Abstract
Renal cell carcinoma (RCC) is not a single entity, but comprises a group of tumors including clear cell RCC, papillary RCC and chromophobe RCC, which arise from the epithelium of renal tubules. The majority of clear cell RCCs, the major histological subtype, have genetic or epigenetic inactivation of the von Hippel-Lindau (VHL) gene. Germline mutations in the MET and fumarate hydratase (FH) genes lead to the development of type 1 and type 2 papillary RCCs, respectively, and such mutations of either the TSC1 or TSC2 gene increase the risk of RCC. Genome-wide copy number alteration analysis has suggested that loss of chromosome 3p and gain of chromosomes 5q and 7 may be copy number aberrations indispensable for the development of clear cell RCC. When chromosome 1p, 4, 9, 13q or 14q is also lost, more clinicopathologically aggressive clear cell RCC may develop. Since renal carcinogenesis is associated with neither chronic inflammation nor persistent viral infection, and hardly any histological change is evident in corresponding non-tumorous renal tissue from patients with renal tumors, precancerous conditions in the kidney have been rarely described. However, regional DNA hypermethylation on C-type CpG islands has already accumulated in such non-cancerous renal tissues, suggesting that, from the viewpoint of altered DNA methylation, the presence of precancerous conditions can be recognized even in the kidney. Genome-wide DNA methylation profiles in precancerous conditions are basically inherited by the corresponding clear cell RCCs developing in individual patients: DNA methylation alterations at the precancerous stage may further predispose renal tissue to epigenetic and genetic alterations, generate more malignant cancers, and even determine patient outcome. The list of tumor-related genes silenced by DNA hypermethylation has recently been increasing. Genetic and epigenetic profiling provides an optimal means of prognostication for patients with RCCs. Recently developed high-throughput technologies for genetic and epigenetic analyses will further accelerate the identification of key molecules for use in the prevention, diagnosis and therapy of RCCs.
Keywords: Renal cell carcinoma, copy number alteration, DNA methylation, precancerous condition, prognostication
Introduction: etiology and pathology
Worldwide about 271,000 cases of kidney cancer have been diagnosed and 116,000 persons have died because of kidney cancer [1]. In the United States, 57,000 cases of kidney cancer have been diagnosed and 14,000 persons have died. The majority of kidney cancers (80-85%) are renal cell carcinomas (RCCs) originating from the renal parenchyma. The remaining 15-20% are mainly urothelial carcinomas of the renal pelvis. Kidney cancer accounts for 2% of all adult malignancies, with a male to female ratio of 3:2 among affected patients [1]. The incidence of RCC peaks in the sixth decade of life, 80% of cases affecting the 40- to 69-year-old age group [2]. The incidence of RCC has been rising steadily each year in Europe and the United States over the last three decades. It is generally highest in Western and Eastern European countries and Scandinavia, as well as in Italy, North America, Australia and New Zealand. The lowest rates are reported in Asia and Africa. This regional variation in the incidence of RCC (more than ten-fold) suggests the strong role of environmental risk factors [3]. However, it is difficult to ascribe a definite and direct cause for this cancer. Smoking and chemical carcinogens such as asbestos and organic solvents are related to renal tumorigenesis. Obesity and hypertension and/or use of antihypertensive medication have been consistently reported to be positively associated with RCC risk [2].
RCC is not a single entity, but comprises a group of tumors that arise from the epithelium of renal tubules [4]. Clear cell RCC is the most common histological subtype (Figure 1A). Typically, the cells have cytoplasm filled with lipids and glycogen, are surrounded by a distinct cell membrane and contain round and uniform nuclei, and show an alveolar, acinar, cystic and solid architecture (Figure 1B). First, based simply on cytologic and histologic criteria, papillary RCCs (Figure 1C) can be divided into two morphologic groups, type 1 and type 2: type 1 papillary RCCs consist of papillae covered with a single or double layer of small cuboid cells with scanty cytoplasm (Figure 1D), and type 2 papillary RCCs consist of papillae covered by large eosinophilic cells arranged in an irregular or pseudo-stratified manner (Figure 1E) [5]. Chromophobe RCC consists of tumor cells with abundant eosinophilic cytoplasm (pale cells and eosinophilic cells with a perinuclear halo) and show mainly a solid structure (Figure 1F to 1H) [5]. Clear cell RCC and papillary RCC are derived from the proximal convoluted tubule, whereas the origin of chromophobe RCC is the distal tubule/collecting tubule. Certain inherited disorders such as von Hippel-Lindau (VHL) disease, hereditary papillary RCC and Birt-Hogg-Dube (BHD) syndrome enhance the risk of acquiring clear cell RCC, papillary RCC and chromophobe RCC, respectively [6].
Figure 1.
Macroscopic (A, C, F and G) and microscopic (B, D, E and H) views of a clear cell RCC (A and B), papillary RCCs (C, D and E) and a chromophobe RCC (F, G and H). A. Clear cell RCCs commonly protrude from the renal cortex as a rounded mass. Their cut surfaces are typically golden yellow, and necrosis and hemorrhage are commonly present. B. Clear cell RCCs typically have cytoplasm filled with lipids and glycogen and show an alveolar architecture. C. Papillary RCCs frequently contain areas of hemorrhage, necrosis and cystic degeneration. D. Type 1 papillary RCCs consist of papillae covered with a single or double layer of small cuboid cells with scanty cytoplasm. E. Type 2 papillary RCCs consist of papillae covered by large eosinophilic cells arranged in an irregular or pseudo-stratified manner. F. Chromophobe RCCs are solid circumscribed tumors with slightly lobulated surfaces. In unfixed specimens, the cut surface is homogeneously light brown or tan. G. Macroscopic view of the same chromophobe RCC after formalin fixation. The cut surface of chromophobe RCCs turns graysh-beige. H. Chromophobe RCCs consist of tumor cells with abundant eosinophilic cytoplasm (pale cells [Pa] and eosinophilic cells with a perinuclear halo [Eo]) and show mainly a solid structure.
Genetic alterations in RCCs
Tumor-related genes and their role in renal carcinogenesis
The World Health Organization (WHO) classification has introduced genetic alterations as a hallmark of certain histological subtypes of RCC, e.g. clear cell RCC is characterized by loss of chromosome 3p and inactivation of the VHL gene at 3p25.3 due to mutation or DNA methylation around the promoter region [7], although the classification of RCC is based largely on histology. The product of VHL is a 3-kDa protein with multiple functions, the best documented of which relates to its role as the substrate-recognition component of the E3-ubiquitin ligase complex. This complex is best known for its ability to target hypoxia-inducible factors (HIFs) for polyubiquitination and proteasomal degradation [8]. Under hypoxic conditions, HIF-1alpha and HIF-2alpha accumulate and form heterodimers with HIF-1beta and translocate to the nucleus where they induce transcription of downstream target genes including vascular endothelial growth factor (VEGF). The absence of wild-type VHL promotes inappropriate activation of downstream target genes and contributes to tumorigenesis [9]. Additionally, VHL protein has functions that are independent of HIF-1alpha and HIF-2alpha and are thought to be important for its tumor-suppressor action, assembly of the extracellular matrix, control of microtubule dynamics, regulation of apoptosis, and possibly stabilization of TP53 proteins [10].
Patients with gain-of-function germline mutations in the MET gene develop type 1 papillary RCC. MET encodes a transmembrane receptor tyrosine kinase whose ligand is hepatocyte growth factor (HGF). Activation of MET by HGF triggers tyrosine kinase activity, which facilitates several transduction cascades resulting in multiple cellular processes such as mitogenesis and migration. However, the incidence of MET mutations in sporadic papillary RCC is not high (about 10%) [11]. Patients with germline mutations in the fumarate hydratase (FH) gene develop type 2 papillary RCC [12]. VHL recognition of HIF requires hydroxylation by HIF prolyl hy-droxylase (HPH), and FH activates HPH. FH mutation promotes tumorigenesis via HIF protein accumulation due to HPH dysfunction. Unlike the gain-of-function mutation of the c-kit (KIT) gene, overexpression of KIT is frequent in chromophobe RCC [13]: KIT is a type III receptor tyrosine kinase that has a role in cell signal transduction. Normally KIT is phosphorylated upon binding to its ligand, stem cell factor. This leads to a phosphorylation cascade ultimately activating various transcription factors. Such activation regulates apoptosis, cell differentiation, proliferation, chemotaxis, and cell adhesion. Although germline mutations of the BHD gene, which encodes folliculin, have been detected in 80% of BHD kindreds, the incidence of the mutation in sporadic chromophobe RCC is very low. Tuberous sclerosis complex (TSC) has been linked to germline inactivating mutations of either of TSC1 (9q34) encoding hamartin or TSC2 (16p13.3) encoding tuberin, and affected patients have an increased risk of developing renal tumors including clear cell RCC, papillary RCC and chromophobe RCC [3]. The TSC1/TSC2 protein complex inhibits mammalian target of rapamycin (mTOR) protein and is involved in signaling pathways that regulate cell growth. Although the Eker rat model with a germline insersion in the Tsc2 gene develops dominantly inherited cancers [14], the role of TSC1 and TSC2 in human sporadic RCC is unclear.
Other known cancer genes that are frequently mutated in adult epithelial cancers, for example RAS, v-raf murine sarcoma viral oncogene homolog B1 (BRAF), TP53, retinoblastoma (RB), cyclin-dependent kinase inhibitor 2A (CDKN2A), phosphoinositide-3-kinase, catalytic alpha polypeptide (PIK3CA), phosphatase and tensin homolog (PTEN), epidermal growth factor receptor (EGFR) and v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 (ERBB2), make only a small contribution to clear cell RCC [15]. Recently somatic truncating mutations in the neurofibromin 2 (NF2) gene, encoding marlin protein that is similar to the ERM (ezrin, radixin, moesin) family members that link cytoskeletal components and the cell membrane, have been reported in clear cell RCCs. Since none of the samples of clear cell RCC with the NF2 mutation harbored a VHL mutation, it has been suggested that somatic NF2 mutations may account for a proportion of cases in this subset [15].
Genetic clustering of clear cell RCCs
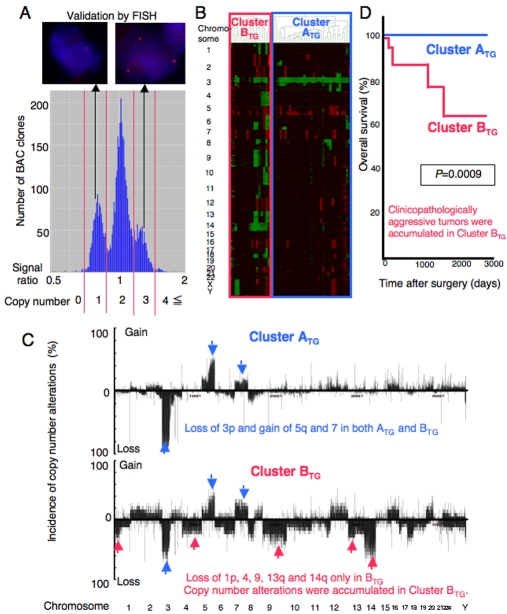
Since the genetic backgrounds of RCCs have not been fully understood to date, we have analyzed copy number alterations by array-comparative genomic hybridization (CGH) using a custom-made bacterial artificial chromosome (BAC) array (MCG Whole Genome Array-4500) harboring 4361 BAC clones throughout chromosomes 1 to 22 and X and Y clones [16] in clinical tissue samples (Figure 2AA), and clarified the genetic clustering of clear cell RCCs [17]. RCC is usually enclosed within a fibrous capsule and well demarcated, and hardly ever contains fibrous stroma between the cancer cells. Therefore, we were able to obtain cancer cells of high purity from surgical specimens, avoiding contamination with both non-cancerous epithelial cells and stromal cells. By unsupervised hierarchical clustering analysis of RCCs based on array-CGH data, clear cell RCCs were clustered into the two subclasses, Clusters ATG and BTG (Figure 2B). In clear cell RCCs, the average number of BAC clones on which loss or gain was detected was significantly higher in Cluster BTG than in Cluster ATG. In both clusters, loss or gain of an entire chromosome or an entire chromosome arm was frequent. Loss of chromosome 3p and gain of chromosomes 5q and 7 were frequent in both Clusters ATG and BTG. On the other hand, loss of chromosome 1p, 4, 9, 13q or 14q was frequent only in Cluster BTG, but not in Cluster ATG (Figure 2C). Gain on 1q31-ter, 3q and 8q was frequent only in Cluster BTG, whereas loss at the same loci was observed in Cluster ATG, although the frequency was rather low. The present genome-wide analysis indicated that loss of chromosome 3p and gain of 5q and 7 may be copy number aberrations that are indispensable for the development of clear cell RCCs, regardless of genetic clustering [17]. Additional loss of chromosome 1p, 4, 9, 13q or 14q may promote the genetic pathway to Cluster BTG [17].
Figure 2.
Genetic clustering of clear cell renal cell carcinomas (RCCs). An example of a histogram of the signal ratios (test signal/reference signal) afforded by array-CGH in a clear cell RCC. The thresholds of the signal ratios for copy numbers of 0, 1, 2, 3 and 4 or more were determined from the troughs (red bars) between the distinct peaks. A. FISH analysis using the same clone validated the results of array-CGH (ref. 17). B. Unsupervised hierarchical clustering analysis based on array-CGH data. Clear cell RCCs were grouped into Clusters ATG and BTG (ref. 17). C. Distinct copy number profiles in Clusters ATG and BTG. LOSS of chromosome 3p and gain of 5q and 7 may promote the development of RCCs belonging to Cluster ATG and showing a favorable outcome. When loss of 1p, 4, 9, 13q or 14q is added, more malignant RCCs in Cluster BTG may develop (ref. 17). D. Kaplan-Meier survival curves based on genetic clustering of clear cell RCCs (Clusters ATG and BTG). None of the patients in Cluster ATG died as a result, and the overall survival rate of patients in Cluster BTG was significantly lower than that of patients in Cluster ATG (Log-rank test, ref. 17).
On the basis of microscopic examination of the entire tumor mass, the presence or absence of vascular involvement was evaluated in the examined clear cell RCCs. Macroscopic observation revealed the presence or absence of renal vein tumor thrombi. Clear cell RCCs in Cluster BTG showed significantly higher histological grades and more frequently showed vascular involvement, renal vein tumor thrombi and higher pathological tumor-node-metastasis (TNM) stages than those in Cluster ATG. Thus, accumulated genetic alterations may play a significant role in the more malignant potential of clear cell RCCs belongingto Cluster BTG.
Even if resection has been considered complete, some RCCs relapse and metastasize to distant organs and can lead to death in middle-aged adults belonging to the working population. Unless relapsed or metastasized tumors are diagnosed early by close follow-up, the effectiveness of any adjuvant therapy is very restricted. Therefore, to assist the close follow-up of patients who have undergone nephrectomy and are still at risk of recurrence and metastasis, prognostic indicators should be explored. Recurrence or metastasis was observed in 40% of patients who underwent curative resection in Cluster BTG, but in only 9% of patients who did so in Cluster ATG [17]. The recurrence-free survival rate of patients in Cluster BTG was significantly lower than that of patients in Cluster ATG. Twenty-four% of the patients in Cluster BTG died as a result, whereas none of the patients in Cluster ATG died [17]. The overall survival rate of patients in Cluster BTG was also significantly lower than that of patients in Cluster ATG (Figure 2D). Multivariate analysis revealed that genetic clustering was a predictor of recurrence-free survival, and was independent of histological grade and pathological TNM stage. In addition, a sufficient quantity of good-quality DNA was obtainable from each nephrectomy specimen. Therefore, use of a mini-array harboring a set of BAC clones that can effectively discriminate Cluster BTG after nephrectomy may be a promising method of prognostication.
Epigentic Alterations in RCCs
Epigenetics and cancers
In addition to genetic events, human cancer cells show drastic epigenetic alterations. DNA methylation, a covalent chemical modification resulting in addition of a methyl group at the carbon 5 position of the cytosine ring in CpG dinucleotides, is one of the most consistent epigenetic changes occurring in human cancers [18-20]. DNA methyltransferases (DNMTs) transfer methyl groups from S-adenosyl-methionine to cytosines. DNA methylation normally promotes a highly condensed heterochromatin structure associated with deacetylation of histones H3 and H4. In addition, methylation of histone H3 lysine 4 (H3K4), H3K36 and H3K79 is connected with transcriptional activation, whereas methylation of H3K9, H3K27 and H4K20 has been connected with transcriptional repression [21]. DNA methylation is a stable modification inherited throughout successive cell divisions, and is essential for X-chromosome inactivation, genome imprinting, silencing of transposons and other parasitic elements, and proper expression of genes [22]. In human cancer cells, DNA hypomethylation induces chromosomal instability through decon-densation of heterochromatin and enhancement of chromosomal recombination [23]. On the other hand, DNA hypermethylation of CpG islands around the promoter regions silences tumor-suppressor genes [24].
Analysis of tissue specimens has revealed that DNA methylation alterations participate in multistage carcinogenesis, even from the early and precancerous stages, especially in association with chronic inflammation and/or persistent infection with viruses or other pathogenic microorganisms, such as hepatitis B or C viruses, Epstein-Barr virus, human papillomavirus and Helicobacter pylori [25-27]. For example, we have observed frequent regional DNA hypermethylation and/or DNMT1 overexpression in non-cancerous liver tissues showing chronic hepatitis or liver cirrhosis with hepatitis virus infection obtained from patients with hepatocel-lular carcinomas (HCCs) [28-32], and in non-cancerous pancreatic tissues showing chronic pancreatitis obtained from patients with pancreatic cancer [33,34]. Unlike cancers derived from such organs, renal tumors are not usually generated from a background of persistent viral infection and/or chronic inflammation. Although several factors such as smoking and obesity have been reported to be possible risk factors for renal tumors as mentioned above, pathologists hardly ever observe any histological change corresponding to such risk factors in non-tumorous renal tissue. Therefore, precancerous conditions in the kidney have been rarely described. Therefore we attempted to clarify the role of DNA methylation alterations during renal carcinogenesis.
Regional DNA hypermethylation in precancerous conditions and RCCs
We focused on C-type CpG islands of the CDKN2A, human MutL homologue 1 (hMLH1) and thrombospondin-1 (THBS-1) genes and the methylated in tumor (MINT)-1, -2, -12, -25 and -31 clones and CpG island of the VHL gene. C-type CpG islands are known to be methylated in a cancer-specific, but not age-related, manner. The cancer phenotype associated with accumulation of DNA methylation on C-type CpG islands is defined as the CpG-island methylator phenotype (CIMP), and such accumulation is generally associated with frequent silencing of tumor-related genes due to DNA hypermethylation only, or a two-hit mechanism involving DNA hypermethylation and loss of heterozygosity in human cancers of various organs [35]. Bisulfite conversion has been carried out using genomic DNA, and this process converts unmethylated cytosine residues to uracil, whereas methylated cytosine residues remain unchanged [36]. The DNA methylation status on CpG islands was determined by methylation-specific PCR (MSP) or combined bisulfite restriction enzyme analysis (COBRA). MSP is based on the principle that the DNA sequences of methylated and unmethylated genomic regions differ after bisulfite conversion and can thus be distinguished by sequence-specific PCR primers. In COBRA, bisulfite-modified DNA was amplified by PCR using primers designed to amplify methylated and unmethylated genomic regions equally. The amplified fragments were digested with restriction enzymes that cleave DNA only if the CpG sites in their recognition sequences are methylated.
Even in non-tumorous renal tissues showing no remarkable histological changes obtained from patients with renal tumors, the average number of methylated CpG islands was significantly higher than in normal renal tissues obtained from patients without any primary renal tumor, regardless of patient age [37]. Stepwise accumulation of DNA methylation on CpG islands from normal renal tissues, to non-tumorous renal tissues showing no remarkable histological changes obtained from patients with renal tumors, and to renal tumors has been clearly shown. Although precancerous conditions in the kidney have been rarely described, as mentioned above, from the viewpoint of altered DNA methylation, we have shown that it is possible to recognize the presence of precancerous conditions even in the kidney [37]. In other words, regional DNA hypermethylation may participate in the early and precancerous stage of multistage renal tumorigenesis.
In renal tumors, the CDKN2A and THBS-1 genes seem to be hot spots of regional DNA hypermethylation during multistage renal tumorigenesis. The incidence of DNA methylation on the MINT 2 clone was low in renal cancers, even though this clone is one of the hot spots of regional DNA hypermethylation in HCCs. The incidence of DNA methylation on the MINT 25 clone was, if anything, high even in normal renal tissues, although it was never observed in normal liver tissues, indicating that MINT 25 may be normally methylated in a renal tissue-specific manner. Thus the DNA methylation profiles of both normal and tumorous tissues tended to be organ-specific.
In clear cell RCCs, correlations between the average number of methylated CpG islands and tumor clinicopathological parameters were evaluated. Clear cell RCCs were classified into three groups on the basis of macroscopic configuration: single nodular type [type 1], single nodular with extranodular growth type [type 2], and contiguous multinodular type [type 3] RCCs [37]. These criteria for macroscopic configuration follow those that have already been established for HCCs: type 2 or 3 HCCs show poorer histological differentiation and a higher incidence of portal vein involvement and intra-hepatic metastasis than type 1 HCCs. Patients with types 2 and 3 HCCs show poorer prognosis than those with type 1 [38]. With respect to clear cell RCCs, accumulation of DNA methylation on CpG islands was significantly correlated with a type 2 or 3 macroscopic configuration, higher histological grade, an infiltrating growth pattern and vascular involvement [37], suggesting that regional DNA hypermethylation is continuously involved in multistage renal tumorigenesis from precancerous conditions to malignant progression. The recurrence-free and overall survival rates of patients with RCCs showing accumulated DNA methylation on 3 or more CpG islands was significantly lower than that of patients with RCCs not showing this feature, indicating that regional DNA hypermethylation may be a biological predictor of patient prognosis. In addition to the above-mentioned genetic clustering, analysis of DNA methylation status in nephrectomy specimens may become a useful tool for prognostication of individual clinical cases.
Surprisingly, the average number of methylated CpG islands in non-tumorous renal tissues obtained from patients with histological grade 3 clear cell RCCs was significantly higher than that in equivalent tissue obtained from patients with histological grade 1 or 2 RCCs [25,37]. These data suggest that precancerous conditions showing regional DNA hypermethylation may generate more malignant RCCs.
Genome-wide DNA methylation profiling in precancerous conditions and RCCs
In order to further clarify the significance of DNA methylation alterations during renal carcinogenesis, we performed genome-wide DNA methylation analysis using BAC array-based methylated CpG island amplification (BAMCA) [39-41] in tissue samples. The promoter regions of specific genes are not the only target of DNA methylation alterations in human cancers. DNA methylation status in genomic regions that do not directly participate in gene silencing, such as the edges of CpG islands, may be altered at precancerous stages before the alterations of the promoter regions themselves occur. Genomic regions in which DNA hypomethylation affects chromosomal instability may not be contained in promoter arrays or CpG island arrays. Moreover, aberrant DNA methylation of large regions of chromosomes, which are regulated in a coordinated manner due to a process of long-range epigenetic silencing, has recently attracted attention in human cancers [42]. Therefore, we again used a custom-made BAC array MCG Whole Genome Array-4500, which may be suitable, not for focusing on specific promoter regions or individual CpG sites, but for overviewing the DNA methylation tendency of individual large regions among all chromosomes [43]. Briefly, test or reference DNA was first digested with the methylation-sensitive restriction enzyme Sma I and subsequently with the methylation-insensitive Xma I. Adapters were ligated to the Xma I-digested sticky ends, and PCR was performed with an adapter primer set. Test and reference PCR products were labeled by random priming with Cy3- and Cy5-dCTP, respectively and applied to the custom-made BAC array. We validated the ability for detecting any tendency for coordinated regulation of DNA methylation at multiple CpG sites in individual large regions of chromosomes of BAMCA by quantitative evaluation of DNA methylation status at each Sma I site on representative BAC clones by py-rosequencing [44].
Non-tumorous renal tissue obtained from patients with papillary RCCs, chromophobe RCCs and oncocytomas did not show any histological changes when compared with both non-tumorous renal tissue obtained from patients with clear cell RCCs and normal renal tissue obtained from patients without any primary renal tumor. However, the average numbers of BAC clones showing DNA hypo- or hypermethylation in non-tumorous renal tissue obtained from patients with chromophobe RCCs and oncocytomas were significantly smaller than the average number in non-tumorous renal tissue obtained from patients with clear cell RCCs [45]. In non-tumorous renal tissue from all examined patients with renal tumors (clear cell RCCs, papillary RCCs, chromophobe RCCs and oncocytomas), biphasic accumulation of DNA methylation alterations was evident. Among such patients, the recurrence-free survival rate of patients showing DNA hypo- or hypermethylation on more BAC clones in their non-tumorous renal tissue was significantly lower than that of patients showing DNA hypo- or hypermethylation on fewer BAC clones [45]. Significant DNA methylation profiles determining the histological subtype (chromophobe RCCs and oncocytomas vs clear cell RCCs) of future developing renal tumors and/or patient outcome (favorable outcome vs poorer outcome) may be already established at the precancerous stage.
In samples of non-cancerous renal tissue from patients with clear cell RCCs, many BAC clones already showed DNA hypomethylation or DNA hypermethylation relative to normal renal tissues. In clear cell RCCs themselves, more BAC clones showed DNA hypomethylation or DNA hypermethylation, the degree of which was increased in comparison with non-cancerous renal tissue samples obtained from patients with clear cell RCCs [46]. In samples of non-cancerous renal tissue from patients with clear cell RCCs, which were already at the precancerous stage with accumulation of DNA methylation on C-type CpG islands in spite of an absence of marked histological changes as mentioned above, genome-wide DNA methylation alterations (both hypo- and hypermethylation) were also confirmed by BAMCA.
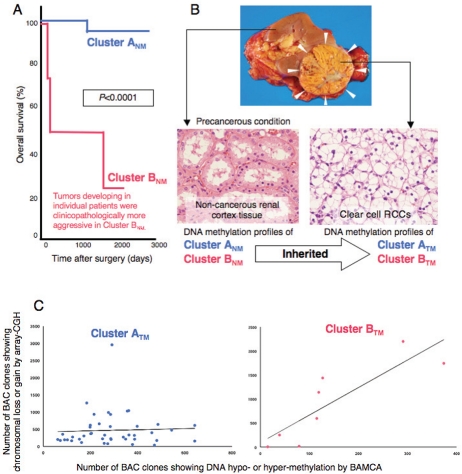
We then performed two-dimensional unsupervised hierarchical clustering analysis based on the genome-wide DNA methylation status (signal ratios by BAMCA) of the non-cancerous renal tissue samples. On the basis of the DNA methylation profiles of their non-cancerous renal tissue samples, the patients with clear cell RCCs were clustered into two subclasses, Clusters ANM and BNM. The corresponding clear cell RCCs of patients in Cluster BNM showed more frequent macroscopically evident multinodular (type 3) growth, vascular involvement and renal vein tumor thrombi, and higher pathological TNM stages than those in Cluster ANM [46]. Our Clusters ANM and BNM in precancerous tissue can be considered clinicopathologically valid, as 60% of the patients in Cluster BNM died of recurrent RCC, compared with only 2% of the patients in Cluster ANM [46]. The overall survival rate of patients in Cluster BNM was significantly lower than that of patients in Cluster ANM (Figure 3A). DNA methylation alterations at the precancerous stage may even determine the outcome of patients with clear cell RCCs.
Figure 3.
DNA methylation profiles in precancerous conditions and clear cell renal cell carcinomas (RCCs). A. Genome-wide DNA methylation profiles in the non-cancerous renal tissue were significantly correlated with clinicopathological parameters of clear cell RCCs developing in individual patients, and also outcome, indicating that DNA methylation alterations at the precancerous stage may generate more malignant cancers and even determine outcome (ref. 46). B. DNA methylation profiles in the non-cancerous renal tissue (Clusters ANM and BNM,see text) were basically inherited by the corresponding clear cell RCCs developing in individual patients as the DNA methylation profiles of Clusters ATM and BTM, respectively (ref. 46). C. In Cluster BTM, the number of clones showing copy number alterations by array-CGH was significantly correlated with that of DNA hypo- and hypermethylation by BAMCA in the same patient, whereas no such significant correlations were observed in Cluster ATM, suggesting that particular DNA methylation profiles may be closely related to chromosomal instability (unpublished data).
Two-dimensional unsupervised hierarchical clustering analysis based on BAMCA data (signal ratios) for clear cell RCCs was able to group patients into two subclasses, Clusters ATM and BTM. Clear cell RCCs in Cluster BTM showed more frequent vascular involvement and renal vein tumor thrombi, and also higher pathological TNM stages than those in Cluster ATM [46]; 37.5% of the patients in Cluster BTM died due to RCC recurrence, compared with only 2.3% of the patients in Cluster ATM [46]. The overall survival rate of patients in Cluster BTM was significantly lower than that of patients in Cluster ATM. Multivariate analysis revealed that our clustering was a predictor of recurrence and was independent of histological grade, macroscopic configuration, vascular involvement or presence of renal vein tumor thrombi. Patients belonging to Cluster BTM were completely discriminated from patients belonging to Cluster ATM based on the DNA methylation status of 14 BAC clones. In other words, the DNA methylation status of the 14 BAC clones was able to determine whether or not patients belonged to Cluster BTM, a significant prognostic indicator, with a sensitivity and specificity of 100% using the appropriate cutoff value of signal ratios [46]. The use of DNA methylation status on such BAC clones as an indicator may be a promising approach for prognostication of clear cell RCCs.
Significance of DNA methylation alterations at the precancerous stage
When we compared the DNA methylation profiles of non-cancerous renal tissue and those of the corresponding clear cell RCC, Cluster BNM was completely included in Cluster BTM. Wil-coxon test revealed that the signal ratios of 1143 BAC clones in non-cancerous renal tissue differed significantly between Clusters ANM and BNM and that the signal ratios of 1111 BAC clones in clear cell RCCs differed significantly between Clusters ATM and BTM. Among the 1143 BAC clones significantly discriminating Cluster BNM from Cluster ANM, 724, i.e. the majority, also discriminated Cluster BTM from Cluster ATM. In 311 of these 724 BAC clones, in which the average signal ratio of Cluster BNM was higher than that of Cluster ANM, the average signal ratio of Cluster BTM was also higher than that of Cluster ATM without exception. In 413 of the 724 BAC clones showing a lower average signal ratio of Cluster BNM than that of Cluster ANM, the average signal ratio of Cluster BTM was also lower than that of Cluster ATM without exception [46]. When we examined each of the representative BAC clones characterizing both Clusters BNM and BTM, the BAMCA signal ratio in the non-cancerous renal tissue was at almost the same level as that in the corresponding clear cell RCC developing in each individual patient. Accordingly, we concluded that the genome-wide DNA methylation profiles of non-cancerous renal tissue are basically inherited by each corresponding clear cell RCC (Figure 3B).
As mentioned above, we examined DNA methylation status on CpG islands for the CDKN2A, hMLH 1, VHL and THBS 1 genes, and the methylated in tumor (MINT)-1, -2, -12, -25 and -31 clones were examined in the same clear cell RCCs. The average number of methylated CpG islands was significantly higher in Cluster BTM (2.75±1.67) than in Cluster ATM. The frequency of CIMP in Cluster BTM (62.5%) was significantly higher than that in Cluster ATM (16%). Genome-wide DNA methylation alterations consisting of both hypo- and hypermethylation of DNA revealed by BAMCA in Cluster BTM are associated with regional DNA hypermethylation on CpG islands [37,46]. Moreover, a subclass of Cluster BNM and BTM based on BAMCA data is completely included in Cluster BTG showing accumulations of copy number alterations [17,46]. Therefore, epigenetic and genetic alterations are not mutually exclusive during renal carcinogenesis. Particular DNA methylation profiles at the precancerous stage may be closely related to, or may be prone to chromosomal instability (Figure 3C). DNA methylation alterations in precancerous conditions, which may not occur randomly but are prone to further accumulation of epigenetic and genetic alterations, can generate more malignant cancers and even determine the outcome for individual patients.
Tumor-related genes silenced by DNA hypermethylation in RCCs
Somatic VHL mutations occur in 50-80% of sporadic clear cell RCCs [47]. Alternative mechanisms of VHL inactivation have therefore been explored, and Herman et al. have demonstrated DNA hypermethylation of the VHL gene in 19% of examined tumors [48]. In a renal cancer cell line, treatment with a DNA demethylating agent, 5-aza-2'-deoxycytidine, resulted in re-expression of the VHL gene. Thus, other than the RB gene, the VHL gene became the second known example of a tumor-suppressor gene silenced by DNA methylation. The list of tumor-related genes silenced by DNA hypermethylation during renal carcinogenesis has recently been increasing. DNA methylation profiling in both VHL-related and VHL-unrelated RCCs has shown that the average number of methylated genes revealed by high-throughput Goldengate analysis in sporadic RCCs of patients with wild-type VHL is higher than in RCCs of patients with mutant VHL [49]. The Ras association domain family member 1 (RASSF1), twist homolog 1 (TWIST1), paired-like homeodomain 2 (PITX2), cadherin 13 (CDH13), heparan sulfate (glucosamine) 3-O -sulfotransferase 2 (HS3ST2), T-cell acute lym-phocytic leukemia 1 (TAL1), Wilms' tumor 1 (WT1), matrix metallopeptidase 2 (MMP2), deleted in colorectal carcinoma (DCC), islet cell autoantigen 1 (ICA1) and tumor suppressor candidate 3 (TUSC3) genes are more frequently methylated in sporadic RCCs of patients with wild-type VHL than in RCCs of patients with mutant VHL, whereas only gamma-aminobutyric acid A receptor, beta 3 (GABRB3) is methylated more frequently in VHL-related RCCs [49].
Frequent DNA methylation of proapoptotic TP53 target genes in stomach and colorectal cancers has recently attracted attention [50]. When examined in RCCs, the apoptotic peptidase activating factor 1 (APAF1) and death-associated protein kinase 1 (DAPK1) proapoptotic genes were frequently silenced due to DNA hyper-methylation, and such DNA hypermethylation had a prognostic impact in affected patients [51]. With respect to Wnt antagonist family genes in RCCs, DNA hypermethylation and/or repressive histone modification have been observed in the secreted frizzled-related protein 1 (SFRP1), SFRP2, SFRP5, WNT inhibitory factor 1 (WIF1) and dickkopf homolog 3 (DKK3) genes. Simultaneous detection of DNA methylation of such Wnt antagonist family genes may be a useful indicator for diagnosis of RCCs [52,53].
Microarray analysis of RCC cell lines treated with 5-aza-2'deoxycytidine has revealed upregulation of the ubiquitin carboxyl-terminal esterase L1 (UCHL1) gene [54]. The UCHL1 gene involved in the regulation of cellular ubiquitin levels plays important roles in different cellular processes. Significant growth inhibition in UCHL1 transfectants suggests that UCHL-1 functions as a potential tumor suppressor gene in RCCs [55]. Moreover, silencing of the UCHL-1 gene due to DNA hypermethylation is reportedly correlated with poor outcome in patients with RCCs [55]. Loss of transforming growth factor beta receptor III (TGFBR3) correlates with loss of TGF-beta responsiveness and dysregulated TGF-beta signaling in RCCs. However, reduced expression of the TGFBR3 gene was shown not to be due to DNA hypermethylation of the promoter region of the TGFBR3 gene itself, but to silencing of the transcriptional factor GATA binding protein 3 (GATA3) due to DNA hypermethylation resulting in reduced expression of TGFBR3 during RCC progression [56]. In addition, silencing due to DNA methylation of a number of genes may play a role in renal carcinogenesis; these include the p53-inducible gene 14-3-3 sigma [57], ABCG2 which is of importance in clinical drug resistance [58], a gap junction molecule connexin 32 [59], actin-binding protein DAL-1/4.1B [60], TIMP3 which participates in cancer invasion [61], the fragile histidine triad (FHIT) gene which encompasses the most common human fragile site FRA3B at 3p14.2 [62], cell adhesion molecule junction plakoglo-bin (JUP) [63], HGF activator inhibitor HAI-2 [64], a member of the homeobox gene family HOXB13 [65], tissue-specific proapoptotic BH3-only protein BCL2-interacting killer (BIK) [66], TU3A which was originally identified as a candidate tumor suppressor gene in RCCs [67] and XAF1 which antagonizes the anticaspase activity of X-linked inhibitor of apoptosis (XIAP) [68].
Recently, the methodology for analysis of DNA methylation on a genome-wide scale has been markedly improved [69], and the use of microar-rays to which bisulfite-converted genomic DNA is applied, has become popular, achieving a resolution as good as a single CpG [70,71]. New-generation sequencing technologies have been introduced for bisulfite-converted genomic DNA or genomic DNA enriched by affinity-based approaches using anti-methyl-cytosine antibody or methyl-binding domain proteins [72]. In addition, a high-throughput technique without bisulfite conversion has been developed based on single-molecule, real-time DNA sequencing [73]. These new technologies will be able to efficiently accelerate the identification of tumor-related genes whose expression is altered due to DNA hypo- or hyper-methylation and reveal the clinical relevance of translational epigenetics.
DNA hypomethylation in RCCs
Unlike the case of DNA hypermethylation, the number of reports addressing DNA hypomethylation of specific genes or elements has been restricted to date. Carbonic anhydrase IX (CA9) is a transmembrane glycoprotein and the only known tumor-associated carbonic anhydrase that may be involved in cell proliferation and transformation. DNA hypomethylation of the CA9 gene has been shown to participate in activation of the promoter activity in RCC cell lines and clinical tissue samples [74,75]. Transposons, proviral DNA and other parasitic elements in the mammalian genome make up the repetitive sequences in the intergenic and introgenic regions of DNA. In general, activation of parasitic elements, such as LINE-1 and HERV-K, can allow for their movement within the genome. However, activation of these parasitic elements due to DNA hypomethylation does not seem to be a major event during renal carcinogenesis [76].
Histone modifications in RCCs
Since techniques for analysis of histone modification in clinical tissue specimens have not been fully established to date, the overall picture of histone modification status in clinical samples of various cancers including RCCs is unclear. However, the results of immunohistochemistry to evaluate histone methylation levels have been reported. Levels of H3K4-monomethyl, -dimethyl and -trimethyl staining were each inversely correlated with the aggressiveness of RCCs. The combined staining score for H3K4 modifications (monomethylation to trimethylation) was shown to be an independent predictor of outcome in patients with RCCs [77].
With respect to cross-talk between genetic alterations and histone modifications, a recent robust analysis of 3544 protein genes in clear cell RCCs has revealed somatic truncating mutations in the SET domain containing 2 (SETD2) gene, which encodes a histone H3K36 methyl-transferase, and also in the lysine-specific de-methylase 5C (KDM5C/JARID1C) gene, which encodes a histone H3K4 demethylase [15]. No mutations were found in either SETD2 or KDM5C in the subset of non-clear cell RCCs. The majority of samples with truncating SETD2 and KDM5C mutations had VHL mutations. Significant (two-fold or less) differences in the expression levels of 298 genes were noted in clear cell RCCs showing the SETD2 mutation relative to those not showing it, whereas KDM5C-mutant RCCs showed significant differences in the expression levels of 18 genes relative to KDM5C-wild-type RCCs [15].
Perspective
Both genetic and epigenetic events appear to accumulate during renal carcinogenesis, reflecting the clinicopathological diversity of RCCs. Loss of chromosome 3p and gain of chromosomes 5q and 7 may be indispensable copy number aberrations for the development of clear cell RCCs. When loss of chromosome 1p, 4, 9, 13q or 14q is added, more malignant RCCs may develop. DNA methylation alterations play significant roles in multistage renal carcinogenesis even in early precancerous stages. Genome-wide DNA methylation profiles in precancerous conditions are basically inherited by the corresponding RCCs developing in individual patients: DNA methylation alterations at the precancerous stage may render cells prone to further epigenetic and genetic alterations, generate more malignant cancers, and even determine patient outcome. Previous attempts have been made to use genetic alterations of VHL and other tumor-related genes as diagnostic indicators in tissue and serum specimens [9,78]. On the other hand, DNA methylation alterations occur earlier than genetic alterations during carcinogenesis and are stably preserved on DNA double strands by covalent bonds, unlike the profiles of mRNA and protein expression, which can be easily affected by the micro-environment of cancer cells or their precursor cells. Genome-wide DNA methylation profiling may provide optimal indicators for early diagnosis of RCCs and prognostication of affected patients.
RCCs are thought to be immunogenic, and immunotherapy including the administration of interferon-alpha or interleukin (IL)-2 has been used as a standard treatment for RCCs for 20 years [79]. However, the success of immuno-therapy is limited because of immuno-escape mechanisms including down-regulation of major histocompatibility complex class I antigens and secretion of immunosuppressive cytokines such as IL10. In addition to traditional surgical approaches and immunotherapy, molecular targeted therapy has recently been introduced. Since the induction of VEGF by HIF activation downstream of VHL inactivation is the most important mechanism determining the hypervas-cularity of RCCs [79,80], VEGF receptor inhibitors such as sunitinib, sorafenib and axitinib, and the VEGF-ligand binding agent bevacizumab, have been introduced for VEGF-targeted therapy. mTOR is another target for treatment of RCCs, and an ester of rapamycin, tersirolimus, has been introduced clinically. However, the mechanisms responsible for refractoriness to molecular targeted therapy are unclear, and the optimal administration regimen for these agents has not been defined [81]. Therefore, recently introduced agents have not accomplished complete anti-tumor effects. Further investigation of the genetic and epigenetic events occurring during renal carcinogenesis is needed to identify more key molecules for use in prevention, diagnosis and therapy.
Acknowledgments
This study was supported by a Grant-in-Aid for the Third Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health, Labor and Welfare of Japan, a Grant-in-Aid for Cancer Research from the Ministry of Health, Labor and Welfare of Japan, and the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NiBio). The authors have no other relevant affiliations or financial involvement with any organization or entity with financial interest in, or financial conflict with, the subject matter or materials discussed in the manuscript, apart from those disclosed.
Glossary
Abbreviations
- BAC
bacterial artificial chromosome
- BAMCA
BAC array-based methylated CpG island amplification
- BHD
Birt-Hogg-Dube
- CGH
comparative genomic hybridization
- CIMP
CpG island methylator phenotype
- DNMT
DNA methyltransferase
- HCC
hepatocellular carcinoma
- HGF
hepatocyte growth factor
- HIF
hypoxia-inducible factor
- HPH
HIF prolyl hydroxylase
- IL
interleukin
- mTOR
mammalian target of rapamycin
- MINT
methylated in tumor
- MSP
me-thylation-specific PCR
- RB
retinoblastoma
- RCC
renal cell carcinoma
- TNM
tumor-node-metastasis
- TSC
tuberous sclerosis complex
- VEGF
vascular endothelial growth factor
- VHL
von Hippel-Lindau
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. doi: 10.1002/ijc.25516. in press; DOI: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Pascual D, Borque A. Epidemiology of kidney cancer. Adv Urol. 2008 doi: 10.1155/2008/782381. 782381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldewijns MM, van Vlodrop IJ, Schouten LJ, Soetekouw PM, de Bruine AP, van Engeland M. Genetics and epigenetics of renal cell cancer. Biochim Biophys Acta. 2008;1785:133–155. doi: 10.1016/j.bbcan.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Beltran A, Carrasco JC, Cheng L, Scarpelli M, Kirkali Z, Montironi R. 2009 update on the classification of renal epithelial tumors in adults. Int J Urol. 2009;16:432–443. doi: 10.1111/j.1442-2042.2009.02302.x. [DOI] [PubMed] [Google Scholar]
- 5.Eble JN, Togashi K, Pisani P. Renal cell carcinoma. In “World Health Organization classification of tumours. Pathology and genetics. Tumours of the urinary system and male genital organs”. Lyon: IARC Press; 2004. pp. 10–43. [Google Scholar]
- 6.Rosner I, Bratslavsky G, Pinto PA, Linehan WM. The clinical implications of the genetics of renal cell carcinoma. Urol Oncol. 2009;27:131–136. doi: 10.1016/j.urolonc.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, Matveev V, Janout V, Kollarova H, Bencko V, Navratilova M, Szeszenia-Dabrowska N, Mates D, Mukeria A, Holcatova I, Schmidt LS, Toro JR, Karami S, Hung R, Gerard GF, Linehan WM, Merino M, Zbar B, Boffetta P, Brennan P, Rothman N, Chow WH, Waldman FM, Moore LE. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14:4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandriota SJ, Turner KJ, Davies DR, Murray PG, Morgan NV, Sowter HM, Wykoff CC, Maher ER, Harris AL, Ratcliffe PJ, Maxwell PH. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- 9.Gossage L, Eisen T. Alterations in VHL as potential biomarkers in renal-cell carcinoma. Nat Rev Clin Oncol. 2010;7:277–288. doi: 10.1038/nrclinonc.2010.42. [DOI] [PubMed] [Google Scholar]
- 10.Frew IJ, Krek W. Multitasking by pVHL in tumour suppression. Curr Opin Cell Biol. 2007;19:685–690. doi: 10.1016/j.ceb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt L, Junker K, Nakaigawa N, Kinjerski T, Weirich G, Miller M, Lubensky I, Neumann HP, Brauch H, Decker J, Vocke C, Brown JA, Jenkins R, Richard S, Bergerheim U, Gerrard B, Dean M, Linehan WM, Zbar B. Novel mutations of the MET proto-oncogene in papillary renal carcinomas. Oncogene. 1999;18:2343–2350. doi: 10.1038/sj.onc.1202547. [DOI] [PubMed] [Google Scholar]
- 12.Pfaffenroth EC, Linehan WM. Genetic basis for kidney cancer: opportunity for disease-specific approaches to therapy. Expert Opin Biol Ther. 2008;8:779–790. doi: 10.1517/14712598.8.6.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamazaki K, Sakamoto M, Ohta T, Kanai Y, Ohki M, Hirohashi S. Overexpression of KIT in chromophobe renal cell carcinoma. Oncogene. 2003;22:847–852. doi: 10.1038/sj.onc.1206153. [DOI] [PubMed] [Google Scholar]
- 14.Hino O. Multistep renal carcinogenesis in the Eker (Tsc 2 gene mutant) rat model. Curr Mol Med. 2004;4:807–811. doi: 10.2174/1566524043359692. [DOI] [PubMed] [Google Scholar]
- 15.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, Teague J, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Forbes S, Jia M, Jones D, Knott H, Kok CY, Lau KW, Leroy C, Lin ML, McBride DJ, Maddison M, Maguire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, O'Meara S, Pleasance E, Rajasingham A, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turrell K, Dykema KJ, Khoo SK, Petillo D, Wondergem B, Anema J, Kahnoski RJ, Teh BT, Stratton MR, Futreal PA. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inazawa J, Inoue J, Imoto I. Comparative genomic hybridization (CGH)-arrays pave the way for identification of novel cancer-related genes. Cancer Sci. 2004;95:559–563. doi: 10.1111/j.1349-7006.2004.tb02486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arai E, Ushijima S, Tsuda H, Fujimoto H, Hosoda F, Shibata T, Kondo T, Imoto I, Inazawa J, Hirohashi S, Kanai Y. Genetic clustering of clear cell renal cell carcinoma based on array-comparative genomic hybridization: its association with DNA methylation alteration and patient outcome. Clin Cancer Res. 2008;14:5531–5539. doi: 10.1158/1078-0432.CCR-08-0443. [DOI] [PubMed] [Google Scholar]
- 18.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 19.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.ChengX, Blumenthal RM. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry. 2010;49:2999–3008. doi: 10.1021/bi100213t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohn F, Schubeler D. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet. 2009;25:129–136. doi: 10.1016/j.tig.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2010;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 25.Kanai Y, Hirohashi S. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state. Carcinogenesis. 2007;28:2434–2442. doi: 10.1093/carcin/bgm206. [DOI] [PubMed] [Google Scholar]
- 26.Kanai Y. Alterations of DNA methylation and clinicopathological diversity of human cancers. Pathol Int. 2008;58:544–558. doi: 10.1111/j.1440-1827.2008.02270.x. [DOI] [PubMed] [Google Scholar]
- 27.Kanai Y. Genome-wide DNA methylation profiles in precancerous conditions and cancers. Cancer Sci. 2010;101:36–45. doi: 10.1111/j.1349-7006.2009.01383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanai Y, Ushijima S, Tsuda H, Sakamoto M, Sugimura T, Hirohashi S. Aberrant DNA methylation on chromosome 16 is an early event in hepatocarcinogenesis. Jpn J Cancer Res. 1996;87:1210–1217. doi: 10.1111/j.1349-7006.1996.tb03135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanai Y, Hui AM, Sun L, Ushijima S, Sakamoto M, Tsuda H, Hirohashi S. DNA hypermethy-lation at the D17S5 locus and reduced HIC-1 mRNA expression are associated with hepatocarcinogenesis. Hepatology. 1999;29:703–709. doi: 10.1002/hep.510290338. [DOI] [PubMed] [Google Scholar]
- 30.Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis -A comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2009;32:970–979. doi: 10.1053/jhep.2000.19797. [DOI] [PubMed] [Google Scholar]
- 31.Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc Natl Acad Sci USA. 2002;99:10060–10065. doi: 10.1073/pnas.152121799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito Y, Kanai Y, Nakagawa T, Sakamoto M, Saito H, Ishii H, Hirohashi S. Increased protein expression of DNA methyltransferase (DNMT) 1 is significantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas. Int J Cancer. 2003;105:527–532. doi: 10.1002/ijc.11127. [DOI] [PubMed] [Google Scholar]
- 33.Peng DF, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kosuge T, Hirohashi S. Increased DNA methyltransferase 1 (DNMT1) protein expression in precancerous conditions and ductal carcinomas of the pancreas. Cancer Sci. 2005;96:403–408. doi: 10.1111/j.1349-7006.2005.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng DF, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kitazawa S, Hirohashi S. DNA methylation of multiple tumor-related genes in association with overexpression of DNA methyltransferase 1 (DNMT1) during multistage carcinogenesis of the pancreas. Carcinogenesis. 2006;27:1160–1168. doi: 10.1093/carcin/bgi361. [DOI] [PubMed] [Google Scholar]
- 35.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 36.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosi-nes. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arai E, Kanai Y, Ushijima S, Fujimoto H, Mukai K, Hirohashi S. Regional DNA hypermethylation and DNA methyltransferase (DNMT) 1 protein overexpression in both renal tumors and corresponding nontumorous renal tissues. IntJ Cancer. 2006;119:288–296. doi: 10.1002/ijc.21807. [DOI] [PubMed] [Google Scholar]
- 38.Kanai T, Hirohashi S, Upton MP, Noguchi M, Kishi K, Makuuchi M, Yamasaki S, Hasegawa H, Takayasu K, Moriyama N, Shimosato Y. Pathology of small hepatocellular carcinoma. A proposal for a new gross classification. Cancer. 1987;60:810–819. doi: 10.1002/1097-0142(19870815)60:4<810::aid-cncr2820600417>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 39.Misawa A, Inoue J, Sugino Y, Hosoi H, Sugimoto T, Hosoda F, Ohki M, Imoto I, Inazawa J. Methylation-associated silencing of the nuclear receptor 1I2 gene in advanced-type neuroblastomas, identified by bacterial artificial chromosome array-based methylated CpG island amplification. Cancer Res. 2005;65:10233–10242. doi: 10.1158/0008-5472.CAN-05-1073. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka K, Imoto I, Inoue J, Kozaki K, Tsuda H, Shimada Y, Aiko S, Yoshizumi Y, Iwai T, Kawano T, Inazawa J. Frequent methylation-associated silencing of a candidate tumor-suppressor, CRABP1, in esophageal squamous-cell carcinoma. Oncogene. 2007;26:6456–6468. doi: 10.1038/sj.onc.1210459. [DOI] [PubMed] [Google Scholar]
- 41.Arai E, Ushijima S, Gotoh M, Ojima H, Kosuge T, Hosoda F, Shibata T, Kondo T, Yokoi S, Imoto I, Inazawa J, Hirohashi S, Kanai Y. Genome-wide DNA methylation profiles in liver tissue at the precancerous stage and in hepatocellular carcinoma. Int J Cancer. 2009;125:2854–2862. doi: 10.1002/ijc.24708. [DOI] [PubMed] [Google Scholar]
- 42.Coolen MW, Stirzaker C, Song JZ, Statham AL, Kassir Z, Moreno CS, Young AN, Varma V, Speed TP, Cowley M, Lacaze P, Kaplan W, Robinson MD, Clark SJ. Consolidation of the cancer genome into domains of repressive chromatin by long-range epigenetic silencing (LRES) reduces transcriptional plasticity. Nat Cell Biol. 2010;12:235–246. doi: 10.1038/ncb2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arai E, Kanai Y. DNA methylation profiles in precancerous tissue and cancers: Carcinogenetic risk estimation and prognostication based on DNA methylation status. Epigenomics. 2010;2:467–481. doi: 10.2217/epi.10.16. [DOI] [PubMed] [Google Scholar]
- 44.Nishiyama N, Arai E, Chihara Y, Fujimoto H, Hosoda F, Shibata T, Kondo T, Tsukamoto T, Yokoi S, Imoto I, Inazawa J, Hirohashi S, Kanai Y. Genome-wide DNA methylation profiles in urothelial carcinomas and urothelia at the precancerous stage. Cancer Sci. 2010;101:231–240. doi: 10.1111/j.1349-7006.2009.01330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arai E, Wakai-Ushijima S, Fujimoto H, Hosoda F, Shibata T, Kondo T, Yokoi S, Imoto I, Inazawa J, Hirohashi S, Kanai Y. Genome-wide DNA methylation profiles in renal tumors of various histological subtypes and non-tumorous renal tissues. Pathobiology. doi: 10.1159/000322072. in press. [DOI] [PubMed] [Google Scholar]
- 46.Arai E, Ushijima S, Fujimoto H, Hosoda F, Shibata T, Kondo T, Yokoi S, Imoto I, Inazawa J, Hirohashi S, Kanai Y. Genome-wide DNA methylation profiles in both precancerous conditions and clear cell renal cell carcinomas are correlated with malignant potential and patient outcome. Carcinogenesis. 2009;30:214–221. doi: 10.1093/carcin/bgn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiss RH, Lin PY. Kidney cancer: identification of novel targets for therapy. Kidney Int. 2006;69:224–232. doi: 10.1038/sj.ki.5000065. [DOI] [PubMed] [Google Scholar]
- 48.Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, Samid D, Duan DS, Gnarra JR, Linehan WM, Baylin SB. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McRonald FE, Morris MR, Gentle D, Winchester L, Baban D, Ragoussis J, Clarke NW, Brown MD, Kishida T, Yao M, Latif F, Maher ER. CpG methylation profiling in VHL related and VHL unrelated renal cell carcinoma. Mol Cancer. 2009;8:31. doi: 10.1186/1476-4598-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki H, Igarashi S, Nojima M, Maruyama R, Yamamoto E, Kai M, Akashi H, Watanabe Y, Yamamoto H, Sasaki Y, Itoh F, Imai K, Sugai T, Shen L, Issa JP, Shinomura Y, Tokino T, Toyota M. IGFBP7 is a p53-responsive gene specifically silenced in colorectal cancer with CpG island methylator phenotype. Carcinogenesis. 2010;31:342–349. doi: 10.1093/carcin/bgp179. [DOI] [PubMed] [Google Scholar]
- 51.Christoph F, Weikert S, Kempkensteffen C, Krause H, Schostak M, Kollermann J, Miller K, Schrader M. Promoter hypermethylation profile of kidney cancer with new proapoptotic p53 target genes and clinical implications. Clin Cancer Res. 2006;12:5040–5046. doi: 10.1158/1078-0432.CCR-06-0144. [DOI] [PubMed] [Google Scholar]
- 52.Urakami S, Shiina H, Enokida H, Hirata H, Kawamoto K, Kawakami T, Kikuno N, Tanaka Y, Majid S, Nakagawa M, Igawa M, Dahiya R. Wnt antagonist family genes as biomarkers for diagnosis, staging, and prognosis of renal cell carcinoma using tumor and serum DNA. Clin Cancer Res. 2006;12:6989–6997. doi: 10.1158/1078-0432.CCR-06-1194. [DOI] [PubMed] [Google Scholar]
- 53.Kawamoto K, Hirata H, Kikuno N, Tanaka Y, Nakagawa M, Dahiya R. DNA methylation and histone modifications cause silencing of Wnt antagonist gene in human renal cell carcinoma cell lines. Int J Cancer. 2008;123:535–542. doi: 10.1002/ijc.23514. [DOI] [PubMed] [Google Scholar]
- 54.Seliger B, Handke D, Schabel E, Bukur J, Lichtenfels R, Dammann R. Epigenetic control of the ubiquitin carboxyl terminal hydrolase 1 in renal cell carcinoma. J Transl Med. 2009;7:90. doi: 10.1186/1479-5876-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kagara I, Enokida H, Kawakami K, Matsuda R, Toki K, Nishimura H, Chiyomaru T, Tatarano S, Itesako T, Kawamoto K, Nishiyama K, Seki N, Nakagawa M. CpG hypermethylation of the UCHL1 gene promoter is associated with pathogenesis and poor prognosis in renal cell carcinoma. J Urol. 2008;180:343–351. doi: 10.1016/j.juro.2008.02.044. [DOI] [PubMed] [Google Scholar]
- 56.Cooper SJ, Zou H, Legrand SN, Marlow LA, von Roemeling CA, Radisky DC, Wu KJ, Hempel N, Margulis V, Tun HW, Blobe GC, Wood CG, Copland JA. Loss of type III transforming growth factor-beta receptor expression is due to methylation silencing of the transcription factor GATA3 in renal cell carcinoma. Oncogene. 2010;29:2905–2915. doi: 10.1038/onc.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang S, Xu Y, Shen G, Zhao X, Zhou J, Li X, Gong F, Ling B, Fang L, Huang C, Wei Y. Gene expression and methylation status of 14-3-3sigma in human renal carcinoma tissues. IUBMB Life. 2008;60:534–540. doi: 10.1002/iub.75. [DOI] [PubMed] [Google Scholar]
- 58.To KK, Zhan Z, Bates SE. Aberrant promoter methylation of the ABCG2 gene in renal carcinoma. Mol Cell Biol. 2006;26:8572–8585. doi: 10.1128/MCB.00650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yano T, Ito F, Kobayashi K, Yonezawa Y, Suzuki K, Asano R, Hagiwara K, Nakazawa H, Toma H, Yamasaki H. Hypermethylation of the CpG island of connexin 32, a candiate tumor suppressor gene in renal cell carcinomas from hemodialysis patients. Cancer Lett. 2004;208:137–142. doi: 10.1016/j.canlet.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 60.Yamada D, Kikuchi S, Williams YN, Sakurai-Yageta M, Masuda M, Maruyama T, Tomita K, Gutmann DH, Kakizoe T, Kitamura T, Kanai Y, Murakami Y. Promoter hypermethylation of the potential tumor suppressor DAL-1/4.1B gene in renal clear cell carcinoma. IntJ Cancer. 2006;118:916–923. doi: 10.1002/ijc.21450. [DOI] [PubMed] [Google Scholar]
- 61.Onay H, Pehlivan S, Koyuncuoglu M, Kirkali Z, Ozkinay F. Multigene methylation analysis of conventional renal cell carcinoma. Urol Int. 2009;83:107–112. doi: 10.1159/000224878. [DOI] [PubMed] [Google Scholar]
- 62.Kvasha S, Gordiyuk V, Kondratov A, Ugryn D, Zgonnyk YM, Rynditch AV, Vozianov AF. Hypermethylation of the 5'CpG island of the FHIT gene in clear cell renal carcinomas. Cancer Lett. 2008;265:250–257. doi: 10.1016/j.canlet.2008.02.036. [DOI] [PubMed] [Google Scholar]
- 63.Breault JE, Shiina H, Igawa M, Ribeiro-Filho LA, Deguchi M, Enokida H, Urakami S, Terashima M, Nakagawa M, Kane CJ, Carroll PR, Dahiya R. Methylation of the gamma-catenin gene is associated with poor prognosis of renal cell carcinoma. Clin Cancer Res. 2005;11:557–564. [PubMed] [Google Scholar]
- 64.Morris MR, Gentle D, Abdulrahman M, Maina EN, Gupta K, Banks RE, Wiesener MS, Kishida T, Yao M, Teh B, Latif F, Maher ER. Tumor suppressor activity and epigenetic inactivation of hepatocyte growth factor activator inhibitor type 2/SPINT2 in papillary and clear cell renal cell carcinoma. Cancer Res. 2005;65:4598–4606. doi: 10.1158/0008-5472.CAN-04-3371. [DOI] [PubMed] [Google Scholar]
- 65.Okuda H, Toyota M, Ishida W, Furihata M, Tsuchiya M, Kamada M, Tokino T, Shuin T. Epigenetic inactivation of the candidate tumor suppressor gene HOXB13 in human renal cell carcinoma. Oncogene. 2006;25:1733–1742. doi: 10.1038/sj.onc.1209200. [DOI] [PubMed] [Google Scholar]
- 66.Sturm I, Stephan C, Gillissen B, Siebert R, Janz M, Radetzki S, Jung K, Loening S, Dorken B, Daniel PT. Loss of the tissue-specific proapoptotic BH3-only protein Nbk/Bik is a unifying feature of renal cell carcinoma. Cell Death Differ. 2006;13:619–627. doi: 10.1038/sj.cdd.4401782. [DOI] [PubMed] [Google Scholar]
- 67.Awakura Y, Nakamura E, Ito N, Kamoto T, Ogawa O. Methylation-associated silencing of TU3A in human cancers. Int J Oncol. 2008;33:893–899. [PubMed] [Google Scholar]
- 68.Kempkensteffen C, Hinz S, Schrader M, Christoph F, Magheli A, Krause H, Schostak M, Miller K, Weikert S. Gene expression and promoter methylation of the XIAP-associated Factor 1 in renal cell carcinomas: correlations with pathology and outcome. Cancer Lett. 2007;254:227–235. doi: 10.1016/j.canlet.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 69.Zuo T, Tycko B, Liu TM, Lin HJ, Huang TH. Methods in DNA methylation profiling. Epigenomics. 2009;1:331–345. doi: 10.2217/epi.09.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bibikova M, Fan JB. GoldenGate assay for DNA methylation profiling. Methods Mol Biol. 2009;507:149–163. doi: 10.1007/978-1-59745-522-0_12. [DOI] [PubMed] [Google Scholar]
- 71.Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou L, Shen R, Gunderson K. Genome-wide DNA methylation profiling using Infinium assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 72.Estecio MR, Issa JP. Tackling the methylome: recent methodological advances in genome-wide methylation profiling. Genome Med. 2009;1:106. doi: 10.1186/gm106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 74.Cho M, Uemura H, Kim SC, Kawada Y, Yoshida K, Hirao Y, Konishi N, Saga S, Yoshikawa K. Hypomethylation of the MN/CA9 promoter and upregulated MN/CA9 expression in human renal cell carcinoma. Br J Cancer. 2001;85:563–567. doi: 10.1054/bjoc.2001.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grabmaier K, de Weijert M, Uemura H, Schalken J, Oosterwijk E. Renal cell carcinoma-associated G250 methylation and expression: in vivo and in vitro studies. Urology. 2002;60:357–362. doi: 10.1016/s0090-4295(02)01711-9. [DOI] [PubMed] [Google Scholar]
- 76.Florl AR, Löwer R, Schmitz-Dräger BJ, Schulz WA. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer. 1999;80:1312–1321. doi: 10.1038/sj.bjc.6690524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ellinger J, Kahl P, Mertens C, Rogenhofer S, Hauser S, Hartmann W, Bastian PJ, Büttner R, Müller SC, von Ruecker A. Prognostic relevance of global histone H3 lysine 4 (H3K4) methylation in renal cell carcinoma. Int J Cancer. 2010 doi: 10.1002/ijc.25250. DOI: 10.1002/ijc.25250. [DOI] [PubMed] [Google Scholar]
- 78.Goessl C, Muller M, Straub B, Miller K. DNA alterations in body fluids as molecular tumor markers for urological malignancies. Eur Urol. 2002;41:668–676. doi: 10.1016/s0302-2838(02)00126-4. [DOI] [PubMed] [Google Scholar]
- 79.Oya M. Renal cell carcinoma: biological features and rationale for molecular-targeted therapy. Keio J Med. 2009;58:1–11. doi: 10.2302/kjm.58.1. [DOI] [PubMed] [Google Scholar]
- 80.Rini BI. New strategies in kidney cancer: therapeutic advances through understanding the molecular basis of response and resistance. Clin Cancer Res. 2010;16:1348–1354. doi: 10.1158/1078-0432.CCR-09-2273. [DOI] [PubMed] [Google Scholar]
- 81.Reeves DJ, Liu CY. Treatment of metastatic renal cell carcinoma. Cancer Chemother Pharmacol. 2009;64:11–25. doi: 10.1007/s00280-009-0983-z. [DOI] [PubMed] [Google Scholar]