunit in the active conformation and the second in an inactive conformation (Lovelace et al., 2007 ▶). High-quality crystals of wt-hTS were previously obtained in the presence of ammonium sulfate because sulfate ions (and phosphate ions) strongly stabilize the inactive conformer, leading to symmetric molecules that are more likely to form well ordered crystals. The inactive conformer, however, does not bind substrate analogues and the crystals obtained using ammonium sulfate are useless for most inhibitor-binding studies.
In all of the subunits present in the structures of form 3, as in the previously studied forms 1 and 2, the eukaryotic insert 1 (loop 107–128) is ordered and the catalytic loop (residues 181–197) is in the active conformation, positioning the catalytic residue Cys195 in the active site. All individual subunits, when superposed, are very similar, with the r.m.s. distance of Cα positions varying in the range 0.24–0.70 Å. Thus, the R163K variant of hTS has the inactive conformer destabilized and its crystals, as reported here, are suitable for studies of inhibitor binding that do not lead to large conformational changes. Inhibitor binding that induces the closure of the C-terminus apparently affects crystal packing and leads to a deterioration in crystal quality.
3.3. hTS complexes with the substrate and inhibitor
Crystals were soaked with either FdUMP/mTHF or dUMP/mTHF mixtures; however, only binary complexes were obtained. The two dimers present in general positions have one subunit with a very high occupancy of the nucleotide, while the other subunit has only partial occupancy of the soaked ligand. In these partially filled subunits strong density corresponding to a phosphate ion (which is present at 3 mM in the crystallization medium) was observed at the active site, indicating that in the absence of a nucleotide a phosphate ion binds. Even though both the FdUMP and dUMP soaks also contained the cosubstrate mTHF, no density for the folate was seen in any of the subunits, while the density for the nucleotides was quite good. This may indicate that diffusion of the nucleotide into the crystal is quick but diffusion of the larger folate is much slower. Birdsall et al. (1996 ▶) were able to grow cocrystals of Lactobacillus casei TS with dUMP and subsequently soak them in mTHF and antifolates over the course of approximately 2 d. Longer soaking times were performed with R163K crystals; however, diffraction by these crystals was poor and data collection was not possible. In contrast, a crystal soaked in FdUMP or dUMP only retained diffracting power. This suggests that the formation of tertiary complexes eventually took place, but the transition to the more compact conformation of these complexes affected crystal packing.
3.3.1. FdUMP complex
At a contour level of σ = 1.0 density that corresponds to FdUMP can be seen in subunits A, B and D; at a lower contouring level of σ = 0.6 density can also be seen in subunit E. Refinement with SHELXL (Sheldrick, 2008 ▶) was used to estimate the occupancy of FdUMP in each subunit. In the first dimer (A/B) subunit A has full occupancy for FdUMP, while the occupancy in subunit B was calculated to be approximately half. The half occupancies calculated with SHELX were likely to be overestimates (because a phosphate ion is likely to be present in the absence of nucleotide), but are consistent with the density observed (Fig. 1 ▶). No covalent bond is seen between the FdUMP and Cys195, thus reflecting the observation that the nucleotide does not form a covalent bond to the catalytic cysteine until a folate binds (Stroud & Finer-Moore, 2003 ▶).
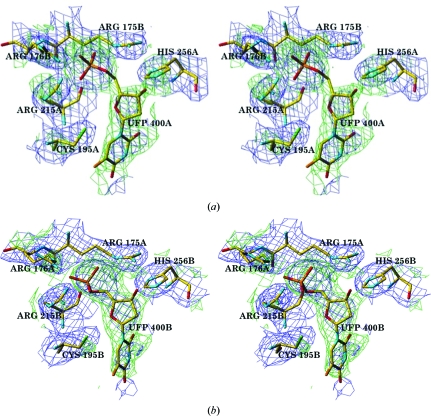
Figure 1.
(a) Stereoview of the hTS active site in subunit A with full occupancy of the ligand FdUMP. (b) Stereoview of the active site in subunit B with partial occupancy of the ligand FdUMP. The F o − F c map (green) is contoured at the 2σ level, with the density of neighboring residues shown in the 2F o − F c map (blue) contoured at the 1σ level.
Ethylene glycol, which was present in the cryosolution, has been modeled into density found in the active sites of subunits A and B. This density is not large enough to represent the much larger mTHF; rather, its presence indicates that at the 3.6 M concentration in the cryosolvent glycol competes with mTHF binding. Even though the subunits that form a dimer do not bind FdUMP equally, superposition of subunits A and B yielded an r.m.s. distance between the Cα positions of only 0.40 Å. Fig. 2 ▶ shows this superposition; it reveals no significant differences at the subunit interface.
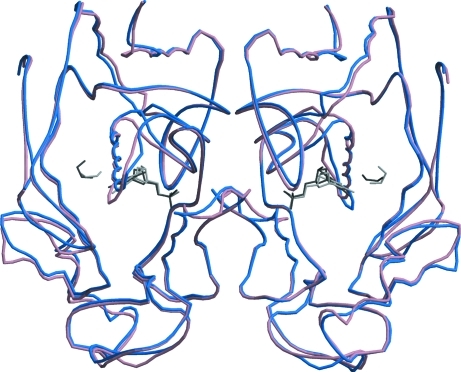
Figure 2.
Superposition of dimer A/B on dimer B/A (shown in blue and pink) of hTS R163K form 3 soaked with FdUMP. The soaked ligands FdUMP and BME are shown in dark gray in dimer A/B, while in dimer B/A they are shown in light gray. FdUMP and BME occupy both active sites, but have full occupancy in subunit A and only partial occupancy in subunit B. This superposition reveals no significant differences at the subunit interface.
Superposition of subunit A of the FdUMP-soaked R163K form 3 structure with subunit A of R163K form 2 (PDB entry 2rd8; Gibson et al., 2008 ▶) yielded an r.m.s. distance between the Cα positions of 0.25 Å, while superposition of subunit B of R163K form 2 yielded an r.m.s. distance of 0.60 Å. This difference, while small, could be attributed to the modification of Cys195 by BME, which is present in subunit B of R163K form 2 but is not present in subunit A.
3.3.2. dUMP complex
In the structure of R163K form 3 soaked with dUMP, full occupancy is seen for dUMP in subunits A and D, while subunits B and E (the complementary halves of the dimers) mostly have phosphate in their active sites (Fig. 3 ▶). There is moderate density for dUMP (more than phosphate) in subunit C, which forms a dimer with a symmetry-equivalent subunit. As in the structure of R163K form 3 with FdUMP, the R163K form 3 structure soaked with dUMP also displays no covalent bond between Cys195 and the nucleotide in the active site. As expected, the structures of R163K form 3 with dUMP and FdUMP in the active site are almost identical; superposition of subunit A from dUMP–R163K with subunit A of the FdUMP–R163K structure gives an r.m.s. distance of only 0.24 Å.
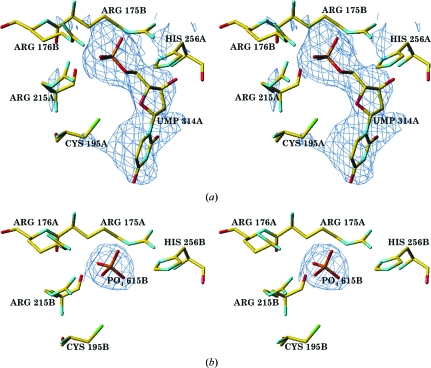
Figure 3.
(a) Stereoview of the F o − F c electron density (shown at the 2σ level) corresponding to dUMP in subunit A with full occupancy and (b) stereoview of the F o − F c electron density (shown at the 2σ level) corresponding to phosphate in subunit B.
3.4. Glutathione complex
R163K form 2 (PDB code 2rd8) displayed two environments for the catalytic cysteine Cys195: (i) unmodified cysteine, in which the thiol is closer to the O atom of the Ser216 backbone and farther from the guanidinium group of Arg215, and (ii) cysteine forming a disulfide bond with a molecule of BME, in which the thiol is closer to the guanidinium group of Arg215 and farther from the O atom of the Ser216 backbone (Gibson et al., 2008 ▶). This variability was correlated with the altered pK a of the thiol in these environments. BME, which is used as a reducing agent in vitro, is not present within cells. The most prevalent nonprotein thiol found in mammalian cells is reduced glutathione (GSH). This tripeptide, consisting of Glu-Cys-Gly, is not only the primary reducing agent and cofactor for many antioxidant enzymes, but also acts as an intermediary in various other physiological reactions, such as being an essential cysteine reservoir, and participates in thiol–disulfide exchange (Franco et al., 2007 ▶). To investigate the interaction of hTS with GSH, we soaked crystals of form 3 with oxidized glutathione (GS-SG), expecting to see modification of either one or both subunits. Somewhat unexpectedly, only one of five subunits contained covalently bound glutathione, while one contained reduced GSH. Apparently, there was a sufficient amount of GSH in the soaking solution, which was likely to have been generated from residual BME, to selectively bind within subunit A. Electron density for the active site of subunit A clearly indicates that glutathione is reduced. In contrast, a covalent bond forms between Cys195 and glutathione in subunit D (Fig. 4 ▶). The 12 October 2010 release of the PDB contains 71 structures that contain glutathione, nine of which are of an enzyme covalently modified by GSH. However, the hTS complex is the only structure that has one subunit with a covalently bound glutathione and another subunit in which a noncovalent adduct is present.
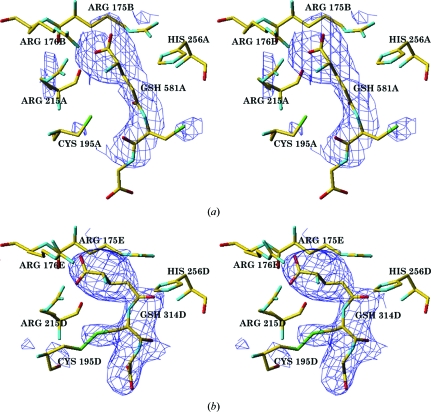
Figure 4.
(a) Stereoview of the F o − F c electron density (shown at the 2σ level) corresponding to nonbonded glutathione (GSH) in the hTS active site in subunit A. (b) Stereoview of the hTS active site in subunit D with a covalently modified Cys195D-linked glutathione; the difference map is contoured at the 2σ level.
The mosaicity of the glutathione-complex crystal was higher than the mosaicities of the crystals soaked in nucleotides. The B factors are also higher and significantly different. Dimer 1 has average B factors of 65.4 Å2 for subunit A and 77.3 Å2 for subunit B. Dimer 2 has average B factors of 92.1 Å2 for subunit D and 92.0 Å2 for subunit E and the overall density for these subunits was not good. It is not unlikely that some other cysteine residues (there are five per subunit) underwent partial derivatization, therefore inducing crystal damage.
Since both reduced and oxidized forms of glutathione are present in the cytosol, it appears that the TS active-site architecture should be such as to prevent entrance of GS-SG to protect its catalytic thiol from oxidation while promoting access of GSH to keep Cys195 reduced. This may be an explanation of our observation that despite a large excess of GS-SG over GSH a complex with GSH was observed.
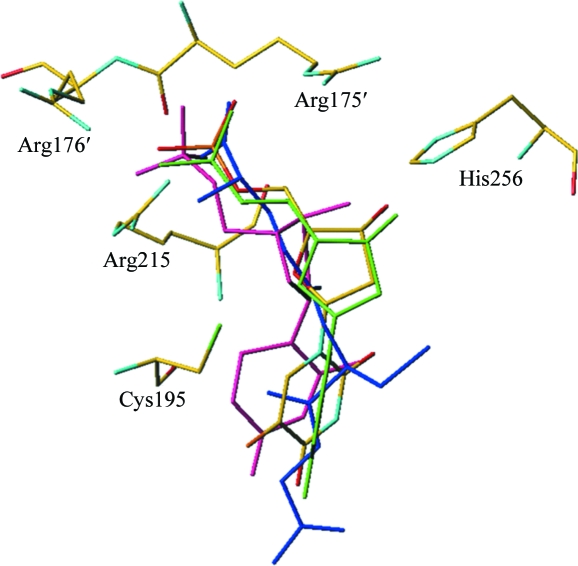
The superposition of R163K form 3 complexes shows that glutathione occupies the same space within the active site as the hTS complexes with substrates and their analogs (Fig. 5 ▶).
Figure 5.
The active site of hTS with superposition of ligands. Comparison of the R163K form 3 complexes with the cocrystallized hTS–dUMP–raltitrexed structure (PDB code 1hvy; Phan, Koli et al., 2001 ▶) shows the soaked ligands occupying the same space within the active site as hTS cocrystallized with ligands. The FdUMP binary complex is shown in atom-type colors, dUMP from the ternary inhibitory complex with raltitrexed (PDB code 1hvy) in pink, dUMP in the binary complex in green and the glutathione noncovalent adduct in blue.
3.5. Communication between the active sites of the hTS dimer
Studies of asymmetric ligand binding in crystals always involve some serendipity. To observe asymmetric ligand binding there must be some correlation between crystal environment and asymmetry, otherwise disorder occurs. The reverse can also happen: a dimer with no subunit cooperativity can crystallize in such a fashion that the crystal environment stabilizes the positions of active-site loops such that ligand binding takes place preferentially in one subunit but not in the other. There is ample evidence for negative cooperativity in TS dimers in solution (Dev et al., 1994 ▶), but the structural information is limited (Anderson et al., 1999 ▶). Clearly, for hTS we observe differences in ligand occupancies between subunits. The partial occupancy that is observed, rather than zero occupancy, may arise from a combination of factors. Firstly, the concentrations of the ligands in the artificial mother liquors were fairly high and this may have overcome the negative cooperativity. Secondly, the correlation between crystal environment and dimer asymmetry may not be sufficient to induce ligand ordering in the lattice. Thirdly, active-site accessibility via diffusion may be an important factor in crystal-soaking experiments.
The observed partial ordering of ligands is not reflected by asymmetry of the dimers. One reason for this is that the residues involved in generating negative cooperativity, and thus asymmetry, are averages of protein structures reflecting ligand occupancies. Also, since the nucleotide-binding constants for the dimer subunits only differ by a factor of six or so, the structural differences between the subunits must be subtle and apparently could not be resolved with the limited data resolution. The differences in nucleotide occupancies between FdUMP and dUMP complexes (Figs. 1 ▶ and 3 ▶) suggest that the observed asymmetric binding is not a result of the diffusion process but rather relates to binding affinities.
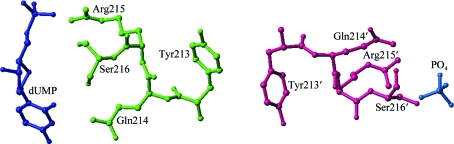
One model that can be proposed for subunit communication involves residues Gln214-Arg215-Ser216, which all interact with the nucleotide. They pull Tyr213, which interacts with Tyr213′ from the other subunit, and affect Gln214′-Arg215′-Ser216′, thus altering the affinity for the nucleotide in the other subunit (Fig. 6 ▶). In addition to its simplicity, the main argument for this model is its analogy to the negative cooperativity mechanism proposed for folate binding in PcTS, in which these residues are conserved. In the PcTS asymmetric dimer, which is composed of a ternary complex in one subunit and a binary complex in the other, the positions of these residues differ by about 0.3 Å. Consequently, the residues have been proposed to be involved in subunit communication (Anderson et al., 1999 ▶). In the structures reported here differences in protein geometry are not observed, but partial occupancies and much smaller structural differences induced by nucleotide binding versus folate binding, as judged by the ratios of K b values, are likely to be responsible for such an outcome. The structures of forms 1 and 2 of R163K (PDB codes 2rda and 2rd8) show asymmetry within the dimer that is associated with phosphate-ion binding; the crystals of form 3 reflect the ability of R163K to asymmetrically bind ligands.
Figure 6.
Potential model for communication between the active sites of the hTS dimer. Subunit A is shown in green, subunit B is shown in pink and dUMP and phosphate are shown in blue. Residues Gln214, Arg215 and Ser216, which all interact with the nucleotide, pull Tyr213, which interacts with its symmetry equivalent Tyr213′ and affects Gln214′, Arg215′ and Ser216′, therefore altering the affinity for the nucleotide in the other subunit.
Supplementary Material
PDB reference: human thymidylate synthase R163K, complex with FdUMP, 3h9k
PDB reference: complex with dUMP, 3hb8
PDB reference: complex with glutathione, 3ob7
Acknowledgments
This work was supported by NIH Grant CA 76560. Data were collected on the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID and 22-BM beamlines at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at http://www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38.
References
- Almog, R., Waddling, C. A., Maley, F., Maley, G. F. & Van Roey, P. (2001). Protein Sci. 10, 988–996. [DOI] [PMC free article] [PubMed]
- Anderson, A. C., O’Neil, R. H., DeLano, W. L. & Stroud, R. M. (1999). Biochemistry, 38, 13829–13836. [DOI] [PubMed]
- Birdsall, D. L., Finer-Moore, J. & Stroud, R. M. (1996). J. Mol. Biol. 255, 522–535. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Carreras, C. W. & Santi, D. V. (1995). Annu. Rev. Biochem. 64, 721–762. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Danenberg, K. D. & Danenberg, P. V. (1979). J. Biol. Chem. 254, 4345–4348. [PubMed]
- Dev, I. K., Dallas, W. S., Ferone, R, Hanlon, M., McKee, D. D. & Yates, B. B. (1994). J. Biol. Chem. 269, 1873–1882. [PubMed]
- Dev, I. K., Yates, B. B., Leong, J. & Dallas, W. S. (1988). Proc. Natl Acad. Sci. USA, 85, 1472–1476. [DOI] [PMC free article] [PubMed]
- Franco, R., Schoneveld, O. J., Pappa, A. & Panayiotidis, M. I. (2007). Arch. Physiol. Biochem. 113, 234–258. [DOI] [PubMed]
- Gibson, L. M., Lovelace, L. L. & Lebioda, L. (2008). Biochemistry, 47, 4636–4643. [DOI] [PMC free article] [PubMed]
- Houghton, P. J. (1999). Antifolate Drugs in Cancer Chemotherapy, edited by A. L. Jackman, pp. 423–436. Totowa: Humana Press.
- Huang, X., Gibson, L. M., Bell, B. J., Lovelace, L. L., Peña, M. M. O., Berger, F. G., Berger, S. H. & Lebioda, L. (2010). Biochemistry, 49, 2475–2482. [DOI] [PMC free article] [PubMed]
- Humphreys, G. K. & Greenberg, D. M. (1958). Arch. Biochem. Biophys. 78, 275–287. [DOI] [PubMed]
- Kabsch, W. (1976). Acta Cryst. A32, 922–923.
- Kleywegt, G. J. & Jones, T. A. (1998). Acta Cryst. D54, 1119–1131. [DOI] [PubMed]
- Kraulis, P. J. (1991). J. Appl. Cryst. 24, 946–950.
- Lovelace, L. L., Gibson, L. M. & Lebioda, L. (2007). Biochemistry, 46, 2823–2830. [DOI] [PMC free article] [PubMed]
- Matthews, D. A., Villafrance, J. E., Janson, C. A., Smith, W. W., Welsh, K. & Freer, S. (1990). J. Mol. Biol. 214, 937–948. [DOI] [PubMed]
- Merritt, E. A. & Bacon, D. J. (1997). Methods Enzymol. 277, 505–524. [DOI] [PubMed]
- Montfort, W. R. & Weichsel, A. (1997). Pharmacol. Ther. 76, 29–43. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Phan, J., Koli, S., Minor, W., Dunlap, R. B., Berger, S. H. & Lebioda, L. (2001). Biochemistry, 40, 1897–1902. [DOI] [PubMed]
- Phan, J., Steadman, D. J., Koli, S., Ding, W. C., Minor, W., Dunlap, R. B., Berger, S. H. & Lebioda, L. (2001). J. Biol. Chem. 276, 14170–14177. [DOI] [PubMed]
- Roussel, A. & Cambillau, C. (1991). Silicon Graphics Geometry Partners Directory, p. 86. Mountain View: Silicon Graphics.
- Schiffer, C. A., Clifton, I. J., Davisson, V. J., Santi, D. V. & Stroud, R. M. (1995). Biochemistry, 34, 16279–16287. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Steadman, D. J., Zhao, P.-S., Spencer, H. T., Dunlap, R. B. & Berger, S. H. (1998). Biochemistry, 37, 7089–7095. [DOI] [PubMed]
- Stroud, R. M. & Finer-Moore, J. S. (2003). Biochemistry, 42, 239–247. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human thymidylate synthase R163K, complex with FdUMP, 3h9k
PDB reference: complex with dUMP, 3hb8
PDB reference: complex with glutathione, 3ob7