Abstract
Excitotoxicity has been implicated as the mechanism of neuronal damage resulting from acute insults such as stroke, epilepsy, and trauma, as well as during the progression of adult-onset neurodegenerative disorders such as Alzheimer’s disease and amyotrophic lateral sclerosis (ALS). Excitotoxicity is defined as excessive exposure to the neurotransmitter glutamate or overstimulation of its membrane receptors, leading to neuronal injury or death. One potential approach to protect against excitotoxic neuronal damage is enhanced glutamate reuptake. The glial glutamate transporter EAAT2 is the quantitatively dominant glutamate transporter and plays a major role in clearance of glutamate. Expression of EAAT2 protein is highly regulated at the translational level. In an effort to identify compounds that can induce translation of EAAT2 transcripts, a cell-based enzyme-linked immunosorbent assay was developed using a primary astrocyte line stably transfected with a vector designed to identify modulators of EAAT2 translation. This assay was optimized for high-throughput screening, and a library of approximately 140,000 compounds was tested. In the initial screen, 293 compounds were identified as hits. These 293 hits were retested at 3 concentrations, and a total of 61 compounds showed a dose-dependent increase in EAAT2 protein levels. Selected compounds were tested in full 12-point dose-response experiments in the screening assay to assess potency as well as confirmed by Western blot, immunohistochemistry, and glutamate uptake assays to evaluate the localization and function of the elevated EAAT2 protein. These hits provide excellent starting points for developing therapeutic agents to prevent excitotoxicity.
Keywords: excitotoxicity, glutamate transporter, EAAT2, high-throughput screen, neurodegeneration
INTRODUCTION
Glutamate is the main excitatory neurotransmitter in the CNS, responsible for fast excitatory neurotransmission. Under normal conditions, glutamate released from the presynaptic neuron activates ionotropic glutamate receptors present on the postsynaptic neuron. This results in the influx of Na+ and Ca2+ ions into the cell, leading to membrane depolarization and generation of action potentials. The concentration of glutamate in the synaptic cleft, as well as the resultant activity of the postsynaptic ionotropic glutamate receptors, is tightly regulated by interplay between glutamate release and glutamate clearance. Glutamate clearance is facilitated by Na+-dependent excitatory amino acid transporters (EAATs), of which 5 mammalian EAATs have been cloned to date: EAAT1 (GLAST), EAAT2 (GLT-1), EAAT3 (EAAC1), EAAT4, and EAAT5.1 EAAT2 is expressed mainly in glial cells throughout the CNS and is a major glutamate transporter in adult tissues.2
Under disease conditions, elevated extracellular glutamate concentrations can occur when the release from presynaptic terminals is augmented or when the reuptake from the synaptic cleft is insufficient, with the latter being caused by a decrease in EAAT2 protein expression. Excessive glutamate can cause overstimulation of glutamate receptors, which gives rise to an increased intracellular concentration of Na+ and Ca2+ ions leading to neuronal injury or death, known as excitotoxicity.3 Ca2+ entry through NMDA and Ca2+-permeable AMPA receptors appears to be the major trigger for excitotoxic neuronal injury by glutamate.4,5 The increase in cytoplasmic Ca2+ activates a number of Ca2+-dependent enzymes involved in the catabolism of proteins, phospholipids, and nucleic acids, as well as in the synthesis of nitric oxide. In addition, mitochondrial dysfunction due to increased Ca2+ uptake in mitochondria and subsequent free radical formation could also contribute to excitotoxic injury or death.6,7
Excitotoxicity has been implicated in the pathogenesis of stroke, neurotrauma, epilepsy, and several neurodegenerative diseases, including Alzheimer’s disease (AD), Parkinson’s disease, amyotrophic lateral sclerosis (ALS), and Huntington’s disease.8–13 Acute elevations of glutamate in the synaptic cleft are thought to induce neuronal damage in conditions such as stroke, status epilepticus, and neurotrauma. More chronic and milder elevations of glutamate are believed to underlie excitotoxicity in neurodegenerative diseases. Modulation of the glutamate neurotransmitter system to reduce excitotoxicity has been a therapeutic target in circumstances of acute and chronic elevations of glutamate in the synapses. There are currently a few drugs targeted at the glutamate neurotransmission process. Riluzole, the only drug that has been proven effective against disease progression in ALS patients, exhibits antiexcitotoxic properties by not only inhibiting the release of glutamate on presynaptic neurons but also by attenuating the effect of glutamate via noncompetitive inhibition of NMDA and AMPA receptors located on postsynaptic neurons.14 Memantine, a noncompetitive NMDA receptor antagonist, can decrease pathological activation of NMDA receptors without affecting physiological NMDA receptor activity.15 Memantine is approved for treating the advanced stages of AD.16 The major problem of glutamate receptor antagonist treatment is the side effects, such as confusion and hallucination. In addition, a growing body of evidence indicates that disruption of the postsynaptic glutamate neurotransmission process may be central to the pathophysiology of conditions such as obesity, schizophrenia, depression, substance abuse, pain disorders, and glaucoma.
Another approach to reduce excitotoxicity is through enhancing glutamate reuptake via increase of EAAT2 protein expression. We previously found that there are 3 forms of human EAAT2 transcripts with 76, 310, and 1091 nucleotides (nts) in their 5′-untranslated regions (5′ UTRs).17 The EAAT2 transcripts with 310 nts as well as with 1091-nt 5′ UTRs are not constitutively translated but require extracellular factors such as retinoic acid, corticosterone, or β-lactam antibiotics to induce translational read-through.17 Moreover, many disease-associated insults affect the efficiency of translation of these EAAT2 transcripts, which suggests that this translational regulation mechanism may play an important role in EAAT2 protein expression under disease conditions.17 In addition, we and other groups have shown that an increase of EAAT2 expression not only results in reduced excitotoxicity but also has a therapeutic effect in several animal models of diseases, including ALS, visceral pain, and focal cerebral ischemia.18–21
One approach to identifying small-molecule activators of expression of EAAT2 would be to screen a reporter gene assay using the EAAT2 promoter to drive expression of a reporter, such as luciferase, as described by Piazza et al.22 However, this approach would not overcome the translational regulation that suppresses EAAT2 protein expression. In an effort to identify novel compounds that can activate translation of EAAT2 as a means of enhancing EAAT2 expression and glutamate uptake, we report the use of a cell-based enzyme-linked immunosorbent assay in a high-throughput screen (HTS). This approach may potentially serve as a therapeutic strategy for protecting against excitotoxic neuronal damage.
MATERIALS AND METHODS
Cell lines and cell culture conditions
PA-EAAT2 cells, a primary astrocyte line that stably expresses human EAAT2 transcripts with the 1091-nucleotides 5′ UTR driven by the cytomegalovirus (CMV) promoter, were generated in our previous study.17 PA-EAAT2 cells at passage 9 were thawed from frozen stocks and grown in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA) containing 25 mM glucose, 1 mM sodium pyruvate, 19.4 μM pyridoxine hydrochloride, and 2 mM glutamine and supplemented with 10% fetal bovine serum (FBS), 700 μg/mL geneticin (Gibco, Los Angeles, CA), and 100 μg/mL penicillin-streptomycin (Sigma, St. Louis, MO). The cells were always maintained at 37°C in the presence of 5% CO2.
To plate cells in reduced serum on 384-well plates for HTS, PA-EAAT2 cells (passage number between 12 and 15) were harvested by trypsinization with 0.25% Trypsin (Gibco), centrifuged at 1000 rpm for 5 min to pellet, and resuspended in DMEM supplemented with 2% FBS, 700 μg/mL geneticin, and 100 μg/mL penicillin-streptomycin. The resuspended cells were passed through a cell strainer, counted with a hemocytometer, and diluted to 160 cells/μL, and 50 μL (8000 cells/well) was added to 384-well white tissue culture–treated plates with white bottoms using a MicroFill (Biotek Instruments, Winooski, VT). The plates were covered in air-permeable membranes and grown for 36 h prior to compound addition (see below). One clear-bottom plate was included to ensure the cells were at 80% to 90% cell density prior to compound addition (see below).
Cell-based enzyme-linked immunosorbent assay optimization
Assay optimization took place in several stages. The first stage was developing the enzyme-linked immunosorbent assay (ELISA) in the 96-well format. In this format, we optimized several conditions, including (1) cell culture conditions: media, seeding density, culture time, and starvation medium time; (2) cell fixation conditions: concentration, incubation time, and fixation reagent; (3) antibody conditions: sources of antibodies and concentration (5 × 5 matrix of 2-fold serial dilutions of primary and secondary antibody from 1/2000 to 1/32,000); (4) detection conditions: exposure time, horseradish peroxidase (HRP) substrate (e.g., luminescence or TMB [3,3′,5,5′-tetramethylbenzidine]). These conditions were evaluated for signal strength, signal (retinoic acid) to background (DMSO only) ratio, coefficient of variation (CV) as a measure of well-to-well variability, and Z′ factor.23 Once a particular condition was chosen as optimal, other conditions were reconfirmed to ensure they were still optimal with the newly confirmed condition.
The next stage consisted of miniaturizing the assay to a 384-well format and determining the new optimal seeding density for the cells. Once determined, various instrumental parameters for the high-throughput format were established by varying conditions such as volumes of buffers for wash steps, tip heights, and number of washes between steps for the EL-405 Biotek Plate Washer (Biotek Instruments), Cybi-well 384- channel simultaneous pipettor (CyBio AG, Jena, Germany), and MicroFill. Once these conditions were established, we evaluated variability across a series of DMSO-treated and retinoic acid–treated plates. Next, 4 compound plates were tested in duplicate to ensure consistent reproducible results.
Compound library
The compound library consisted of approximately 140,000 small molecules, including compounds approved by the Food and Drug Administration (FDA), a purified natural products library, and compounds purchased from Peakdale (High Peak, UK), Maybridge Plc. (Cornwall, UK), Cerep (Paris, France), Bionet Research Ltd. (Cornwall, UK), Prestwick (Ilkich, France), Specs and Biospecs (CP Rijswijk, the Netherlands), ENAMINE (Kiev, Ukraine), Life Chemicals, Inc. (Burlington, Canada), MicroSource Diversity System’s NINDS customs collection (Gaylordsville, CT), Chemical Diversity Labs (San Diego, CA), ChemBridge (San Diego, CA), and small molecules procured from various academic institutions. Compounds were selected from the different vendors by applying a series of filters, including for clogP and predicted solubility. All of the small molecules generally adhere to Lipinski’s rules (i.e., molecular weight <500, H-bond donors >5, H-bond acceptors >10, and logP <5) and contain a low proportion of known toxicophores (i.e., Michael acceptors and alkylating agents) and unwanted functionalities (i.e., imines, thiol, and quaternary amines) and have been optimized to maximize molecular diversity.
Compound source plates were prepared by spotting 0.4 μL of 1.67 mM compound in DMSO in each well of columns 1 to 22 of a Greiner 384-well plate, with columns 23 and 24 spotted with neat DMSO for positive and negative controls. The plates were immediately sealed with aluminum plate seals and stored at −20°C. Once the assay was ready for HTS, plates were stored for the short term at 4°C. The day of the screen, the plates were warmed to room temperature for 2 h, and then 40 μL of serum-free DMEM supplemented with 700 μg/mL geneticin (DMEM/gen) was added to each well of the first 23 columns of the compound plates using the MicroFill, diluting the compounds from 1.67 mM to an intermediate concentration 13.3 μM. Moreover, 40 μL of 7.5 μg/mL retinoic acid (positive control) in DMEM/gen was added to each well of column 24 using a Multidrop 384 (Thermo Fisher Scientific, Waltham, MA).
HTS for EAAT2 activators
Assaying a particular set of compound plates (40 compound plates; approximately 14,000 compounds) took place over a period of 7 days (Fig. 1B). Cell plating and compound plate preparation were described above. Compounds and controls were added to cell plates 36 h after plating the cells. Prior to adding compounds, cell plates were gently washed thrice with 50 μL of DMEM/gen using the plate washer to replace the plating media with serum-free media to reduce background stimulation of EAAT2 by components of FBS and penicillin. This plate washer was used for all subsequent washing steps. Then, 10 μL of compound or controls was transferred from compound plates to the cell plate using the CyBio and immediately covered with air-permeable membranes. The final concentration of compounds was an average of 2.4 μM based on an average molecular weight of 500 g/mol in the compound library. The final concentration of retinoic acid and DMSO was 1.5 μg/mL and 0.20%, respectively. Cells were exposed to compound for 72 h. The ELISA protocol began on day 6 by fixing the cells for 2 h in 4% formaldehyde (20 μL of 14% formaldehyde in phosphate-buffered saline [PBS] added to each well containing 60 μL media with the multidrop). Unless otherwise indicated, the plates were washed 3 times with 80 μL/well wash buffer (0.1% Triton X-100 in PBS) between each subsequent step. The plates were then quenched for 2 h by adding 40 μL/well of quenching buffer (0.1% NaN3, 1% H2O2 in washing buffer) using the MicroFill. Nonspecific antibody binding was reduced by blocking at room temperature for 2 h by adding 40 μL of blocking buffer (5% milk in washing buffer). Then, 15 μL/well of 1:2000 dilution of 10 antibodies (rabbit anti-EAAT2 polyclonal antibody18) in antibody dilution buffer (3% FractionV BSA in wash buffer) was added using the CyBio. The plates were immediately sealed with aluminum plate seals and placed at 4°C overnight. The next morning, the plates were warmed to room temperature and washed thrice in wash buffer, and 15 μL of 1:6000 dilution of 20 antibody (anti-rabbit IgG-HRP linked Antibody; Cell Signaling, Danvers, MA) in antibody dilution buffer was added using the CyBio. The plates were incubated for 2 h at room temperature, then washed thrice with 80 μL wash buffer and then thrice with 80 μL PBS before adding 8 μL each of chemiluminescent substrate and hydrogen peroxide (SuperSignal ELISA Femto Maximum Sensitivity Substrate, Pierce, Rockford, IL) using the CyBio. The luminescent signal was recorded using an LJL Analyst plate reader (Molecular Devices, Sunnyvale, CA) 40 min after substrate addition. The data points were recorded as luminescent counts/s (or signal intensity) and transformed to percent activation over average signal intensity in the DMSO-treated control wells by using the following equation: % activation = [(signal intensity in test compound well/average signal intensity in DMSO control well) − 1) × 100].
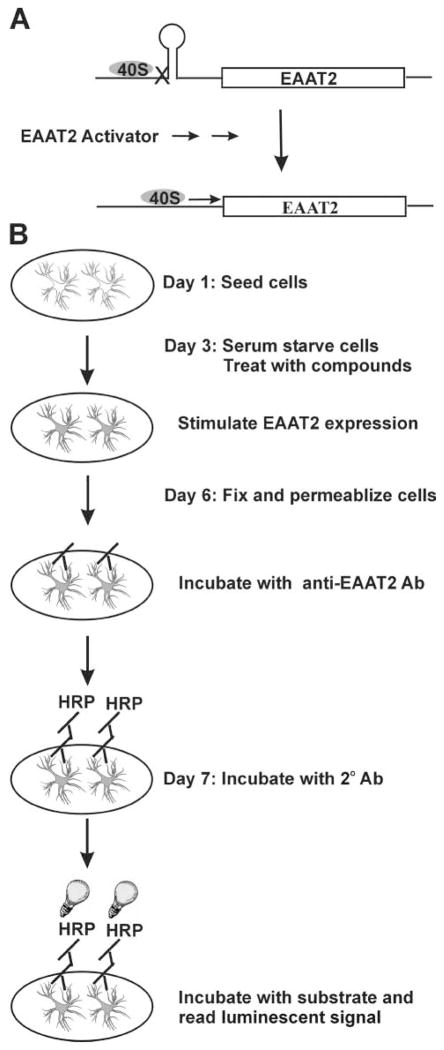
FIG. 1.
(A) The EAAT2 transcripts with long 5′ UTR are not constitutively translated but require extracellular factors to induce translational read-through. The 5′ UTR may form secondary structure, or stem-loops, which negatively affect translation by impeding the binding or migration of 40S ribosomal subunits. (B) A schematic illustration of the enzyme-linked immunosorbent assay procedures.
A compound was considered a “hit” and selected for validation if it increased activation >3 times the standard deviation of the sample wells. To confirm the activity of compounds identified as hits in the initial screen, compounds were tested in quadruplicate at 3 concentrations. Then, 3 μL of each compound selected for validation was pulled from stock solutions of approximately 10 mM in DMSO and diluted with 3 μL DMSO into 96-well plates, followed by 2 subsequent 10-fold dilutions (5, 0.5, and 0.05 mM, respectively) using a Beckman Biomek FX system (Beckman Coulter, Fullerton, CA) with span 8 pipetting head. Next, 0.4 μL of each dose of each compound was quad-mapped onto 384-well plates, then diluted with 40 μL DMEM/gen, and 10 μL of compounds was added as per HTS technique. The final concentrations for the validation assay were 10 μM, 1 μM, and 0.1 μM, respectively. Finally, selected confirmed hits were reordered from the commercial suppliers, and a 12-point dose-response series of 3-fold dilutions ranging from 0.01 to 30 μM final concentration was tested in quadruplicate in the HTS assay to generate EC50 values for each compound.
Evaluation of toxicity of compounds to PA-EAAT2 cells in culture
In parallel to the 12-point dose ELISA experiments, toxicity of the compounds to the PA-EAAT2 cells was evaluated using the CellTiterGLO Luminescent viability assay from Promega (Madison, WI). Cells were cultured and exposed to compound as described above. Following a 72-h exposure to compound, 20 μL of the luminescent cell viability detection reagent was added to the culture media, incubated for 10 min at room temperature, and the luminescent counts per second measured using an LJL Analyst plate reader (Molecular Devices). Toxicity was assessed at compound concentrations ranging from 0.1 nM to 30 μM to determine the LD50 of each compound.
Determination of protein level by immunoblotting
PA-EAAT2 cells grown on 6-well plates were cultured in DMEM and treated with compound for 72 h and then harvested for immunoblotting. Immunoblotting was performed as described previously.24 Briefly, the harvested samples were sonicated in 1× PBS containing complete protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN), assayed for protein concentration, resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE; 8% PA), and transferred onto PVDF membranes. The following primary antibodies were used: rabbit anti-EAAT2 polyclonal antibodies (1:4000) and goat anti-actin (1:2000; Santa Cruz Biotechnology, Santa Cruz, CA). The immunoreactive bands were detected using the SuperSignal West Pico Chemiluminescent Substrate (Pierce Biotechnology, Rockford, IL) according to the manufacturer’s directions. Band intensities were analyzed with ImageJ software.
[3H]glutamate uptake assay
PA-EAAT2 cells grown on 24-well plates were cultured in DMEM and treated with compound for 72 h and then subjected to [3H]glutamate uptake assay. Uptake of radiolabeled glutamate was monitored as described previously.25 Briefly, cultured cells grown on 24-well plates were washed with uptake sample buffer (320 mM sucrose in 50 mM Tris-HCl, pH 7.4) and then incubated for 10 min at 37°C with L-[3H]glutamate (0.5 μCi; Amersham Biosciences, Piscataway, NJ) in either Na+-containing or Na+-free Kreb’s buffer supplemented with 40 μM unlabeled glutamate. The cells were washed with ice-cold 1× PBS and lysed in 1 mM NaOH. The amount of radiolabeled glutamate was measured using a Beckman Coulter LS6500 Multi-Purpose Scintillation Counter. Na+-dependent L-[3H] glutamate uptake was calculated by subtracting Na+-independent L-[3H]glutamate uptake (in Na+-free Kreb’s buffer) from the total L-[3H]glutamate uptake (in Na+-containing Kreb’s buffer).
Determination of cellular localization by immunofluorescence staining
Immunofluorescence staining was performed as described previously.26 Briefly, cells that were grown on glass coverslips were fixed with 2% paraformaldehyde, 50 mM sucrose, and 400 mM CaCl2 in 100 mM phosphate buffer for 30 min at room temperature, followed by thorough rinsing with PBS+ (0.1% saponin, 0.02% sodium azide in PBS). Coverslips were then blocked with PBS + BSA (10 mg/mL BSA in PBS+) for 30 min at room temperature and then incubated with primary antibody solution (rabbit anti-EAAT2 polyclonal antibodies [1:500] in PBS + BSA) overnight at room temperature. After rinsing, the coverslips were then incubated with secondary antibody solution (Alexa Fluor 594-goat anti-rabbit IgG [1:1000] in PBS + BSA) for 60 min at room temperature. The coverslips were then rinsed and mounted onto glass slides with ImmuMount (Shandon Lipshaw, Pittsburgh, PA). Images were obtained using a Zeiss Axioskop 2 inverted microscope (Carl Zeiss, Inc., Thornwood, NY).
Measurement of mRNA level by real-time RT-PCR
Total RNA from PA-EAAT2 cells was isolated with TRIzol (Invitrogen), and first-strand cDNA was synthesized with M-MLV reverse transcriptase (Invitrogen) using an EAAT2-specific primer (5′-ACGCTGGGGAGTTTATTCAAGAAT-3′). β-Actin was used as an internal control (primer: 5′-TGTCA AAGAAAGGGTGTAAAACGCAGC-3′). Real-time PCR was performed using SYBR GREEN PCR Master Mix (Applied Biosystems, Foster City, CA). PCR primers (5′-TAACTC TGGCGGCCAATGGAAAGT-3′ and 5′-ACGCTGGGGAGTT TATTCAAGAAT-3′) were used for EAAT2 cDNA, and primers (5′-TGGATCAGCAAGCAGGAGTACGA-3′ and 5′-TGTCAAA GAAAGGGTGTAAAACGCAGC-3′) were used for actin cDNA. PCR conditions were as follows: 95°C for 10 min, 95°C for 15 s, and 60°C for 1 min for 40 cycles.
RESULTS
Screening strategy
The goal of this screen was to identify compounds that can induce translation of EAAT2 transcripts with long 5′ UTRs. As discussed in the Introduction, EAAT2 transcripts with long 5′ UTRs are tightly regulated at the translational level and require extracellular factors such as retinoic acid for translational read-through. In addition to the role of the 5′ UTR in the translational regulation of EAAT2 transcripts, the coding region of EAAT2 also plays a role, as translational regulation is lost when the EAAT2 coding sequence is replaced by that of luciferase.17 This added complexity to the translational regulation of EAAT2 transcripts eliminates the potential for the development of a screening strategy using a luminescent reporter gene assay.
Our laboratory has generated a rabbit polyclonal antibody against the C-terminus of EAAT2, which shows low background and high specificity toward EAAT2 on Western blot analysis and immunohistochemistry.18 We reasoned that these antibodies could be used to develop an ELISA capable of determining quantitative differences in EAAT2 expression levels in a cell-based system. We previously established a primary astrocyte line that did not express endogenous EAAT2 but constantly expressed ectopic human EAAT2 transcripts with the 1091-nts 5′ UTR, driven by the CMV promoter, referred to as PA-EAAT2.17 In the absence of serum, these cells express very low amounts of EAAT2 protein, although high levels of the recombinant EAAT2 mRNA are expressed (translational silencing); however, EAAT2 protein levels are significantly increased when an activator (such as retinoic acid) is added to the culture (translational activation) (schematically illustrated in Fig. 1A). Importantly, this translational regulation of EAAT2 occurs in vivo (i.e., both in primary cortical neurons-astrocytes mixed cultures and in mice), indicating that the PA-EAAT2 is an appropriate cell line for HTS to identify EAAT2 translation activators.17 We therefore decided to use PA-EAAT2 cells to develop a cell-based ELISA for measuring expression levels of EAAT2 protein.
Establishment of ELISA
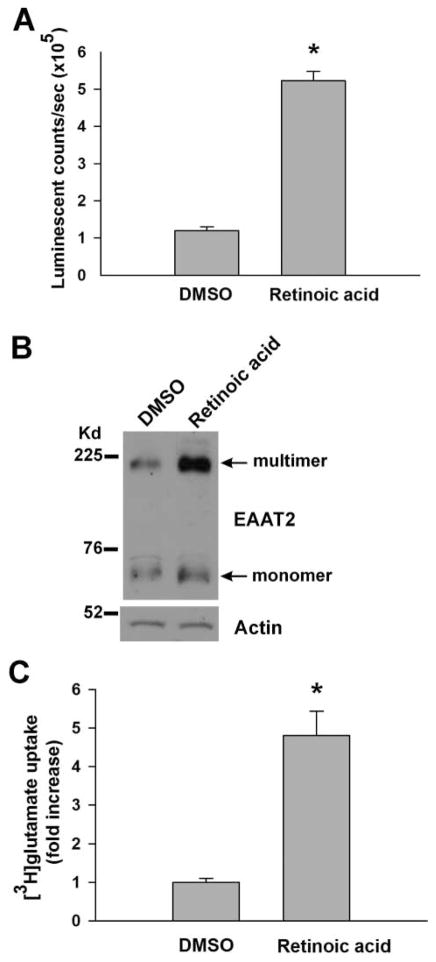
We initially developed the ELISA procedures in 96-well plates using retinoic acid (1.5 μg/mL) as a stimulus to assess responsiveness. Figure 1B schematically illustrates the ELISA procedures. We optimized the conditions for each individual step, including cell culture conditions, fixative conditions, antibodies conditions, and detection conditions. The optimized conditions are described in the Materials and Methods. The ELISA results were validated by Western blot analysis and [3H] glutamate uptake assay. As shown in Figure 2, the magnitude of retinoic acid–induced EAAT2 protein expression as measured by ELISA (Fig. 2A) was highly correlated with that measured by Western blot analysis (Fig. 2B) or the functional increase measured in glutamate uptake assays (Fig. 2C).
FIG. 2.
Validation of the enzyme-linked immunosorbent assay (ELISA). PA-EAAT2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) and treated with retinoic acid (1.5 μg/mL) for 72 h and then subjected to (A) ELISA, (B) Western blot analysis, or (C) [3H]glutamate uptake assay. The rate of retinoic acid–induced EAAT2 protein expression as measured by ELISA (n = 192 for each; *p < 0.0001) was correlated with that measured by Western blotting or glutamate uptake (n = 12 for each; *p < 0.0001). For glutamate uptake data, the fold increase of the retinoic acid–treated cells was normalized to the DMSO-treated cells.
After the ELISA procedure was established, we proceeded to evaluate the suitability of the procedure for HTS. We prepared test plates in the 384-well format to measure signal strength, interwell consistency of signal, and reproducibility in retinoic acid–treated and vehicle (DMSO)–treated cells. Cell density in the range of 7000 to 8000 per well was found to be optimal. The PA-EAAT2 cell signal increased ~2- to 3.5-fold with retinoic acid (1.5 μg/mL). The well-to-well variation was ~10%. We measured Z′ factor to assess whether changes in signal intensity and interwell variation can accurately distinguish “hits” in a large number of test compounds. The average Z′ factor calculated for our test plates was 0.3. A Z′ factor >0.5 is typically considered suitable for HTS. The Z′ factor is a commonly used statistical parameter for HTS assays to reflect the quality of the data within an assay plate. However, in assays such as this where activation is measured and the positive control does not reflect the maximal possible signal window and is instead an indicator as to the responsiveness of the cells to stimulation of the signal being measured, the Z′ factor may not be the only parameter by which to judge the data. If the signal window between the control cells and the positive control is relatively small, the Z′ factor will be low, even if the variability in the signal is within an acceptable range. In addition, a multistage assay such as an ELISA is expected to have some variability because of the number of wash steps. The combination of these circumstances resulted in lower than typical Z′ factors for the assay (below 0.5). In this situation, an alternative measure of quality of data is the %CV of the positive and vehicle controls. As mentioned above, the %CV for our test plates was ~10%, which was acceptable.
We then performed a preliminary screen, which were 4 plates containing 1400 FDA-approved compounds/natural products. A final concentration of 2.4 μM, based on an average molecular weight of the compound library of 500 g/mol, was chosen in this preliminary screen. Final approximate compound concentrations of 1 to 3 μM are typically used in cell-based screens to reduce the potential for compound toxicity and to limit the effects of DMSO on the cells. In addition, because it is desirable to identify compounds that have activity in or below the low micromolar range, it is reasonable to select a compound concentration within this range for screening. The final DMSO concentration in test and control wells was 0.16%. These plates were tested in duplicate to ensure consistent reproducible results. The Z′ factor average for the 8 plates tested was 0.3. We previously found that corticosterone is a translational activator of EAAT2.17 There were 15 corticosteroid compounds in this screen. When we used “>70% activation above DMSO control” as the hit selection criterion, 22 compounds were considered hits, and importantly, all 15 corticosteroid compounds were on the list of hits. The results of this preliminary screen indicated that although the Z′ factor is relatively low, the procedure still identified the candidates efficiently. This led us to continue the HTS.
HTS for EAAT2 activators
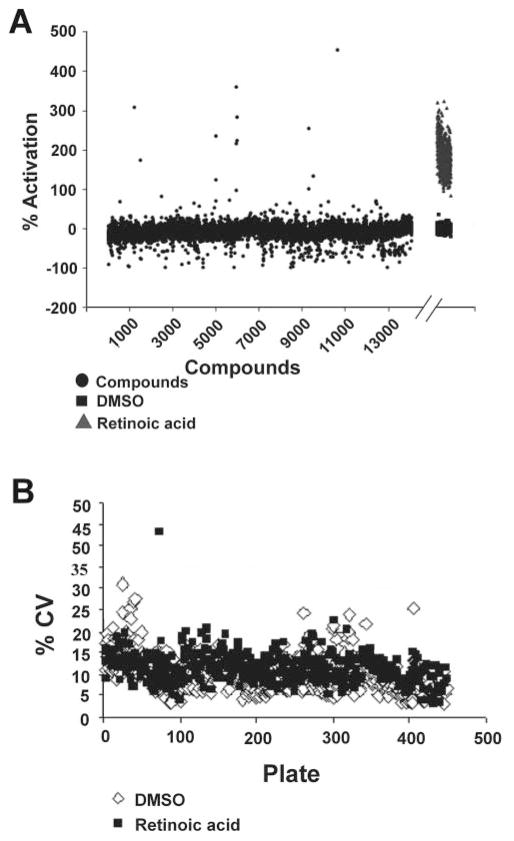
We screened the 140,000-compound library assembled by the Laboratory of Drug Discovery in Neurodegeneration (LDDN). The conditions were the same as that used in our preliminary screen (the final compound concentration was 2.4 μM and the final DMSO concentration was 0.16%). For comparison, all data points were transformed from signal intensity to percent activation over average signal intensity in the DMSO-treated control wells. Figure 3A show the results of the ~14,000 compounds tested as well as the 640 DMSO and retinoic acid control wells from the same plates, which are representative of the entire screen. The %CV for the DMSO controls and the retinoic acid–positive controls were evaluated as a means of judging the quality of the data for each assay plate based on the signal variability within the controls (Fig. 3B). The control %CVs for the assay were generally below 20% (the average for the screen was 9.8%). Those plates that were higher than 20% were repeated.
FIG. 3.
Screen results. (A) Percent activation for 14,000 compounds screened at 2.4 μM concentration. Data are representative of the ~140,000 compounds screened. Note: DMSO and retinoic acid control data appear after the break. (B) Percent coefficients of variation (%CVs) for DMSO vehicle control and retinoic acid–treated wells for each plate screened.
Next, we selected realistic limits to define hits for further characterization. We defined a hit as a molecule that increased EAAT2 protein expression by 70% over DMSO controls. This threshold represented values that were >3-fold the standard deviation for the plates, ensuring that the compounds identified as hits were well outside the scatter of the assay. This criterion resulted in selection of 293 compounds as hits, for an overall 0.2% hit rate. These compounds were then retested for dose dependence of response at 0.1, 1, and 10 μM in quadruplicate (not shown). Of the 293 compounds selected for retesting, a total of 61 compounds showed a dose-dependent increase of activation, for a confirmation rate of 21%.
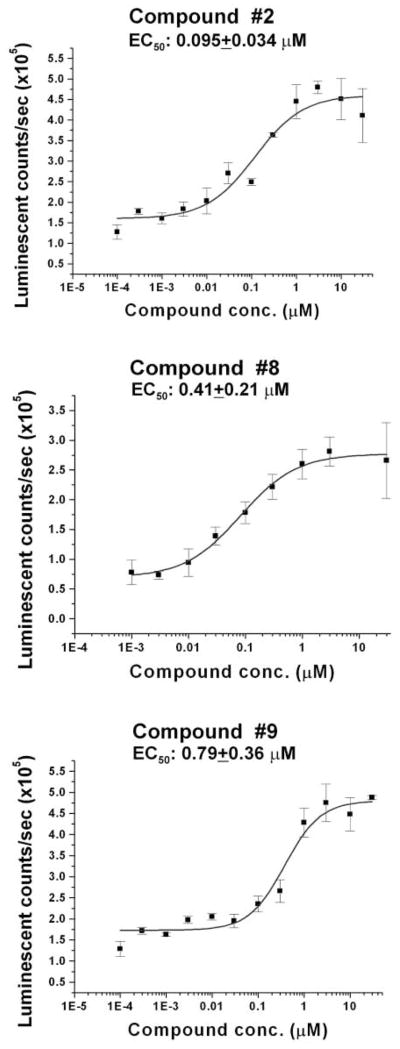
Compounds were prioritized based on the dose dependence of activation of EAAT2 protein expression and on chemical structure. Eleven confirmed compounds (compounds 1-11; Table 1) were evaluated in a full 12-point dose-response assay with concentrations ranging from 1 nM to 30 μM to obtain EC50 values. Figure 4 shows representative dose-response curves for 3 compounds: compound 2, compound 8, and compound 9. Furthermore, we assessed toxicity at compound concentrations ranging from 0.1 nM to 30 μM to determine the LD50 of each compound. The results of EC50, fold activation, and LD50 for the 11 compounds tested are summarized in Table 1.
Table 1.
Summary of Results from Confirmation Studies of Selected Hits
| Compound | % Activation in Primary Screen | EC50, μM | Maximum Activation (Fold DMSO) | LD50, μM |
|---|---|---|---|---|
| 1 | 126 | 0.24 ± 0.3 | 3 ± 1.5 | 30 |
| 2 | 225 | 0.095 ± 0.04 | 2.9 ± 1 | >30 |
| 3 | 80 | 15 | 2.3 ± 0.3 | 1.2 ± 0.21 |
| 4 | 72 | 0.19 ± 0.27 | 3.9 ± 0.8 | >30 |
| 5 | 240 | 0.12 ± 0.06 | 3.2 ± 1.7 | 0.37 ± 0.13 |
| 6 | 119 | 5.6 ± 6.3 | 2.25 ± 0.5 | 1.1 ± 0.15 |
| 7 | 151 | 25 ± 8.6 | 1.3 ± 0.2 | 6.1 ± 2.2 |
| 8 | 307 | 0.4 ± 0.2 | 1.75 ± 0.3 | 7.6 ± 2.5 |
| 9 | 73 | 0.8 ± 0.4 | 2.8 ± 0.7 | >30 |
| 10 | 165 | 22.5 ± 10 | 1.65 ± 0.2 | >30 |
| 11 | 146 | 16.2 ± 9 | 1.8 ± 0.9 | 5.0 ± 1.6 |
EC50, fold activation, and LD50 values represent the mean ± SD of values determined from at least 3 separate experiments.
FIG. 4.
The enzyme-linked immunosorbent assay (ELISA) 12-point dose responses for 3 compounds. Compounds for this assay were applied to quadruplicate wells at concentrations ranging from 0.1 nM to 30 μM. Error bars represent the standard deviation. EC50s are provided on the inset of each graph.
Hit confirmation
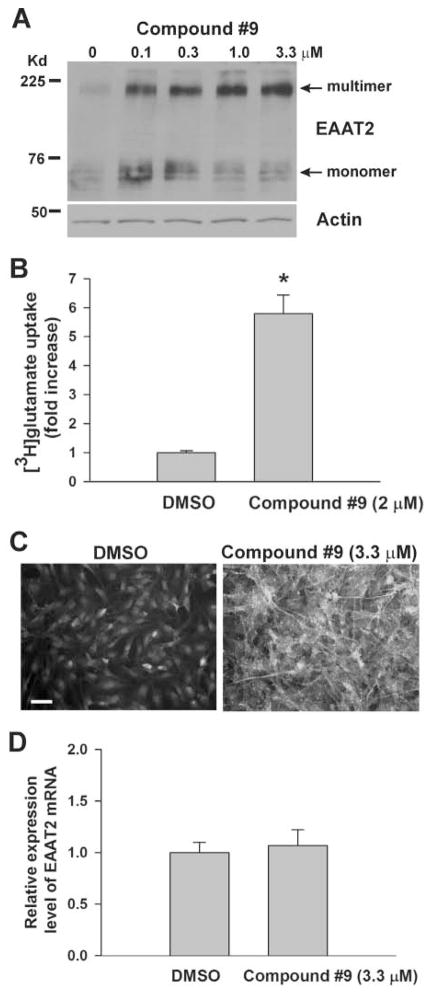
The next step was to evaluate the effects of these compounds on EAAT2 protein levels by Western blot analysis, glutamate transport functions, EAAT2 protein localization, and EAAT2 mRNA levels. PA-EAAT2 cells were treated with varying concentrations of compound for 72 h and then harvested for analysis. The Western blot results were generally consistent with the 12-point dose-response results (Fig. 5A shows results for compound 9). [3H]-glutamate uptake activity was consistent with the induced EAAT2 protein levels, indicating that the induced EAAT2 protein had glutamate uptake activity (Fig. 5B shows results for compound 9). Immunofluorescent labeling of the cells showed that the induced EAAT2 was properly localized to the plasma membrane (Fig. 5C shows results for compound 9). PA-EAAT2 cells endogenously express the other glial glutamate transporter, EAAT1. We found that these compounds did not increase EAAT1 protein expression (not shown), indicating the specificity of their effect. Our screening assay was designed to identify compounds that are able to stimulate translation of EAAT2 transcripts. As expected, EAAT2 mRNA levels were not increased as measured by quantitative real-time RT-PCR analysis (Fig. 5D shows the result for compound 9), demonstrating that the increased EAAT2 protein expression resulted from an increase in translation.
FIG. 5.
Confirmation of the hits. PA-EAAT2 cells were treated with indicated concentrations of compound 9 in Dulbecco’s modified Eagle’s medium (DMEM) for 72 h and then harvested (A) for determining EAAT2 protein levels by Western blotting (equal protein loading was confirmed by Ponceau S staining and also by reprobing the blots with antiactin antibodies), (B) for measuring glutamate transport function by [3H]-glutamate uptake assay, (C) for determining induced EAAT2 protein cellular localization by immunofluorescent staining using anti-EAAT2 antibodies (scale bar, 50 μm), and (D) for measuring EAAT2 mRNA levels by real-time RT-PCR analysis. For glutamate uptake and RT-PCR data, the fold increase of the compound treated cells was normalized to the DMSO-treated cells.
DISCUSSION
Glutamate-mediated excitotoxicity has been implicated in many neuropathological states. Enhanced glutamate reuptake by increased EAAT2 protein expression is one potential approach to protect against excitotoxic neuronal damage. EAAT2 protein expression is regulated at both transcriptional and translational levels. Ceftriaxone, a β-lactam antibiotic, was found to increase transcription of the GLT-1 gene (EAAT2 gene) through an NF-κB-mediated mechanism, subsequently increasing EAAT2 protein expression.19,27 Ceftriaxone is currently in clinical trials. However, several reports indicate that ceftriaxone only slightly induces EAAT2 protein expression or glutamate uptake activity in mice by intraperitoneal injection.28 The assay reported here is unique in that it is designed to identify compounds that increase the translation of EAAT2 transcripts that are silenced by long 5′ UTRs.
We developed and used a cell-based ELISA capable of screening large compound libraries for molecules that can induce translational read-through of silenced EAAT2 transcripts under the control of long 5′ UTRs. Of the 140,000 compounds screened, 293 were identified as hits, with 61 compounds showing a dose-dependent increase in EAAT2 expression. Of these 61 compounds, 3 have been selected as lead compounds based on potency (EC50 <1 μM), lack of toxicity, and chemical tractability for medicinal chemistry. These lead compounds are currently undergoing chemical optimization. Importantly, the rate of compound-induced EAAT2 protein expression as measured by ELISA correlated strongly with that measured by Western blot analysis (Fig. 5A), indicating that the assay reliably determines expression levels of EAAT2 in the presence of compound. In addition, proper localization and function of the induced EAAT2 was determined by immunofluorescent staining (Fig. 5C) and [3H]-glutamate uptake assay, respectively (Fig. 5B). Initial data also indicate that some of these compounds can increase EAAT2 protein expression in brain and spinal cord in vivo (publication in preparation).
Collectively, these data demonstrate that further development of the lead compounds identified in this study should lead to the development of not only useful pharmacological tools to study the translational silencing of EAAT2 transcripts with long 5′ UTRs but also drugs capable of preventing excitotoxicity observed in many neurodegenerative diseases as well as other neurological disorders.
Acknowledgments
This work was supported by NIH grants (U24 NS049339 and R01 NS064275), the Alzheimer’s Association, and the Neuroscience Education and Research Foundation.
References
- 1.Danbolt NC. Glutamate uptake. Prog Neurobiol. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 2.Rothstein JD, Dykes-Hoberg M, Pardo CA, Bristol LA, Jin L, Kuncl RW, et al. Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron. 1996;16:675–686. doi: 10.1016/s0896-6273(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 3.Sattler R, Tymianski M. Molecular mechanisms of glutamate receptor-mediated excitotoxic neuronal cell death. Mol Neurobiol. 2001;24:107–129. doi: 10.1385/MN:24:1-3:107. [DOI] [PubMed] [Google Scholar]
- 4.Choi DW. Ionic dependence of glutamate neurotoxicity. J Neurosci. 1987;7:369–379. doi: 10.1523/JNEUROSCI.07-02-00369.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Den Bosch L, Vandenberghe L, Klaassen H, Van Houtte E, Robberecht W. Ca(2+)-permeable AMPA receptors and selective vulnerability of motor neurons. J Neurol Sci. 2000;180:29–34. doi: 10.1016/s0022-510x(00)00414-7. [DOI] [PubMed] [Google Scholar]
- 6.Carriedo SG, Sensi SL, Yin HZ, Weiss JH. AMPA exposures induce mitochondrial Ca(2+) overload and ROS generation in spinal motor neurons in vitro. J Neurosci. 2000;20:240–250. doi: 10.1523/JNEUROSCI.20-01-00240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urushitani M, Nakamizo T, Inoue R, Sawada H, Kihara T, Honda K, et al. N-methyl-D-aspartate receptor-mediated mitochondrial Ca(2+) overload in acute excitotoxic motor neuron death: a mechanism distinct from chronic neurotoxicity after Ca(2+) influx. J Neurosci Res. 2001;63:377–387. doi: 10.1002/1097-4547(20010301)63:5<377::AID-JNR1032>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 8.Arundine M, Tymianski M. Molecular mechanisms of glutamate-dependent neurodegeneration in ischemia and traumatic brain injury. Cell Mol Life Sci. 2004;61:657–668. doi: 10.1007/s00018-003-3319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujikawa DG. Prolonged seizures and cellular injury: understanding the connection. Epilepsy Behav. 2005;7(suppl 3):S3–S11. doi: 10.1016/j.yebeh.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Estrada Sanchez AM, Mejia-Toiber J, Massieu L. Excitotoxic neuronal death and the pathogenesis of Huntington’s disease. Arch Med Res. 2008;39:265–276. doi: 10.1016/j.arcmed.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Hazell AS. Excitotoxic mechanisms in stroke: an update of concepts and treatment strategies. Neurochem Int. 2007;50:941–953. doi: 10.1016/j.neuint.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 12.Koutsilieri E, Riederer P. Excitotoxicity and new antiglutamatergic strategies in Parkinson’s disease and Alzheimer’s disease. Parkinsonism Relat Disord. 2007;13(suppl 3):S329–S331. doi: 10.1016/S1353-8020(08)70025-7. [DOI] [PubMed] [Google Scholar]
- 13.Van Den Bosch L, Van Damme P, Bogaert E, Robberecht W. The role of excitotoxicity in the pathogenesis of amyotrophic lateral sclerosis. Biochim Biophys Acta. 2006;1762:1068–1082. doi: 10.1016/j.bbadis.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Doble A. The pharmacology and mechanism of action of riluzole. Neurology. 1996;47:S233–S241. doi: 10.1212/wnl.47.6_suppl_4.233s. [DOI] [PubMed] [Google Scholar]
- 15.Danysz W, Parsons CG. The NMDA receptor antagonist memantine as a symptomatological and neuroprotective treatment for Alzheimer’s disease: preclinical evidence. Int J Geriatr Psychiatry. 2003;18:S23–S32. doi: 10.1002/gps.938. [DOI] [PubMed] [Google Scholar]
- 16.Reisberg B, Doody R, Stoffler A, Schmitt F, Ferris S, Mobius HJ. Memantine in moderate-to-severe Alzheimer’s disease. N Engl J Med. 2003;348:1333–1341. doi: 10.1056/NEJMoa013128. [DOI] [PubMed] [Google Scholar]
- 17.Tian G, Lai L, Guo H, Lin Y, Butchbach ME, Chang Y, et al. Translational control of glial glutamate transporter EAAT2 expression. J Biol Chem. 2007;282:1727–1737. doi: 10.1074/jbc.M609822200. [DOI] [PubMed] [Google Scholar]
- 18.Guo H, Lai L, Butchbach ME, Stockinger MP, Shan X, Bishop GA, et al. Increased expression of the glial glutamate transporter EAAT2 modulates excitotoxicity and delays the onset but not the outcome of ALS in mice. Hum Mol Genet. 2003;12:2519–2532. doi: 10.1093/hmg/ddg267. [DOI] [PubMed] [Google Scholar]
- 19.Rothstein JD, Patel S, Regan MR, Haenggeli C, Huang YH, Bergles DE, et al. Beta-lactam antibiotics offer neuroprotection by increasing glutamate transporter expression. Nature. 2005;433:73–77. doi: 10.1038/nature03180. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y, Tian G, Roman K, Handy C, Travers JB, Lin CL, et al. Increased glial glutamate transporter EAAT2 expression reduces visceral nociceptive response in mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G129–G134. doi: 10.1152/ajpgi.90556.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu K, Lee ST, Sinn DI, Ko SY, Kim EH, Kim JM, et al. Pharmacological induction of ischemic tolerance by glutamate transporter-1 (EAAT2) upregulation. Stroke. 2007;38:177–182. doi: 10.1161/01.STR.0000252091.36912.65. [DOI] [PubMed] [Google Scholar]
- 22.Piazza G, White E, Sneed B, Kushner N, Harwell J, Jones A. Development and implementation of a HTS Assay for inducers of the excitatory amino acid transporter-2 (EAAT-2) www.southernresearch.org/pdf/EEAT-2-Poster-9-2-05.pdf.
- 23.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 24.Guo H, Lai L, Butchbach ME, Lin CL. Human glioma cells and undifferentiated primary astrocytes that express aberrant EAAT2 mRNA inhibit normal EAAT2 protein expression and prevent cell death. Mol Cell Neurosci. 2002;21:546–560. doi: 10.1006/mcne.2002.1198. [DOI] [PubMed] [Google Scholar]
- 25.Butchbach ME, Tian G, Guo H, Lin CL. Association of excitatory amino acid transporters, especially EAAT2, with cholesterol-rich lipid raft micro-domains: importance for excitatory amino acid transporter localization and function. J Biol Chem. 2004;279:34388–34396. doi: 10.1074/jbc.M403938200. [DOI] [PubMed] [Google Scholar]
- 26.Butchbach ME, Guo H, Lin CL. Methyl-beta-cyclodextrin but not retinoic acid reduces EAAT3-mediated glutamate uptake and increases GTRAP3-18 expression. J Neurochem. 2003;84:891–894. doi: 10.1046/j.1471-4159.2003.01588.x. [DOI] [PubMed] [Google Scholar]
- 27.Lee SG, Su ZZ, Emdad L, Gupta P, Sarkar D, Borjabad A, et al. Mechanism of ceftriaxone induction of excitatory amino acid transporter-2 expression and glutamate uptake in primary human astrocytes. J Biol Chem. 2008;283:13116–13123. doi: 10.1074/jbc.M707697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melzer N, Meuth SG, Torres-Salazar D, Bittner S, Zozulya AL, Weidenfeller C, et al. A beta-lactam antibiotic dampens excitotoxic inflammatory CNS damage in a mouse model of multiple sclerosis. PLoS ONE. 2008;3:e3149. doi: 10.1371/journal.pone.0003149. [DOI] [PMC free article] [PubMed] [Google Scholar]