Abstract
The mechanism by which the inner cell mass (ICM) and trophectoderm (TE) become specified is poorly understood. Considerable species variation is evident in the expression of lineage-specific and embryonic stem cell (ESC) regulatory markers. We sought to investigate localization patterns of these markers in rhesus macaque compact morulae and blastocysts. NANOG protein was restricted to the ICM of blastocysts. In contrast to a previous report, the expression of CDX2 was detected in the primate blastocyst, localized specifically to the TE. Unlike the mouse embryo, OCT4 protein was detected using two different antibodies in both the ICM and TE. The ubiquitous pattern of OCT4 expression is consistent with observations in human, cow, and pig embryos. Significantly, lack of restricted OCT4 protein, and ICM localization of NANOG in primate blastocysts, suggests that NANOG may determine inner cell mass fate more specifically during primate development or may be less susceptible to culture artifacts. These results contrast markedly with current mechanistic hypotheses, although other factors may lie upstream of NANOG to constitute a complex interactive network. This difference may also underlie observations that regulatory mechanisms in ESC differ between mice and primates.
Introduction
Maintenance of pluripotency in embryonic stem cells (ESC) is regulated by specific transcription factors that are activated during preimplantation embryonic development. Following fertilization, the cleaving zygote undergoes the first lineage decision, forming the outer trophectoderm (TE) cells that enclose the inner cell mass (ICM). Long-standing models of how the embryo regulates the differentiation of the ICM and TE propose that cell position drives cell fate, the “inside outside” hypothesis [1]; or, conversely, that cell fate drives cell position, the cell polarity hypothesis [2] (reviewed by [3]).
The prevailing molecular model of lineage specification (Fig. 1A; [4]) highlights the importance of the POU domain transcription factor OCT4 (also known as OCT3/4 and POU5F1). OCT4 is expressed throughout the early embryo until the blastocyst stage, when its expression becomes restricted to the ICM in the mouse [5]. While OCT4 null mouse embryos appear to form normal blastocysts, with both TE- and ICM-like cell compartments, the embryos die around the time of implantation, possessing only TE-like cells [6]. These results suggest that OCT4 is required for ICM maintenance, but is not essential for initial specification. In contrast, CDX2 (caudal-related transcription factor 2) is restricted to the TE by the late morula stage in the mouse [7,8]. In the absence of CDX2, the blastocyst forms, but a functional TE is not established and the embryo dies prior to implantation [8], with OCT4 and NANOG expression detected throughout the embryo. These data suggest that CDX2 plays a role in overriding the ICM fate, but is not required for TE specification. Reciprocal inhibition of OCT4 and CDX2 was evident in a stem cell model of early development. Specifically, an increase in OCT4 lead to decreased CDX2 expression, while overexpression of CDX2 reduced OCT4 expression [7]. These data have supported the model in which OCT4 and CDX2 act as “selector genes” for ICM and TE fates and negatively regulate each other to promote the segregation of the two lineages. However, recent studies report CDX2 expression and TE specification appear to be regulated by the transcriptional regulator TEAD4 [9,10].
FIG. 1.
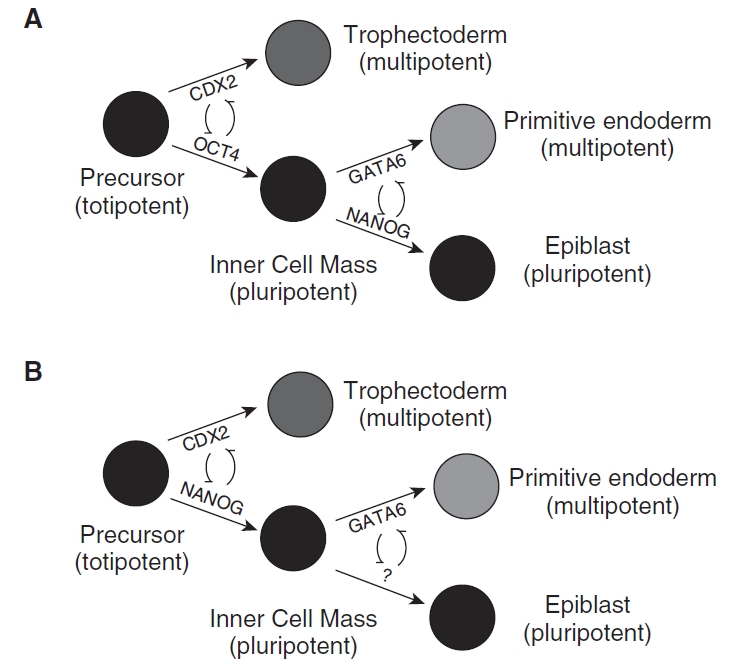
Proposed models of early lineage specification in the mouse (A; adapted from [4]) and non-human primate (B).
Stem cells derived from either lineage likewise express the respective markers. The expression of OCT4 is commonly used as a measure of ESC pluripotency [11,12], while CDX2 is a marker for trophoblast stem cells (TSC) [13]. NANOG is a second ICM-specific transcription factor identified in ESC [14,15]. Loss of NANOG expression in ESCs is associated with loss of pluripotency, and differentiation toward primitive endoderm [15]. Mutant embryos fail to develop an ICM [14] and form only TE and primitive endoderm, supporting a role of NANOG in regulating epiblast cell fate. KLF4 has also been identified as a necessary regulator of stem cell maintenance [16], as have a number other factors; however, its regulation during preimplantation development has not been investigated. Evidence suggests that OCT4 expression in non-murine embryos, including the human, is not restricted to the ICM [17–19] in vitro or in vivo, possibly reflecting differences in the mechanism responsible for formation of the ICM. Interestingly, a recent report [20] described the derivation of TSC from rhesus macaque blastocysts that lack CDX2 expression, which is surprising, as human embryos express CDX2 [21,22], and other species of embryos display a similar TE-specific localization [23]. However, a more recent review [24] suggests that CDX2 is likely localized to the TE in rhesus blastocysts.
Little is known about the expression of lineage-specific markers in rhesus blastocysts, with the exception of OCT3/4 [25], which appears to display localization similar to that of the mouse. Previous studies have not combined detection of multiple markers of pluripotency and TE-specific CDX2 expression in primates. Therefore, we sought to examine the expression patterns of markers of lineage specification and/or ESC maintenance in rhesus macaque morulae and blastocysts during the period of lineage divergence.
Materials and Methods
Unless otherwise stated, chemicals were obtained from Sigma-Aldrich (St. Louis, MO).
Controlled rhesus macaque hormonal ovarian stimulation
All procedures were performed according to the Institutional Animal Care and Animal Use Committee protocols approved at the Oregon National Primate Research Center (ONPRC) and the Caribbean Primate Research Center (CPRC), Puerto Rico. Ovarian stimulation was carried out as previously described [26]. In brief, rhesus macaques received a sequential regimen of recombinant human gonadotropins to support follicular growth. Female macaques at ONPRC received intramuscular (IM) injections of 30 IU recombinant human follicule-stimulating hormone (FSH) (Organon, Oss, The Netherlands) twice daily for 7 days, followed by 2 days of 30 IU each of recombinant human FSH and recombinant human luteinizing hormone (LH) (EMD Serono, Rockland, MA) twice daily. During the final 3 days of the recombinant human FSH treatment, animals also received the GnRH antagonist acycline (0.075 mg/kg animal body weight) to prevent a spontaneous LH surge. Ovarian stimulation of rhesus macaques situated at the CPRC was performed by administration of recombinant human FSH (37.5 IU per injection, twice daily, IM; Organon, Roseland, NY) for 8 days, followed by 2 days of recombinant human FSH and recombinant hCG EMD, Serono; 37.5 IU recombinant human FSH and 100 IU recombinant hCG per injection, twice daily, IM). At both primate centers, on the final day of recombinant human FSH treatment, 32–33 h before follicular aspiration, oocyte maturation was induced with a single injection of recombinant hCG (750–1,000 IU, IM).
Rhesus macaque oocyte and sperm collection, insemination, and embryo culture
Procedures for oocyte recovery, sperm collection, insemination, and embryo culture have been described previously [27–29]. Briefly, follicular fluid aspirates were collected in TALP-HEPES containing 0.3% bovine serum albumin (BSA). The aspirates were sifted through a cell strainer (Becton-Dickinson, Franklin Lakes, NJ), rinsed, and oocytes were collected from the resulting suspension. After cumulus removal using hyaluronidase (0.03%), oocytes were rinsed and placed in culture drops of TALP medium supplemented with BSA [30] and incubated in 5% CO2 in air at 37°C until insemination. Semen was obtained by electroejaculation [31]. Seminal plasma was removed according to standard protocols and obtained spermatozoa were activated with 1 mM each of cyclic AMP and caffeine. Sperm were used for insemination at a final concentration of 1.5–2.0 × 105 sperm per milliliter [28,32]. The presence of pronuclei was assessed 18–20 h after insemination. Presumptive zygotes were transferred to culture drops of amino acid-supplemented Hamster Embryo Culture Medium 6 (HECM-6aa) [33] and incubated at 5% CO2 in air at 37°C. The culture medium was refreshed every 48 h with HECM-9aa supplemented with 5% fetal bovine serum (FBS).
Embryonic stem cell culture
To confirm the specificity of the antibodies used, rhesus macaque ESC were cultured as previously described [34] and stained as outlined below for each marker. Briefly, Ormes 22 ESCs cultured as previously described were grown on mitotically inactivated mouse embryonic fibroblast (MEF) feeder cells in Dulbecco’s Modified Eagle Medium (DMEM/F12) (Invitrogen, Grand Island, NY) supplemented with 15% FBS (Hyclone, Logan, UT), 0.1 mM β-mercaptoethanol, 1% nonessential amino acids (Invitrogen, Grand Island, NY), 2 mM l-glutamine (Invitrogen, Grand Island, NY), and 4 ng/mL FGF2 (Sigma), at 37°C under a 5% CO2-balanced air atmosphere.
Trophoblast cell culture
Mouse TSC isolated from E3.5 blastocysts or E6.5 extraembryonic ectoderm provided as a gift by Dr. Janet Rossant were cultured as previously described [13] and plated on slides for immunofluorescent localization of CDX2 to confirm antibody specificity.
Immunofluorescence
Immunofluorescence of compact morulae and various blastocyst stages was carried out as described elsewhere [35]. At least 10 embryos (from a minimum of five replicates) were analyzed from each stage. Compact morulae were collected at 120 h post-insemination and blastocysts were collected at 164 and 214 h post-insemination, fixed in 4% paraformaldehyde (PFA), and stained immediately, or stored in 0.4% PFA at 4°C for no longer than 1 week prior to staining. Primary antibodies were obtained from R&D Systems (NANOG, OCT3/4, KLF4; Minneapolis, MN), BioGenex (CDX2; San Ramon, CA), and Santa Cruz Biotechnology (OCT4; Santa Cruz, CA). Embryos were subsequently incubated with Cy-3-conjugated donkey anti-goat IgG, or donkey anti-mouse IgG. Embryos were counterstained for 1 min with 10 µg/mL DAPI (Calbiochem, La Jolla, CA) and mounted in Vectoshield (Vector Laboratories, Burlingame, CA). Mounted embryos were examined using an Olympus BX41 fluorescence microscope equipped with a DP71 color camera. Controls were performed by omission of the primary antibody and use of the species-specific IgG.
Results and Discussion
Maintenance of pluripotency in ESC is regulated by a core group of transcription factors whose expression is activated during preimplantation embryonic development. While the interplay between OCT4 and CDX2 is thought to regulate ICM and TE fate, the mechanism of lineage specification in mammalian embryos remains poorly understood. A large proportion of our understanding comes from studies of mouse development, leading to the current model of lineage specification regulated in part by the transcription factors OCT4 and CDX2 (Fig. 1A). Genetic ablation of OCT4 or CDX2 prevents implantation, resulting from the lack of the establishment of a functional ICM or TE, respectively. However, in both cases nonfunctional ICM or TE formation is initiated with deficient expression of the opposing transcription factor, suggesting that each factor is required for subsequent maintenance rather than initial specification.
Consistent with this view, OCT4 expression was not exclusively localized to the ICM of primate blastocysts at 164 h post-insemination (Fig. 2D). To determine whether OCT4 localization became more specific at a later stage, embryos were cultured to 214 h post-insemination. OCT4 localization became more restricted at 214 h (Fig. 2G), although weak staining was still present in the TE in all embryos examined. Blastocysts examined at 164 and 214 h post-insemination displayed an ICM-specific localization pattern for NANOG (Fig. 3). CDX2 protein was localized to the TE at both 164 and 214 h post-insemination (Fig. 4). KLF4 protein expression was not specific to either lineage at any stage (Fig. 5). OCT4, NANOG, CDX2, and KLF4 were detected ubiquitously in compact morula stage embryos (Figs. 2–5). Examination of protein expression during cleavage stages demonstrated nuclear localization of OCT4 only in 16+ cell embryos (Supplementary Fig. 1; Supplementary materials are available online at http://www.liebertpub.com). Antibody specificity was confirmed by analyzing monkey ESC (Figs. 2, 3, and 5), which displayed nuclear localization of OCT4, NANOG, and KLF4, respectively. Non-pluripotent MEFs were not labeled by these antibodies. Mouse TSC expressed CDX2 in some, but not all, nuclei (Fig. 4). Negative controls stained without primary antibody, or using the corresponding species of non-immune IgG, were negative for staining (Supplementary Fig. 1).
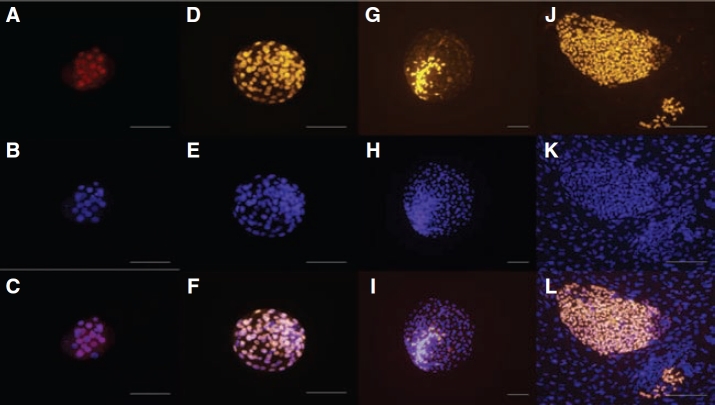
FIG. 2.
Localization of OCT4 in nuclei ofrhesus macaque compact morulae (A–C), and blastocysts cultured to 164 h (D–F) or 214 h (G–I) post-insemination. A Cy-3-conjugated antibody was used to detect protein expression (A, D, and G). Nuclei were counterstained with DAPI (B, E, and H). Merged images are presented (C, F, and I). Nuclear localization of OCT4 in Ormes 22 rhesus macaque embryonic stem cells (J–L). Surrounding mouse embryonic fibroblasts remained negative. Bars = 200 µm.
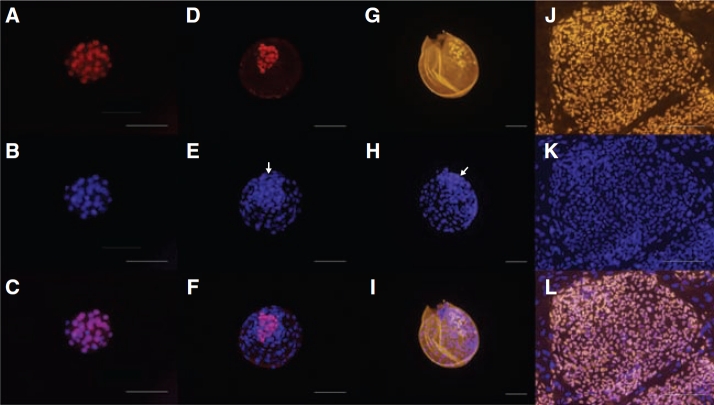
FIG. 3.
NANOG localization in rhesus macaque compact morulae (A–C), and the inner cell mass (ICM; arrowheads) of blastocysts cultured to 164 h (D–F) or 214 h (G–I) post-insemination. A Cy-3-conjugated antibody was used to detect protein expression (A, D, and G). Nuclei were counterstained with DAPI (B, E, and H). Merged images are presented (C, F, and I). Nuclear localization of NANOG in Ormes 22 rhesus macaque embryonic stem cells (J–L). Surrounding mouse embryonic fibroblasts remained negative. Bars = 200 µm.
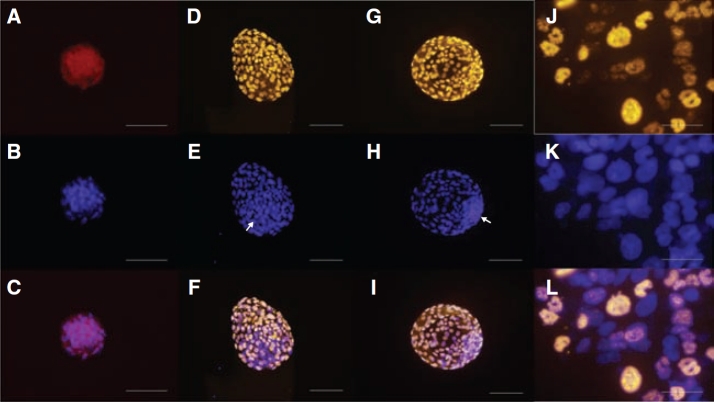
FIG. 4.
Nuclear localization of CDX2 in rhesus macaque compact morula (A–C), and blastocysts cultured to 164 h (D–F) and 214 h (G–I) post-insemination. A Cy-3-conjugated secondary antibody was used to detect protein expression (A, D, and G). Nuclei were counterstained with DAPI (B, E, and H). Arrowheads indicate location of the ICM. Merged images are presented (C, F, and I). Nuclear localization of CDX2 in mouse trophoblast stem cells (J–L). Bars = 200 µm.
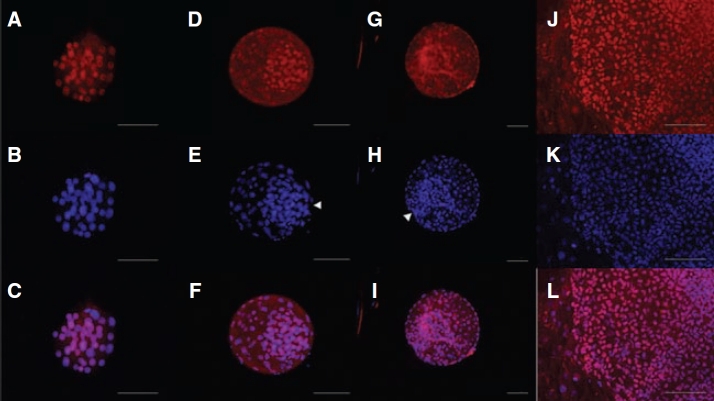
FIG. 5.
Nuclear localization of KLF4 in rhesus macaque compact morulae (A–C) and blastocysts cultured to 164 h (D–F) and 214 h (G–I) post-insemination. A Cy-3-conjugated secondary antibody was used to detect protein expression (A, D, and G). Nuclei were counterstained with DAPI (B, E, and H). Merged images are presented (C, F, and I). Arrowheads indicate location of the ICM. Nuclear localization of KLF4 in Ormes 22 rhesus macaque embryonic stem cells (J–L). Surrounding mouse embryonic fibroblasts remained negative. Bars = 200 µm.
Previous studies in several species have documented both TE and ICM localization of OCT4, including early stages of mouse preimplantation embryo development [5], and in human preimplantation embryos [22,36]. Table 1 summarizes the distribution of OCT4, NANOG, CDX2, and KLF4 in preimplantation embryos of several mammalian species found in this and prior reports. TE and ICM localization of OCT4 has been demonstrated in both bovine and porcine blastocysts derived in vivo or cultured in vitro [19,37], as well as in human blastocysts [17,18]. However, these results contrast with those of Mitalipov et al. [25] where OCT4 expression in rhesus macaque expanded and hatched blastocysts, cultured to 164 h post-insemination, displayed an ICM-specific localization. This result was never observed in the present study, despite use of the same culture methods and antibody (Santa Cruz, CA), although Mitalipov et al. [25] utilized intracytoplasmic sperm injection for fertilization, while this study relied on standard in vitro fertilization methods. Unrestricted expression of OCT4 protein during in vitro embryo culture may reflect an inability to respond to appropriate environmental stimuli, including growth factors, although this remains to be examined. Slight alterations in the number of embryos expressing OCT4 (and other) transcripts has been shown in human embryos associated with differences in growth factor supplementation of culture media [22]. Results of the present study suggest that OCT4 may be more sensitive to culture conditions, while NANOG is less affected, localizing specifically to the ICM regardless of the culture environment. Furthermore, all embryos analyzed may have exhibited inappropriate OCT4 expression predictive of poor developmental ability. This may be reflected in the rates of ESC derivation reported for the monkey (27%) [11], and has been correlated with ESC derivation success in the mouse [38]. It should be noted that mouse in vitro-cultured embryos can display an unrestricted pattern of OCT4 expression (TE and ICM) during blastocyst formation, compared with equivalent in vivo-collected embryos [5], perhaps indicating suboptimal culture conditions. This may be the case in the rhesus macaque where culture to the blastocyst stage still relies on the use of serum. However, in vivo produced porcine embryos also display OCT4 expression throughout both the ICM and TE compartments [23]. The ratio of OCT4:CDX2 may be important for the proper delineation of the TE and maintenance of pluripotency within the ICM, but may not be essential for the initial specification of the ICM. In mice, TEAD4 appears to provide the signal that specifies TE prior to blastocyst formation upstream of CDX2 [9,10]. A comparable protein has not been identified for ICM specification, perhaps indicating that ICM is the default state.
Table 1.
Summary of Lineage-Specific Protein Detection in Preimplantation Embryos of Different Species
| Marker | Species | Origin | Stages analyzed | Localization (stage) | References |
|---|---|---|---|---|---|
| OCT3/4 | Mouse | In vivo | GV-HB | Detected 8c+ | [25] |
| eB-XB: ICM+ weak TE | |||||
| HB: ICM | |||||
| In vivo | eB-XB | eB-B: ICM + weak TE | [19] | ||
| XB: ICM | |||||
| In vivo | MII-XB | eB: ICM+TE | [5] | ||
| BL: ICM, weak TE | |||||
| XB: ICM | |||||
| Pig | In vitro | CM-XB | CM: +ve | [23] | |
| In vivo | CM: absent | ||||
| In vitro + in vivo | BL-XB: TE+ICM | ||||
| In vitro + in vivo | XB | TE+ICM | [19] | ||
| Cow | In vitro | GV-HB | 8c+: ICM+TE | [37] | |
| In vitro + in vivo | XB | ICM+TE | [19] | ||
| Rhesus | In vitro | GV-HB | XB-HB: ICM | [25] | |
| 16c-HB | CM-HB: ICM+TE | This study | |||
| HB | ICM, TE weak | [20] | |||
| Human | In vitro | Zyg-XB | 8c+; ICM+TE | [53] | |
| GV-hB | 4c+; ICM+TE | [17] | |||
| NANOG | Mouse | In vitro | 16c-eB | eB: ICM | [8] |
| Pig | In vitro + in vivo | M+BL | undetectable | [23] | |
| Cow | In vitro | XB | ICM only | [23] | |
| Rhesus | In vitro | 16c-HB | eB-HB: ICM | This study | |
| Human | In vitro | GV-XB | EB+, ICM+TE | [36] | |
| CDX2 | Mouse | In vitro | 8c+ | Detected M+ | [8] |
| XB: TE only | |||||
| Pig | In vitro | XB | TE only | [23] | |
| Cow | In vitro | XB | TE only | [23] | |
| Rhesus | In vitro | HB | TE only | [24] | |
| 16c-HB | eB-HB: TE | This study | |||
| KLF4 | CM-HB | ICM+TE | This study |
Abbreviations: 4c/8c/16c, four/eight/sixteen cell embryo; BL, blastocyst; CM, compact morula; eB, early blastocyst; GV, germinal vesicle; HB, hatched blastocyst; ICM, inner cell mass; MII, metaphase II oocyte; M, morula; ND, not determined; +ve, positive; TE, trophectoderm; XB, expanded blastocyst.
Unlike OCT4, NANOG expression was restricted to the ICM, suggesting that localization of NANOG precedes that of OCT4 in the primate. A recent study of human preimplantation embryos by Cauffman et al. [36] documented similar, although variable, ICM-specific localization of NANOG at the expanded blastocyst stage. Kimber et al. [22] also demonstrate ICM-specific localization of NANOG in human blastocysts. Expression of NANOG was not detected prior to blastocyst expansion [36], contrasting that observed in rhesus compact morulae in the present study. SOX2 expression in human embryos was localized largely to ICM nuclei of expanded human blastocysts [36], but was detected in all blastomeres in early blastocysts [22,36] and in early stages [36]. Considering previous localization of OCT4 protein expression in human embryos to both the TE and ICM, results suggest that both SOX2 and NANOG are specified in the ICM upstream of OCT4 in primates. However, a cooperative interaction between OCT4 and SOX2 has been proposed to regulate NANOG [39], based on a study performed in mouse ESC. Other studies show cooperation between all three transcription factors [40]; however, OCT4 appears not to initiate the signaling cascade in primate embryos. The regulatory importance of NANOG is supported by mouse knockout studies, in which OCT4 ablation results in the formation of an ICM, albeit with TE properties [6], while NANOG ablation results in the complete absence of an ICM [14]. However, Dietrich and Hiiragi [41] have reported mosaic NANOG expression in mouse embryos. ESC deficient in NANOG fail to maintain pluripotency and undergo differentiation [15]. Additionally, selection of induced pluripotent stem cells was improved by the use of NANOG instead of Fbx15 [42,43]. Recently, upstream regulators of NANOG have been found in ESC, including Med12 (mediator complex 12) [44] and SMAD signaling [45]. Whether these factors play a role in regulating lineage specification in mammalian embryos requires investigation. Recent studies of early lineage specification have largely focused on the involvement of cell polarity in regulating cell position [46], as a potential activator of transcription factor responses.
Niwa et al. [7] demonstrated that the ratio of OCT4 to CDX2 was critical in mice for determining the differentiation of TE and ICM. Whether this is also true for non-human primates requires further examination. Primate embryo lineage specification may instead rely on a reciprocal relationship between NANOG and CDX2, with upstream effectors up-regulating these factors. In contrast to a previous report [20], CDX2 was detected in the TE of developing primate embryos in the present study (as cited by [24]). Significantly, Vandevoort et al. [20] reported the derivation of rhesus TSC lacking CDX2 at both the transcript and protein level. While the authors tested two antibodies, which also failed in the present study (data not shown), the antibody that successfully detected CDX2 has been used extensively in other reports [7,41,46,47]. A recent review by Vandevoort et al. supports our current findings, documenting CDX2 localization in the TE of rhesus macaque blastocysts [24]. Therefore, of the transcription factors associated with lineage specification analyzed in the present study, only NANOG and CDX2 displayed lineage-specific localization in rhesus blastocysts.
KLF4 expression has been associated with the maintenance of pluripotency in human ES cells [16], and has been used as one of the four transcription factors required to induce pluripotency in somatic cells [43]. Its localization during early embryo development has not been reported previously. Here we demonstrate that KLF4 does not localize to a specific compartment within the primate embryo. KLF4 cooperates with OCT4 and SOX2 to regulate pluripotency [48]. However, the function of KLF4 during early development and its specific role in regulating pluripotency are less clear. Based on its lack of cell specificity, KLF4 may play a role in regulating both embryonic and TSC populations, or it could participate in the potentiation of genes that regulate pluripotency. Whether KLF4 controls later differentiation steps, or in combination with other KLFs regulates the core transcription factors, should be explored with the use of knockout mouse models. Rather than an activating role in reprogramming, KLF4 may act to repress transcription [49] in support of a nonspecific localization pattern in preimplantation embryos. Examination of additional transcription factors, including GATA6 and SOX2, could delineate the regulatory mechanisms that control lineage specification in the primate.
The dynamic process of cell fate specification is modulated by regulatory gene networks. Results of this study demonstrate, for the first time, TE-specific localization of CDX2 in primate blastocysts. Restricted ICM-specific localization of NANOG suggests that it is a potent indicator of ICM pluripotency. The unrestricted expression pattern of OCT4 at the blastocyst stage in primates may represent a key regulatory distinction between the human and mouse that is also reflected by differences in ESC maintenance, whereby mouse ESC are dependent on leukemia inhibitory factor (LIF) and bone morphogenic protein (BMP) [50], while these factors are not sufficient to maintain pluripotency, or result in differentiation, of human ESC [51]. Recent analysis of mouse epiblast stem cells (EpiSC) isolated under human ESC culture conditions (in the presence of bFGF and activin A) has documented higher expression levels of both NANOG and OCT4 [52] than observed in mouse ESC, as well as an ability to generate TE that has not been achieved from mouse ESC [52]. Likewise, EpiSC lack expression of Rex1 and alkaline phosphatase, known markers of ESC and ICM. Whether these markers are expressed in rhesus ICM is yet to be determined, but raises the question as to whether primate ESC truly represent their ICM counterparts or are epiblast in nature.
Our data suggest that the murine model of lineage determination may not accurately mirror other species. NANOG and CDX2, rather than OCT4 and CDX2, may interact to repress each other in each respective lineage in the primate (Fig. 1B). This is supported by a lack of ICM-specific localization of OCT4, preceded by ICM- and TE-specific localization of NANOG and CDX2, respectively. In this respect, NANOG may antagonize the activity of CDX2, and vice versa, to maintain ICM and TE lineages. Downstream specification of primitive endoderm and epiblast may be mediated by GATA6, although this was not examined in the present study. Further molecular analysis is required to elucidate the interactive network and upstream mediators of this first differentiation step.
Supplementary Material
Contributor Information
A.J. Harvey, Department of Physiology, Wayne State University, Detroit, Michigan.; Department of Obstetrics and Gynecology, Wayne State University, Detroit, Michigan.
D.R. Armant, Department of Obstetrics and Gynecology, Wayne State University, Detroit, Michigan. Department of Anatomy and Cell Biology, Wayne State University, Detroit, Michigan. National Institutes of Health, NICHD, Reproductive Biology and Medicine Branch, Bethesda, Maryland.
B.D. Bavister, Department of Obstetrics and Gynecology, Wayne State University, Detroit, Michigan. Caribbean Primate Research Center, Sabana Seca, Puerto Rico.
S.M. Nichols, Reproductive Biology Program, Caribbean Primate Research Center, Unit of Comparative Medicine, University of Puerto Rico, Sabana Seca, Puerto Rico.
C.A. Brenner, Department of Physiology, Wayne State University, Detroit, Michigan. Department of Obstetrics and Gynecology, Wayne State University, Detroit, Michigan.
Acknowledgments
Preliminary data relating to this study were presented at the International Society for Stem Cell Research, Philadelphia, PA, 2008, and the Society for Gynecological Investigation, San Diego, CA, 2008.
We thank Dr. Janet Rossant at the Hospital for Sick Children and University of Toronto for generously providing the TSC line used in this study. Supported, in part, by the intramural research program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, DHHS and NIH Grants HD045966, RR015395, RR021881, and HD046553. Support was also provided by Wayne State University through the President’s Research Enhancement Program.
Author Disclosure Statement
There are no conflicts of interest relating to this work for any of the authors.
References
- 1.Tarkowski AK, Wroblewska J. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J Embryol Exp Morphol. (1967);18:155–180. [PubMed] [Google Scholar]
- 2. Johnson MH, Pratt HP, Handyside AH. (1981). The generation and recognition of positional information in the preimplantation mouse embryo. In: Cellular and Molecular Aspects of Implantation Glasser SR, Bullock DW. Plenum Press; New York: 55 74 [Google Scholar]
- 3.Yamanaka Y, Ralston A, Stephenson RO, Rossant J. Cell and molecular regulation of the mouse blastocyst. Dev Dyn. (2006);235:2301–2314. doi: 10.1002/dvdy.20844. [DOI] [PubMed] [Google Scholar]
- 4.Ralston A, Rossant J. Genetic regulation of stem cell origins in the mouse embryo. Clin Genet. (2005);68:106–112. doi: 10.1111/j.1399-0004.2005.00478.x. [DOI] [PubMed] [Google Scholar]
- 5.Palmieri SL, Peter W, Hess H, Scholer HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol. (1994);166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- 6.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. (1998);95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 7.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. (2005);123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 8.Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. (2005);132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 9.Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, Buonanno A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. (2007);134:3827–3836. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- 10.Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. (2008);125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Mitalipov S, Kuo HC, Byrne J, Clepper L, Meisner L, Johnson J, Zeier R, Wolf D. Isolation and characterization of novel rhesus monkey embryonic stem cell lines. Stem Cells. (2006);24:2177–2186. doi: 10.1634/stemcells.2006-0125. [DOI] [PubMed] [Google Scholar]
- 12.Pal R, Mandal A, Rao HS, Rao MS, Khanna A. A panel of tests to standardize the characterization of human embryonic stem cells. Regen Med. (2007);2:179–192. doi: 10.2217/17460751.2.2.179. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. (1998);282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 14.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. (2003);113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 15.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. (2003);113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 16.Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. (2008);10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 17.Cauffman G, Van de Velde H, Liebaers I, Van Steirteghem A. Oct-4 mRNA and protein expression during human preimplantation development. Mol Hum Reprod. (2005);11:173–181. doi: 10.1093/molehr/gah155. [DOI] [PubMed] [Google Scholar]
- 18.Hansis C, Grifo JA, Krey LC. Oct-4 expression in inner cell mass and trophectoderm of human blastocysts. Mol Hum Reprod. (2000);6:999–1004. doi: 10.1093/molehr/6.11.999. [DOI] [PubMed] [Google Scholar]
- 19.Kirchhof N, Carnwath JW, Lemme E, Anastassiadis K, Scholer H, Niemann H. Expression pattern of Oct-4 in preimplantation embryos of different species. Biol Reprod. (2000);63:1698–1705. doi: 10.1095/biolreprod63.6.1698. [DOI] [PubMed] [Google Scholar]
- 20.Vandevoort CA, Thirkill TL, Douglas GC. Blastocyst-derived trophoblast stem cells from the rhesus monkey. Stem Cells Dev. (2007);16:779–788. doi: 10.1089/scd.2007.0020. [DOI] [PubMed] [Google Scholar]
- 21.Adjaye J, Huntriss J, Herwig R, BenKahla A, Brink TC, Wierling C, Hultschig C, Groth D, Yaspo ML, Picton HM, Gosden RG, Lehrach H. Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells. (2005);23:1514–1525. doi: 10.1634/stemcells.2005-0113. [DOI] [PubMed] [Google Scholar]
- 22.Kimber SJ, Sneddon SF, Bloor DJ, El-Bareg AM, Hawkhead JA, Metcalfe AD, Houghton FD, Leese HJ, Rutherford A, Lieberman BA, Brison DR. Expression of genes involved in early cell fate decisions in human embryos and their regulation by growth factors. Reproduction. (2008);135:635–647. doi: 10.1530/REP-07-0359. [DOI] [PubMed] [Google Scholar]
- 23.Kuijk EW, Du Puy L, Van Tol HT, Oei CH, Haagsman HP, Colenbrander B, Roelen BA. Differences in early lineage segregation between mammals. Dev Dyn. (2008);237:918–927. doi: 10.1002/dvdy.21480. [DOI] [PubMed] [Google Scholar]
- 24.Douglas GC, Vandevoort C, Kumar P, Chang TC, Golos TG. Trophoblast stem cells: models for investigating trophectoderm differentiation and placental development. Endocr Rev. (2009);30:228–240. doi: 10.1210/er.2009-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitalipov SM, Kuo HC, Hennebold JD, Wolf DP. Oct-4 expression in pluripotent cells of the rhesus monkey. Biol Reprod. (2003);69:1785–1792. doi: 10.1095/biolreprod.103.019455. [DOI] [PubMed] [Google Scholar]
- 26.Dupont C, Froenicke L, Lyons LA, Bavister BD, Brenner CA. Chromosomal instability in rhesus macaque preimplantation embryos. Fertil Steril. (2008);91:1230–1237. doi: 10.1016/j.fertnstert.2008.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bavister BD, Boatman DE, Collins K, Dierschke DJ, Eisele SG. Birth of rhesus monkey infant after in vitro fertilization and nonsurgical embryo transfer. Proc Natl Acad Sci U S A. (1984);81:2218–2222. doi: 10.1073/pnas.81.7.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bavister BD, Boatman DE, Leibfried L, Loose M, Vernon MW. Fertilization and cleavage of rhesus monkey oocytes in vitro. Biol Reprod. (1983);28:983–999. doi: 10.1095/biolreprod28.4.983. [DOI] [PubMed] [Google Scholar]
- 29.Wolf DP, Vandevoort CA, Meyer-Haas GR, Zelinski-Wooten MB, Hess DL, Baughman WL, Stouffer RL. In vitro fertilization and embryo transfer in the rhesus monkey. Biol Reprod. (1989);41:335–346. doi: 10.1095/biolreprod41.2.335. [DOI] [PubMed] [Google Scholar]
- 30.Bavister BD, Yanagimachi The effects of sperm extracts and energy sources on the motility and acrosome reaction of hamster spermatozoa in vitro. Biol Reprod. (1977);16:228–237. doi: 10.1095/biolreprod16.2.228. [DOI] [PubMed] [Google Scholar]
- 31.Mastroianni L, Manson WA. Collection of monkey semen by electroejaculation. Proc Soc Exp Biol Med. (1963);112:1025–1027. doi: 10.3181/00379727-112-28242. [DOI] [PubMed] [Google Scholar]
- 32.Boatman DE, Bavister BD. Stimulation of rhesus monkey sperm capacitation by cyclic nucleotide mediators. J Reprod Fertil. (1984);71:357–366. doi: 10.1530/jrf.0.0710357. [DOI] [PubMed] [Google Scholar]
- 33.McKiernan SH, Bavister BD. Culture of one-cell hamster embryos with water soluble vitamins: pantothenate stimulates blastocyst production. Hum Reprod. (2000);15:157–164. doi: 10.1093/humrep/15.1.157. [DOI] [PubMed] [Google Scholar]
- 34.Byrne JA, Mitalipov SM, Clepper L, Wolf DP. Transcriptional profiling of rhesus monkey embryonic stem cells. Biol Reprod. (2006);75:908–915. doi: 10.1095/biolreprod.106.053868. [DOI] [PubMed] [Google Scholar]
- 35.Harvey AJ, Kind KL, Pantaleon M, Armstrong DT, Thompson JG. Oxygen-regulated gene expression in bovine blastocysts. Biol Reprod. (2004);71:1108–1119. doi: 10.1095/biolreprod.104.028639. [DOI] [PubMed] [Google Scholar]
- 36.Cauffman G, De Rycke M, Sermon K, Liebaers I, Van de Velde H. Markers that define stemness in ESC are unable to identify the totipotent cells in human preimplantation embryos. Hum Reprod. (2009);24:63–70. doi: 10.1093/humrep/den351. [DOI] [PubMed] [Google Scholar]
- 37.van Eijk MJ, van Rooijen MA, Modina S, Scesi L, Folkers G, van Tol HT, Bevers MM, Fisher SR, Lewin HA, Rakacolli D, Galli C, de Vaureix C, Trounson AO, Mummery CL, Gandolfi F. Molecular cloning, genetic mapping, and developmental expression of bovine POU5F1. Biol Reprod. (1999);60:1093–1103. doi: 10.1095/biolreprod60.5.1093. [DOI] [PubMed] [Google Scholar]
- 38.Tielens S, Verhasselt B, Liu J, Dhont M, Van Der Elst J, Cornelissen M. Generation of embryonic stem cell lines from mouse blastocysts developed in vivo and in vitro: relation to Oct-4 expression. Reproduction. (2006);132:59–66. doi: 10.1530/rep.1.00887. [DOI] [PubMed] [Google Scholar]
- 39.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. (2005);280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 40.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. (2005);122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. (2007);134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- 42.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. (2007);448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. (2006);126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 44.Tutter AV, Kowalski MP, Baltus GA, Iourgenko V, Labow M, Li E, Kadam S. A role for med12 in regulation of nanog and nanog target genes. J Biol Chem. (2008);284:3709–3718. doi: 10.1074/jbc.M805677200. [DOI] [PubMed] [Google Scholar]
- 45.Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, Thomson JA. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. (2008);3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jedrusik A, Parfitt DE, Guo G, Skamagki M, Grabarek JB, Johnson MH, Robson P, Zernicka-Goetz M. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev. (2008);22:2692–2706. doi: 10.1101/gad.486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ralston A, Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol. (2008);313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 48.Nakatake Y, Fukui N, Iwamatsu Y, Masui S, Takahashi K, Yagi R, Yagi K, Miyazaki J, Matoba R, Ko MS, Niwa H. Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol Cell Biol. (2006);26:7772–7782. doi: 10.1128/MCB.00468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem. (2007);282:33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- 50.Chambers I. The molecular basis of pluripotency in mouse embryonic stem cells. Cloning Stem Cells. (2004);6:386–391. doi: 10.1089/clo.2004.6.386. [DOI] [PubMed] [Google Scholar]
- 51.Xu RH, Peck RM, Li DS, Feng X, Ludwig T, Thomson JA. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat Methods. (2005);2:185–190. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- 52.Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. (2007);448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 53.Cauffman G, Liebaers I, Van Steirteghem A, Van de Velde H. POU5F1 isoforms show different expression patterns in human embryonic stem cells and preimplantation embryos. Stem Cells. (2006);24:2685–2691. doi: 10.1634/stemcells.2005-0611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.