Abstract
Transposable elements can be viewed as natural DNA transfer vehicles that, similar to integrating viruses, are capable of efficient genomic insertion. The mobility of class II transposable elements (DNA transposons) can be controlled by conditionally providing the transposase component of the transposition reaction. Thus, a DNA of interest (be it a fluorescent marker, a small hairpin (sh)RNA expression cassette, a mutagenic gene trap or a therapeutic gene construct) cloned between the inverted repeat sequences of a transposon-based vector can be used for stable genomic insertion in a regulated and highly efficient manner. This methodological paradigm opened up a number of avenues for genome manipulations in vertebrates, including transgenesis for the generation of transgenic cells in tissue culture, the production of germline transgenic animals for basic and applied research, forward genetic screens for functional gene annotation in model species, and therapy of genetic disorders in humans. Sleeping Beauty (SB) was the first transposon shown to be capable of gene transfer in vertebrate cells, and recent results confirm that SB supports a full spectrum of genetic engineering including transgenesis, insertional mutagenesis, and therapeutic somatic gene transfer both ex vivo and in vivo. The first clinical application of the SB system will help to validate both the safety and efficacy of this approach. In this review, we describe the major transposon systems currently available (with special emphasis on SB), discuss the various parameters and considerations pertinent to their experimental use, and highlight the state of the art in transposon technology in diverse genetic applications.
Transposons as genetic tools
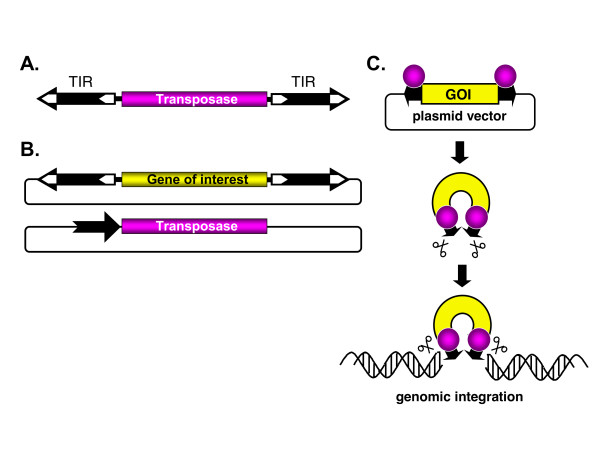
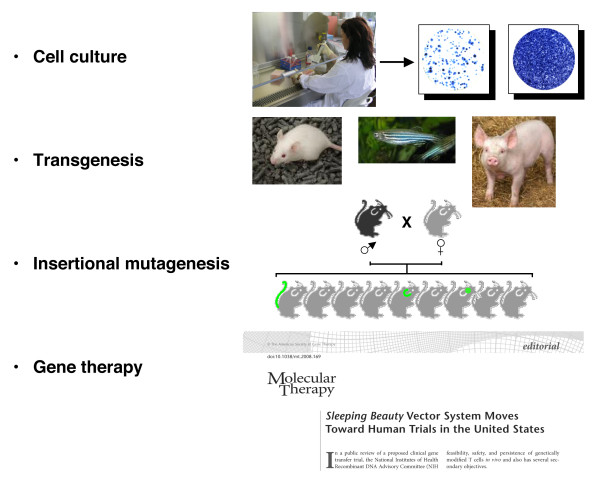
DNA transposons are discrete pieces of DNA with the ability to change their positions within the genome via a 'cut and paste' mechanism called transposition. In nature, these elements exist as single units containing the transposase gene flanked by terminal inverted repeats (TIRs) that carry transposase binding sites (Figure 1A). However, under laboratory conditions, it is possible to use transposons as bi-component systems, in which virtually any DNA sequence of interest can be placed between the transposon TIRs and mobilized by trans-supplementing the transposase in the form of an expression plasmid (Figure 1B) or mRNA synthesized in vitro. In the transposition process, the transposase enzyme mediates the excision of the element from its donor plasmid, followed by reintegration of the transposon into a chromosomal locus (Figure 1C). This feature makes transposons natural and easily controllable DNA delivery vehicles that can be used as tools for versatile applications, ranging from somatic and germline transgenesis to functional genomics and gene therapy (Figure 2).
Figure 1.
General organization and use of class II transposable elements as gene vectors. (A) Autonomous transposable elements consist of terminal inverted repeats (TIR; black arrows) that flank the transposase gene. (B) Bi-component transposon vector system for delivering transgenes that are maintained in plasmids. One component contains a DNA of interest between the transposon TIRs carried by a plasmid vector, whereas the other component is a transposase expression plasmid, in which the black arrow represents the promoter driving expression of the transposase. (C) The transposon carrying a DNA of interest is excised from the donor plasmid and is integrated at a chromosomal site by the transposase.
Figure 2.
Broad applicability of transposon-based gene vectors in vertebrate genetics.
Transposons have been successfully used in plants and in invertebrate animal models, including Arabidopsis, rice, Caenorhabditis elegans [1-3] and Drosophila [4-6] for transgenesis and insertional mutagenesis, but until recently, there was no known transposon that was sufficiently active to be tailored as a tool for such purposes in vertebrates. This is because transposons tend to have limitations with respect to the species in which they can jump. In 1997, the Sleeping Beauty (SB) transposon system was engineered by molecular reconstruction of an ancient, inactive Tc1/mariner-type transposon found in several fish genomes [7]. This newly reactivated element allowed highly efficient transposition-mediated gene transfer in major vertebrate model species without the potential risk of cross-mobilization of endogenous transposon copies in host genomes. This is because the genomes of major models lack endogenous transposon sequences with sufficient sequence similarity for mobilization by an exogenously supplied SB transposase. Indeed, SB has been successfully used as a tool for genetic modifications of a wide variety of vertebrate cell lines and species including humans [8-10].
During the past decade, other elements have been shown to catalyze efficient transposition in vertebrate model organisms. For example, the insect elements piggyBac [11,12] and Minos [13,14] catalyze efficient transposition in mammalian cells. Minos was also shown to be active in the basal chordate Ciona intestinalis [15]. Moreover, the reconstructed amphibian element Frog Prince [16], the reconstructed human Hsmar1 element [17], the reconstructed zebrafish transposon Harbinger3_DR [18], and the Tol1 [19] and Tol2 [20] elements isolated from the medaka fish have been found to be active in vertebrate species. Passport, a Tc1-family transposon isolated from a fish (Pleuronectes platessa), is active in a variety of vertebrate cells [21], and the Ac/Ds transposon originally discovered in maize by McClintock undergoes efficient transposition in zebrafish embryos [22]. Thus, the piggyBac, Minos and Ac/Ds elements appear to have a significantly wider possible host range than most other transposons. The basic criteria for the applicability of a transposon in any given model organism are 1) a sufficient level of transpositional activity in the given species, and 2) target site selection properties of the transposon, which are discussed below.
Hyperactive transposon systems
In evolutionary terms, the SB transposon represents a successful element that was able to colonize several fish genomes millions of years ago [7]. However, even successful transposons have not been selected for the highest possible activity in nature, because unlike viruses, they have to coexist with their hosts and, consequently, there is strong selective pressure to avoid insertional mutagenesis of essential genes. Indeed, although the resurrected SB element was sufficiently active to be mobilized in vertebrate cells, its relatively limited transpositional activity still presented a bottleneck for some applications. For example, requirements for transfection of primary cells and other hard-to-transfect cell types, or for remobilization of transposons from chromosomally resident single-copy donor sites, demanded an enzyme with more robust activity. Thus, enhancing transpositional activity has been one of the main targets for transposon vector development. To date, almost every single amino acid in the SB transposase has been changed in an attempt to increase its activity. Three main strategies have been applied to derive hyperactive mutants of the SB transposase: 'importing' amino acids and small blocks of amino acids from related transposases [23-25], systematic alanine scanning [26], and rational replacement of selected amino acid residues [24]. Together, these studies have yielded several, single amino acid replacements, each resulting in a relatively modest increase in transpositional activities. Unfortunately, the hyperactivity of most of the SB transposase mutants selected in immortalized cell lines did not translate to efficient stable gene transfer in primary cells in vivo [25,26].
Earlier studies established that certain combinations of amino acid replacements, each leading to hyperactivity, can yield a further enhancement in transpositional activity of the SB transposase [23-26], but guessing the correct combinations of variants out of the millions that are possible is like finding the correct combinations of numbers in a lottery. A high-throughput, PCR-based, DNA-shuffling strategy and screening of 2000 gene variants in mammalian cells produced a variant of SB that was 100-fold more potent in chromosomal insertion of a transgene than the originally reconstructed protein [27]. The use of SB100X demonstrated that it is possible to establish a transposon-based, non-viral vector system that is capable of stable gene transfer coupled with long-term gene expression at an efficiency comparable with that of viral strategies [27]. Thus, the hyperactive SB100X transposase holds great promise of offering broad utility in gene therapy and functional genomics.
Integration site preference
The insertion pattern of most transposons is nonrandom, showing characteristic preferences for insertion sites at the primary DNA sequence level, and 'hotspots' and 'cold regions' on a genome-wide scale. For example, for the primary DNA sequence, the Tol2 element does not appear to exhibit a pronounced preference for any sequence for insertion [28]. By contrast, the Harbinger3_DR transposon is highly specialized to integrate into the palindromic AAACACCWGGTCTTT consensus sequence [18], the piggyBac transposon targets the sequence TTAA, and all Tc1/mariner transposons, including SB, Frog Prince, Minos and Hsmar1, target their integration into TA dinucleotides. In the case of SB, this preference has been studied in detail, and palindromic AT repeats found to be the preferred sites for integration [29]. However, computational analyses revealed that target selection is determined primarily at the DNA structure level, not by specific base-pair interactions. For example, protein-induced deformability was shown to be associated with preferred SB insertion sites, whereas piggyBac and Tol2 integration sites lack such consistent, clear-cut structural patterns [30,31]. This suggested that integrations of SB will occur into any DNA available, depending on these preferences only, but this is not the case. In the context of chromatin, Tc1/mariner elements have no or weak preference for transcription units, the 5' regulatory regions are not favored, and most hits in genes are localized within introns [29,32]. By contrast, piggyBac shows a greater propensity to integrate into transcription units, with a preference for insertion around transcription start sites [12,33-35], and the Tol2 transposon also shows a pronounced preference for integration close to transcriptional start sites [28]. This control of integration at the chromatin level is poorly understood. One possible explanation for this is the interaction of the transposase with unknown, chromatin-associated factors. Supporting this hypothesis, it has been shown that a host-encoded protein, lens epithelium derived growth factor (LEDGF), is involved in directing integration of human immunodeficiency virus (HIV) into active genes [36]. Taken together, the preferences of particular elements to integrate into expressed genes versus non-coding DNA, and their preferences for integration sites within genes are expected to be substantially different.
Integration site preference can greatly influence the utility of transposon vectors for different applications. For example, human gene therapy protocols require application of transposon vectors showing the least preference for target genes, for obvious safety reasons. The SB system (which shows close-to-random insertion site distribution) appears to best satisfy these needs, whereas the piggyBac and Tol2 systems (which prefer genes and their upstream regulatory regions for insertion) appear to be less favorable for potential therapeutic applications. Nevertheless, a systematic assessment of potential genotoxic effects associated with genomic integration of transposon vectors will need to be performed either in cell-based assays and/or in animal models to provide clinically relevant data.
Unlike in therapeutic applications, hitting genes by insertional elements is the goal with forward mutagenesis screens. However, the insertional biases associated with vector systems represent the main limitation to full genome coverage with individual transposon-based vectors. Thus, in this respect, the utility of transposons for mutagenesis is greatly enhanced by the availability of multiple alternative vector systems with distinct preferences for insertion, such as SB, Tol2 and piggyBac. Indeed, the propensity of Tol2 to insert close to transcriptional start sites of genes might be particularly advantageous for enhancer trapping [37,38], while the propensity of piggyBac to insert into transcription units supports genome-wide mutagenesis with gene trap cassettes [39].
Local hopping
'Local hopping' describes a phenomenon of chromosomal transposition in which transposons have a preference for landing into cis-linked sites in the vicinity of the donor locus. Local hopping seems to be a shared feature of 'cut and paste' transposons. However, the actual extent of hopping to linked chromosomal sites and the interval of local transposition varies. For example, the P-element transposon of Drosophila prefers to insert within ~100 kb of the donor site at a rate ~50-fold higher than in regions outside that interval [40]. Similarly, in germline mutagenesis screens in mice using SB, 30-80% of the transposons re-insert locally on either side of the transposon donor locus [41-43]. In contrast to the P-element, SB seems to have a much larger local transposition interval between 5 and 15 Mb [42].
The local hopping feature not only differs between different transposons, but a given transposon may show great variations in local hops in different hosts, and in different donor loci even in the same host. For example, about 50-60% of the reinserted Ac elements were found to be distributed within a 5-cM distance of the donor site in maize [44,45], and the frequency of local hopping greatly varies in Arabidopsis and tobacco, depending on the chromosomal location of the donor site [46-48]. Moreover, local hopping of the Ac element in tomato seems overall to be less prevalent than in maize [49,50], and there are species-specific differences in its tendency for local hopping out of different transposon donor loci [51]. This variation in local hopping of the same element could possibly be explained by varying affinity of the transposase to unknown, chromatin-associated factors in different hosts [52].
Local hopping can play a significant role in mutagenesis using chromosomally resident transposons. In practical terms, local hopping limits the chromosomal regions accessible to a transposon jumping out of a given chromosomal site [53]. To circumvent this limitation, establishing numerous 'launch pads' to initiate transposition out of different loci can be a viable strategy to increase coverage of gene mutations. On the other hand, local hopping can be useful for saturation mutagenesis within limited chromosomal regions.
Transposons and functional genomics
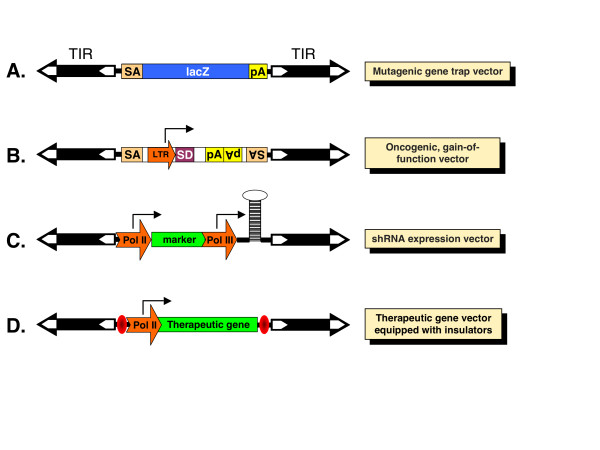
The post-genomic era presented the scientific community with the new challenge of functional annotation of every gene and identification of elaborate genetic networks. Diverse methods have been employed to address this task, including mutational analysis, which proved to be one of the most direct ways to decipher gene functions. There are versatile strategies for creating mutations, including insertional mutagenesis by discrete pieces of foreign DNA, which has the advantage that the inserted DNA fragment can serve as a molecular tag that allows rapid, usually PCR-based, identification of the mutated allele. Because the function of the gene in which the insertion has occurred is often disturbed, such loss-of-function insertional mutagenesis is frequently followed by functional analysis of mutant phenotypes. In many instances, retroviral vectors were used to introduce mutagenic cassettes into genomes, but their chromosomal insertion bias does not allow full coverage of genes [54]. The random integration pattern of the SB transposon, combined with its ability to efficiently integrate versatile transgene cassettes into chromosomes established this system as an extremely useful tool for insertional mutagenesis in both embryonic stem cells (ESCs) [34,55] and in somatic [56,57] and germline tissues [41,42,53,58-63] in animal models (Figure 2). There are several types of mutagenic cassettes that can be efficiently combined with transposon-based gene delivery for insertional mutagenesis. 5' gene-trap cassettes include splice acceptors and polyadenylation sequences so that transcription of genes can be disrupted upon vector insertion into introns (Figure 3A) [54]. Often, such cassettes are also equipped with a reporter gene (usually, a fluorescent protein, β-galactosidase or antibiotic resistance) whose expression is dependent on correct splicing between exons of the trapped gene and the splice acceptor site carried by the transposon vector [64,65].
Figure 3.
Major types of expression and mutagenic cassettes delivered by transposon vectors for versatile applications. (A) A gene-trap transposon contains a splice acceptor (SA) sequence followed by a promoterless reporter gene such as lacZ and a poly-A (pA) signal. The reporter gene is expressed only when its transcription is initiated from the promoter of a disrupted, actively transcribed endogenous transcription unit. Therefore, the expression pattern of the transposon reporter reflects that of the endogenous gene harboring the transposon insertion. (B) Oncogene-trap transposons include a strong viral promoter/enhancer long terminal repeat (LTR) sequence that can drive transcription toward the outside of the transposon and a splice donor (SD) sequence, so that forced transcription and splicing to downstream exons will result in overexpression of a product of a gene into which the transposon has inserted. Classic oncogene trap vectors also contain SA and pA sites to induce gene truncations that have dominant phenotypes. (C) Knockdown expression cassette including a polymerase (Pol) II promoter that drives expression of a marker gene and a Pol III promoter that drives expression of a short hairpin (sh)RNA. (D) A typical therapeutic expression cassette contains a ubiquitous or tissue-specific enhancer/Pol II promoter that drives expression of a therapeutic gene. To enhance the safety of such a vector, the expression cassette might be flanked by insulator elements that will block transactivation of endogenous promoters by the transposon insertion, and simultaneously protect the expression of the therapeutic gene from position effects.
Insertional mutagenesis can be applied to cultured, germline-competent stem cells including ESCs and spermatogonial stem cells (SSCs) [39,66]. One advantage of this approach is the ability to perform preselection of modified ESC clones before generating mutant animals, and to differentiate selected clones into many different tissue types in vitro. It is possible to perform large-scale, transposon-based, insertional mutagenesis screens in ESCs and SSCs by simply transfecting or electroporating transposon donor and transposase expression plasmids into the cells. The amounts of the delivered plasmids can be adjusted to obtain the desired insertion frequencies per cell. In addition, transposons can also be remobilized from chromosomally resident loci and reintegrated somewhere else in the genome by transiently providing the transposase source; such excision-re-integration events can be monitored using double selection systems, in which excision activates the first and re-integration activates the second selection marker [43].
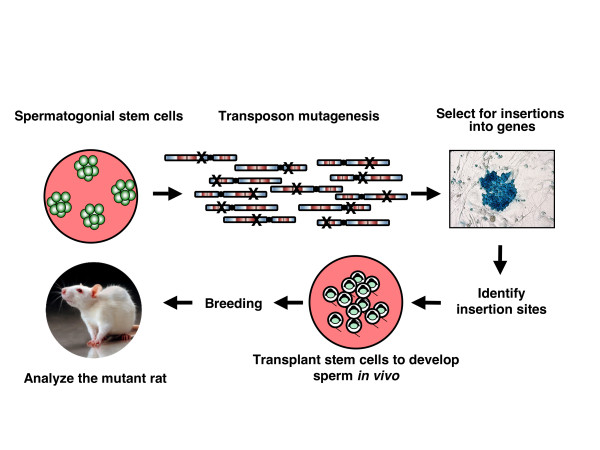
Because several aspects of physiology in rats have evolved to be more similar to humans than to mice, it would be desirable to use rat models in the process of functionally annotating the human genome by identifying the causative relationships between genes and disease phenotypes. As an important step towards this goal, an approach of establishing SB transposon-mediated insertional mutagenesis in rat SSCs was recently reported [66]. SB transposition can be used to tag and simultaneously mutate thousands of genes in culture, using gene-trap cassettes. Importantly, culture conditions maintain the potential of genetically manipulated SSCs to produce viable sperm cells. In that study, spermatogonial clones were transplanted to repopulate the testes of sterilized, wild-type recipient male rats. The stem cell genome was then passed on to transgenic offspring upon crossing of the recipient males with wild-type females (Figure 4). Although transposition events in a given target gene occur by chance, the tissue culture conditions allow screening for a large number of events. Transposition-mediated gene insertion and cell culture conditions thus allow generation of libraries of gene knockouts in rat SSCs (Figure 4). This technology has the potential to develop powerful genomic tools for use in the rat, offering the opportunity to create a bridge between physiology and genomics.
Figure 4.
Generation of knockout rats by insertional mutagenesis with gene trap transposons in spermatogonial stem cells. Cultured stem cells are transfected with gene trap transposon and transposase constructs that will lead to thousands of transposon insertions covering all chromosomes. Those cells in which insertions have occurred in expressed genes can be selected based on activation of the gene trap marker, and the insertion sites can be mapped. Cell clones or polyclonal insertion libraries can be transplanted into the testes of sterile males, in which the spermatogonial step cells will undergo spermatogenesis. These transplanted males are crossed with wild-type females to pass the insertions through the germline and generate transgenic/knockout animals.
Another method in which transposons are used for insertional mutagenesis in animal models employs a 'jump-starter and mutator' scheme [42,58,61]. In this arrangement, mutator transgenic lines carry SB transposon-based gene-trapping vectors in the form of multicopy concatemers, whereas a jump-starter line expresses the transposase preferentially in the male germline [41,64]. Crossing of the two lines results in transposition in the germline of the F1 double-transgenic males, which are then repeatedly crossed with wild-type females to segregate the transposition events that occurred in their sperm cells to separate F2 animals. In the mouse system, a single sperm cell of an F1 male contains, on average, two transposon insertions [58], and up to 90% of the F2 progeny can carry transposon insertions [61]. The applicability of this approach has been demonstrated by the identification of mouse genes with either ubiquitous or tissue-specific expression patterns [42,64,67,68]. Recently, a similar system for SB insertional mutagenesis was also established in rats [62,63].
One cautionary note of launching transposition out of transposon arrays is that recombination between newly transposed transposon copies and the donor concatemer could lead to unwanted genomic rearrangements, as observed by Geurts et al. [68]. The most likely explanation for the rearrangements is that transposition out of a concatemer generates new transposase binding sites, linked either in cis (in the case of local hops) or in trans (in the case of transposition onto other chromosomes). However, because some transposon copies remain at the original donor locus, transposase can recombine chromosomal sequences that are located between the individual transposon units by hybrid element transposition (that is, the end of one transposon pairs with the opposite end of another transposon at a different location) [69], leading to deletions and translocations. Such chromosomal rearrangements are unlikely to occur if a single-copy donor is used. Thus, transposon systems sufficiently active for efficient transposition out of single-copy donors might eliminate the need for concatemeric donor sites in animal breeding schemes. Indeed, the Tol2 element was demonstrated to show transposition at reasonable efficiencies when launched from singly-copy donor sites in transgenic zebrafish [70]. In this context, the newly developed SB100X hyperactive system might also prove useful in future genetic screens.
Forward genetic screens do not necessarily need to depend on the breeding scheme described above; in some cases, a reasonable throughput in generation of transposon insertion mutants can be achieved by introducing the mutagenic transposon into individual animals, such as in zebrafish. A gene trap mutagenesis screen was recently employed to uncover genetic determinants of nicotine response in zebrafish, through a behavioral genetic screening paradigm [71]. Using standard transposase-mediated transgenesis protocols, Tol2-based mutagenic vectors were co-injected into early zebrafish embryos by to generate a pool of mosaic F0 founder animals, which then underwent two successive rounds of crossing to generate homozygous mutant animals. Segregation of mutant animals from wild-type siblings was carried out using fluorescent reporters built into the gene trap cassettes. After profiling nicotine response in mutant versus wild-type fish, two mutants were identified out of a total of 102 fish lines screened [71]. This study emphasizes the utility of transposons for the discovery and functional annotation of genes relevant to human health in forward, phenotype-driven genetic screens in model species.
Transposon-based screens for cancer gene discovery
To induce gain-of-function mutations, transposon vectors can be equipped with oncogene trap cassettes that contain strong viral enhancers/promoters that can drive transcription outwards from the vector, thereby leading to overexpression of a full-length or truncated protein product of the trapped gene, as well as splice acceptor and polyA sites that lead to gene truncation with dominant phenotypes (Figure 3B) [9,72]. SB vectors harboring oncogene traps have been successfully used in large-scale cancer gene discovery screens in experimental animals (Figure 5) [10,73,74]. In these studies, SB transposons were somatically mobilized from donor chromosomal concatemers, which contained either low (25) [56] or high (150-350) [57] numbers of the oncogene trap transposon. Dominant mutations in somatic tissues of double transgenic mice carrying a transposase source and the mutagenic transposons resulted in the generation of experimental tumors in cancer-predisposed [56] and wild-type [57] animals. In a follow-up study, Collier et al. demonstrated that a combination of low-copy oncogene trap lines with the SB11 transposase (an early-generation hyperactive SB variant) expressed from the Rosa26 locus could achieve whole-body transposon mobilization at rates sufficient to promote penetrant tumorogenesis without complications of embryonic lethality or genomic instability [75]. Thus, this approach can be successfully employed not only to identify novel cancer genes, but also combinations of cancer genes that act together to transform a cell.
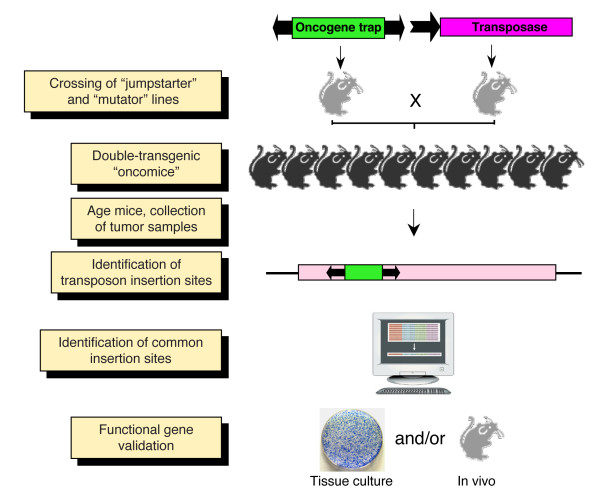
Figure 5.
Transposon-mediated cancer gene discovery screen. Breeding of 'jumpstarter' and 'mutator' stocks induces transposition in the soma of double-transgenic animals ('oncomice'). In the case of tissue-specific screens, a third genotype containing a tissue-specific Cre allele has to be crossed in to the backgrounds. The crosses can be made either in wild-type or in specific cancer-predisposed genetic backgrounds. Transposition in somatic cells leads to random insertional mutations, and animals are monitored for tumor development. Transposon insertions are cloned from genomic DNA isolated from tumor samples, and are subsequently mapped and the mutagenized genes identified. Those genes repeatedly mutated in multiple, independent tumors are designated as common insertion sites (CIS). These candidate cancer genes are then functionally validated.
Current efforts are concentrating on customized, tissue-specific screens for cancer development studies. The strategies employed to achieve this goal focus on establishing mouse lines that either conditionally express the transposase from tissue-specific promoters, or rely on generation of Cre recombinase-inducible transposase alleles that can be used in conjunction with mice that express Cre in a tissue-specific manner [76-78]. For example, this approach was addressed by Dupuy and co-workers [72], who were able to experimentally modify the spectrum of tumors by creating a Cre-inducible SB transposase allele (RosaSBaseLsL). With this strategy, they managed to overcome the obstacle of high embryonic lethality associated with ubiquitous SB transposase expression in the presence of the pT2/Onc2 oncogene trap [76,79], and to generate a model of germinal center B-cell lymphoma. They achieved this by activating SB transposase expression with an AidCre allele that drove Cre-mediated recombination in germinal center B-cells. In another approach, ubiquituous expression of the SB transposase was combined with the novel T2/Onc3 oncogene trap transposon vector. In that study, the MSCV (mouse stem cell virus) 5' long terminal repeat that was previously used to drive oncogene expression was replaced by the ubiquitously active CAGGS promoter, resulting in removal of the bias towards inducing mostly lymphomas and in reducing embryonic lethality. This strategy emphasizes that the change in the design of the mutagenic transposon (e.g. promoter choice) can have profound effects on the tumor types induced by transposition. Notably, this approach resulted in production of nearly 200 independent tumors of more than 20 types, and identification of novel, candidate cancer genes, suggesting that the combination of tissue-specific promoters and inducible transposase alleles could provide a fine mechanism of control in tumorogenesis studies.
Transposons as vectors for stable transgene integration and expression
The classic approaches to stable expression of foreign genes in vertebrates rely on physical methods of delivering gene constructs into cultured cells, such as transfection, electroporation and sonoporation, or microinjection into oocytes or fertilized eggs to generate germline transgenic animals. The main drawbacks of these approaches are the low rates of genomic integration and the unstable expression of the chromosomally integrated gene construct, which is believed to be associated with the phenomenon of concatemerization of the injected DNA before genomic integration [80]. Another particular problem in transgenic animals is that founders that develop from the injected oocytes or eggs are predominantly mosaic for the transgene, because integration generally occurs relatively late during embryonic development. In principle, all of these drawbacks can be circumvented by transposition-mediated gene delivery, as it can increase the efficiency of chromosomal integration and facilitates single-copy insertion events. Single units of expression cassettes are presumably less prone to transgene silencing than are the concatemeric insertions created by classic methods.
Transposon-based technologies can be exploited for gene transfer in cultured cells and in primary cell types, including stem cells (Figure 2). For example, transposons can be harnessed to integrate plasmid-based shRNA expression cassettes into chromosomes to obtain stable knockdown cell lines by RNA interference (Figure 3C) [81]. Such technologies have been evaluated as a potential approach to the therapy of acquired immunodeficiency syndrome by stable RNA interference with SB vectors knocking down the CCR5 and CXCR4 cell surface co-receptors that are required for viral entry as a first step to confer resistance to HIV [82]. Both the SB and the piggyBac systems were shown to support efficient transposition in mouse [33,43] and human [83,84] ESCs. In a recent, elegant study, piggyBac-derived transgene vectors were introduced into human ESCs for the purpose of driving ESC differentiation toward a specific cell type [84]. The vectors included loss-of-function shRNA expression cassettes that could simultaneously knock down the expression of pluripotency genes and of genes that contribute to endodermal and mesodermal differentiation, plus a gain-of-function construct expressing Sox1 to direct differentiation towards neuroectoderm. What makes such complex gene-transfer experiments possible is the observation that several transgene constructs maintained on separate transposon vectors can be delivered simultaneously in 'multiplex' transposition reactions, in which the different constructs are simply mixed together and cotransfected into cells [85].
The recent discovery of induced pluripotent stem cells (iPSCs) by the expression of four key genes (Oct4, Sox2, Klf4 and c-Myc) in differentiated somatic cells holds enormous promise for future regenerative medicine [86]. Transposons are attractive vehicles for reversible production of iPSCs, because the excision step of the transposition reaction produced by transient re-expression of the transposase offers removal of the transgenes after completion of reprogramming, allowing subsequent differentiation of the iPSCs into various lineages in vitro [87]. Transposition-mediated generation of mouse and human iPSCs, and removal of the reprogramming factors from the pluripotent cells have already been achieved by the piggyBac system [88]. What makes piggyBac transposons especially attractive vectors for the production of transgene-free iPSCs is a special feature of these transposons: excision fully restores the sequence of the original wild-type locus [89], thereby allowing traceless removal of transgenes from the genome.
In vivo, co-injection of engineered transposons with transposase mRNA into fertilized oocytes can facilitate early integration events that potentiate successful transmission of the transgene through the germline to the next generation (Figure 2). This method has been employed to generate transgenic zebrafish with Tc3 [90], Mos1 [91], Tol2 [20] and SB [92]; transgenic Xenopus with SB [93] and Tol2 [94]; and transgenic mice with SB [27,95-97], piggyBac [11] and Tol2 [98]. In this context, an important step towards transgenesis with bacterial artificial chromosomes (BACs) has been made by the delivery of a ~70-kb BAC construct into zebrafish and mouse embryos with Tol2 [99]. Thus, transposons can evidently be used to stably deliver large transgene constructs together with complex regulatory regions, without the complications of DNA rearrangements and silencing associated with classic methods.
Any transgene vector system should provide long-term expression of transgenes. Transgenes delivered by non-viral approaches often form long, repeated arrays (concatemers) that are targets for transcriptional silencing by heterochromatin formation. In addition, long-term expression of transgenes delivered by retroviruses has been shown to be compromised by transcriptional silencing [100]. It was recently shown that the zinc finger protein ZFP809 bridges the integrated proviral DNA of the murine leukaemia virus and the tripartite motif-containing 28 transcriptional co-repressor in embryonic stem cells [101]. Thus, sequence elements in the vector itself can predispose the cargo for silencing. The cut and paste mechanism of DNA transposition results in a single copy of the transgene per insertion locus, thus concatemer-induced gene silencing is unlikely to be an issue with transposition-mediated gene transfer. Indeed, Grabundzija et al. found that transposon insertions delivered by the SB, Tol2 and piggyBac systems only rarely (< 4% of all insertions) undergo silencing in HeLa cells [28]. Furthermore, stable transgene expression observed in hundreds of independent insertions in this study suggests that these three transposon systems rarely target heterochromatic chromosomal regions for insertion, and that it is unlikely that certain sequence motifs in the transposon vectors are recognized by mediators of silencing in the cell. An additional factor that may provoke transgene silencing is the cargo DNA, particularly the type of promoter used to drive expression of the gene of interest. Indeed, it was previously shown that transgene constructs delivered into mouse cells using SB transposition can be subject to epigenetic regulation by CpG methylation and that a determinant of epigenetic modifications of the integrating transposon vector is the cargo transgene construct, with the promoter playing a major role [102]. However, with careful promoter choice, several studies have established that SB-mediated transposition provides long-term expression in vivo. For example, stable transgene expression from SB vectors was seen in mice after gene delivery in the liver [103-106], lung [107,108], brain [109] and blood after hematopoietic reconstitution in vivo [27,110]. Thus, although our understanding of all the factors that will ultimately determine the expressional fate of an integrated transposon is still rudimentary, it appears that transposon vectors have the capacity to provide long-term expression of transgenes both in vitro and in vivo.
Transposons as vectors for gene therapy
Considerable effort has been devoted to the development of gene delivery strategies for the treatment of inherited and acquired disorders in humans. A desirable gene therapy approach should 1) achieve delivery of therapeutic genes at high efficiency specifically into the relevant cells, 2) be adaptable to changing needs in terms of vector design, 3) minimize the risk of genotoxicity, and 4) be cost-effective.
Adapting viruses for gene transfer is a popular approach; for example, γ-retroviral and lentiviral vectors are efficient at integrating foreign DNA into the chromosomes of transduced cells and have enormous potential for lifelong gene expression [111]. A major concern of using retroviral vectors is the potential for mutagenic effects at the sites of genomic integration [112-114]. Indeed, insertional mutagenesis has been observed in clinical trials using a retroviral vector for gene therapy of X-linked severe combined immunodeficiency [112,114,115]. The clinical use of retroviral vectors can be curtailed because of the limited size of the payload, as multiple or large transgenes compromise the efficiency of viral reverse transcription and packaging. Finally, regulatory issues and the high costs associated with manufacture of clinical-grade retrovirus hamper their widespread translation into clinical practice. An ideal therapeutic vector would combine the favorable attributes of integrating viral vectors (that is, stable chromosomal insertion) while significantly reducing the potential for adverse events. Transposons could potentially offer such an alternative (Figure 2).
The advantage of SB transposon-based gene delivery is that, owing to stable genomic insertion of expression cassettes, it can lead to both long-term and efficient transgene expression in preclinical animal models [116]. Thus, the SB plasmid-based transposon system combines the advantages of viral vectors with those of naked DNA molecules. However, in contrast to viral vectors, transposon vectors can be maintained and propagated as plasmid DNA, which makes them simple and inexpensive to manufacture, an important issue for the implementation of future clinical trials. Further advantages of the SB system include its reduced immunogenicity [103], no strict limitation of the size of expression cassettes [24] and improved safety and toxicity profiles [87,117-119]. Because the transposition mechanism does not involve reverse transcription, DNA-based transposon vectors are not prone to incorporating mutations and can tolerate larger and more complex transgenes, including those containing repeat DNA motifs. Moreover, the use of SB-based gene delivery eliminates the risk of rearrangements of the expression cassette that, as part of a transposing unit of DNA, integrates into chromosomal DNA in an intact form [120]. Compared with retroviral systems, the SB vectors have an inherently low enhancer/promoter activity [117,118]. Inserting insulator sequences flanking the transcription units of the cargo to prevent accidental trans-activation of promoters of neighboring genes further increased the safety features of the SB system (Figure 3D) [117]. Notably, the transposase can be provided as messenger RNA, thereby reducing the risk of 'rehopping' of the transposon-based vector [96]. Chromosomal integration of SB transposons is precise and random (see above), and no SB-associated adverse effects have been reported [116,120,121]. Of note, a precise integration mechanism, random integration pattern and negligible promoter/enhancer activity do not appear to be general features of all recombinase/transposon systems. For example, integration promoted by the bacteriophage-derived PhiC31 system was reported to generate chromosomal rearrangements [122,123]. The 5' TIR of the piggyBac transposon exhibits significant promoter activity in mammalian cells [124], and its genomic integration profile resembles that of integrating viral vectors [12], as described above.
The past few years have seen a steady growth in interest in applying the SB system for the treatment of several conditions including haemophilia A and B [103,104,106,107,125], junctional epidermolysis bullosa [126], tyrosinemia I [127], Huntington disease [128] sickle cell disease [129], mucopolysaccharidosis [105,130], cancer [109,131] and type 1 diabetes [132]. In addition, important steps have been made towards SB-mediated gene transfer in the lung for potential therapy of α-1-antitrypsin deficiency, cystic fibrosis and a variety of cardiovascular diseases [108,133]. Thus, the establishment of non-viral, integrating vectors has generated considerable interest in developing efficient and safe vectors for human gene therapy [116,120,134-136].
The SB100X hyperactive transposon system yields efficient stable gene transfer after non-viral gene delivery into therapeutically relevant primary cell types, including stem or progenitor cells. For example, the use of the SB100X system yielded robust gene transfer efficiencies into human hematopoietic progenitors [27,110], mesenchymal stem cells, muscle stem/progenitor cells (myoblasts) and iPSCs [137]. These cells are relevant targets for stem cell biology and for regenerative medicine and gene- and cell-based therapies of complex genetic diseases. Importantly, expression of the SB100X hyperactive transposase did not adversely influence the differentiation or function of these adult stem/progenitor cells, nor was there any evidence of any cytogenetic abnormalities [137]. In the context of iPSC technology, the ability to coax the differentiation of pluripotent stem cells into clinically relevant, transplantable cell types is a key step towards their ultimate use in clinical applications, especially because undifferentiated iPSCs pose an intrinsic tumorigenic risk [138]. It was recently demonstrated that SB transposon-mediated delivery of the myogenic PAX3 transcription factor into iPSCs coaxed their differentiation into MyoD+ myogenic progenitors and multinucleated myofibers [137], suggesting that PAX3 may serve as a myogenic 'molecular switch' in iPSCs, a finding that has implications for cell therapy of congenital degenerative muscle diseases, including Duchenne muscular dystrophy.
The first clinical application of the SB system is currently ongoing using autologous T cells genetically modified to redirect specificity for B-lineage malignancies [139]. Lymphocytes are a suitable initial platform for testing new gene transfer systems, as there have been hundreds of infusions of clinical-grade T cells genetically modified using viral and non-viral approaches without apparent genotoxicity [140]. The SB transposon tested in the first human application carries a chimeric antigen receptor (CAR) to render the T cells specifically cytotoxic toward CD19-positive lymphoid tumors [141,142]. The advantage of using the SB system for the genetic modification of T cells includes the reduced cost associated with manufacturing of clinical-grade DNA plasmids compared with recombinant viral vectors. This is particularly important when one considers that trials infusing CAR-positive T cells are only now beginning to demonstrate anti-tumor effects [143,144]. The higher enzymatic activity of SB100X might enable integration efficiencies comparable with that of retroviral vectors to be achieved for next-generation trials.
Conclusions
Transposon-based technologies have enormous potential to develop powerful genomic tools with the vision of creating a bridge between physiology and genetics and to establish safe and inexpensive protocols for clinical gene transfer. Simple, plasmid-based vectors matched by a corresponding transposase source offer an easy and efficient method for germline transgenesis in laboratory animals and in large animal species for biotechnology. Furthermore, it is now both accessible and practical to generate highly complex libraries of gene knockouts in model species with a view to establishing new models of human disease for the annotation of disease pathways and for therapeutic and pharmaceutical intervention. The recently developed SB100X hyperactive transposon system yields highly efficient stable gene transfer after non-viral gene delivery into therapeutically relevant primary cell types, including stem cells, and thus may facilitate the clinical implementation of ex vivo and in vivo gene therapies. The next phase of preclinical research will focus on further refinement in large animal models to undertake SB-mediated transposition in vivo and to improve the safety profile of SB vectors by target-selected transgene integration into genomic 'safe harbors'. Although it remains to be seen whether the first clinical application of the SB system will result in a therapeutic effect, this trial will help validate the safety of this approach. The ongoing investigations will certainly prompt new ideas and new designs to be developed in this (ever) expanding universe of transposon technologies for genetic and cell engineering.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Both authors contributed to drafting, reading and approving the final manuscript.
Contributor Information
Zoltán Ivics, Email: zivics@mdc-berlin.de.
Zsuzsanna Izsvák, Email: zizsvak@mdc-berlin.de.
Acknowledgements
Work in the authors' laboratories was supported by EU FP6 (INTHER) and EU FP7 (PERSIST and InduStem), grants from the Deutsche Forschungsgemeinschaft SPP1230 'Mechanisms of gene vector entry and persistence', and from the Bundesministerium für Bildung und Forschung (NGFN-2, NGFNplus, iGene, InTherGD and ReGene).
References
- Rushforth AM, Saari B, Anderson P. Site-selected insertion of the transposon Tc1 into a Caenorhabditis elegans myosin light chain gene. Mol Cell Biol. 1993;13:902–910. doi: 10.1128/mcb.13.2.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaal RR, Broeks A, van Meurs J, Groenen JT, Plasterk RH. Target-selected gene inactivation in Caenorhabditis elegans by using a frozen transposon insertion mutant bank. Proc Natl Acad Sci USA. 1993;90:7431–7435. doi: 10.1073/pnas.90.16.7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessereau JL, Wright A, Williams DC, Schuske K, Davis MW, Jorgensen EM. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature. 2001;413:70–74. doi: 10.1038/35092567. [DOI] [PubMed] [Google Scholar]
- Spradling AC. P element-mediated transformation in Drosophila. A Practical Approach. 1986. pp. 175–198. IRL Press.
- Cooley L, Kelley R, Spradling A. Insertional mutagenesis of the Drosophila genome with single P elements. Science. 1988;239:1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, Killpack K, Laufer A, Mazzotta J, Smith RD, Stevens LM, Stuber C, Tan LR, Ventura R, Woo A, Zakrajsek I, Zhao L, Chen F, Swimmer C, Kopczynski C, Duyk G, Winberg ML, Margolis J. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/S0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Miskey C, Izsvak Z, Kawakami K, Ivics Z. DNA transposons in vertebrate functional genomics. Cell Mol Life Sci. 2005;62:629–641. doi: 10.1007/s00018-004-4232-7. [DOI] [PubMed] [Google Scholar]
- Mates L, Izsvak Z, Ivics Z. Technology transfer from worms and flies to vertebrates: transposition-based genome manipulations and their future perspectives. Genome Biol. 2007;8(Suppl 1):S1. doi: 10.1186/gb-2007-8-s1-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Li MA, Mates L, Boeke JD, Nagy A, Bradley A, Izsvak Z. Transposon-mediated genome manipulation in vertebrates. Nat Methods. 2009;6:415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Wilson MH, Coates CJ, George AL Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- Zagoraiou L, Drabek D, Alexaki S, Guy JA, Klinakis AG, Langeveld A, Skavdis G, Mamalaki C, Grosveld F, Savakis C. In vivo transposition of Minos, a Drosophila mobile element, in mammalian tissues. Proc Natl Acad Sci USA. 2001;98:11474–11478. doi: 10.1073/pnas.201392398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit T, Dekker S, Maas A, Breedveld G, Knoch TA, Langeveld A, Szumska D, Craig R, Bhattacharya S, Grosveld F, Drabek D. Tagged mutagenesis by efficient Minos-based germ line transposition. Mol Cell Biol. 2010;30:68–77. doi: 10.1128/MCB.00913-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakura Y, Awazu S, Chiba S, Satoh N. Germ-line transgenesis of the Tc1/mariner superfamily transposon Minos in Ciona intestinalis. Proc Natl Acad Sci USA. 2003;100:7726–7730. doi: 10.1073/pnas.1230736100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskey C, Izsvak Z, Plasterk RH, Ivics Z. The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 2003;31:6873–6881. doi: 10.1093/nar/gkg910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskey C, Papp B, Mates L, Sinzelle L, Keller H, Izsvak Z, Ivics Z. The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR Protein on transposon ends. Mol Cell Biol. 2007;27:4589–600. doi: 10.1128/MCB.02027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinzelle L, Kapitonov VV, Grzela DP, Jursch T, Jurka J, Izsvak Z, Ivics Z. Transposition of a reconstructed Harbinger element in human cells and functional homology with two transposon-derived cellular genes. Proc Natl Acad Sci USA. 2008;105:4715–4720. doi: 10.1073/pnas.0707746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga A, Cheah FS, Hamaguchi S, Yeo GH, Chong SS. Germline transgenesis of zebrafish using the medaka Tol1 transposon system. Dev Dyn. 2008;237:2466–2474. doi: 10.1002/dvdy.21688. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci USA. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KJ, Carlson DF, Leaver MJ, Foster LK, Fahrenkrug SC. Passport, a native Tc1 transposon from flatfish, is functionally active in vertebrate cells. Nucleic Acids Res. 2009;37:1239–47. doi: 10.1093/nar/gkn1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov A, Gao Y, Naqvi NI, Parinov S. Trans-kingdom transposition of the maize dissociation element. Genetics. 2006;174:1095–1104. doi: 10.1534/genetics.106.061184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, Bell JB, Largaespada DA, Hackett PB. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/S1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Zayed H, Izsvak Z, Walisko O, Ivics Z. Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Baus J, Liu L, Heggestad AD, Sanz S, Fletcher BS. Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol Ther. 2005;12:1148–1156. doi: 10.1016/j.ymthe.2005.06.484. [DOI] [PubMed] [Google Scholar]
- Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mutational analysis of the N-terminal DNA-binding domain of sleeping beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mates L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, Ma L, Samara-Kuko E, Gysemans C, Pryputniewicz D, Miskey C, Fletcher B, VandenDriessche T, Ivics Z, Izsvák Z. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- Grabundzija I, Irgang M, Mates L, Belay E, Matrai J, Gogol-Doring A, Kawakami K, Chen W, Ruiz P, Chuah MK, VandenDriessche T, Izsvák Z, Ivics Z. Comparative analysis of transposable element vector systems in human cells. Mol Ther. 2010;18:1200–1209. doi: 10.1038/mt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigdal TJ, Kaufman CD, Izsvak Z, Voytas DF, Ivics Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J Mol Biol. 2002;323:441–452. doi: 10.1016/S0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- Geurts AM, Hackett CS, Bell JB, Bergemann TL, Collier LS, Carlson CM, Largaespada DA, Hackett PB. Structure-based prediction of insertion-site preferences of transposons into chromosomes. Nucleic Acids Res. 2006;34:2803–2811. doi: 10.1093/nar/gkl301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett CS, Geurts AM, Hackett PB. Predicting preferential DNA vector insertion sites: implications for functional genomics and gene therapy. Genome Biol. 2007;8(Suppl 1):S12. doi: 10.1186/gb-2007-8-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D, Wang X, Bradley A, Liu P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Kong J, Stalker J, Bradley A. Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis. 2009;47:404–408. doi: 10.1002/dvg.20508. [DOI] [PubMed] [Google Scholar]
- Galvan DL, Nakazawa Y, Kaja A, Kettlun C, Cooper LJ, Rooney CM, Wilson MH. Genome-wide mapping of PiggyBac transposon integrations in primary human T cells. J Immunother. 2009;32:837–844. doi: 10.1097/CJI.0b013e3181b2914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- Parinov S, Kondrichin I, Korzh V, Emelyanov A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev Dyn. 2004;231:449–459. doi: 10.1002/dvdy.20157. [DOI] [PubMed] [Google Scholar]
- Kondrychyn I, Garcia-Lecea M, Emelyanov A, Parinov S, Korzh V. Genome-wide analysis of Tol2 transposon reintegration in zebrafish. BMC Genomics. 2009;10:418. doi: 10.1186/1471-2164-10-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Bradley A, Huang Y. A piggyBac transposon-based genome-wide library of insertionally mutated Blm-deficient murine ES cells. Genome Res. 2009;19:667–673. doi: 10.1101/gr.085621.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J, Karpen GH, Craig N, Spradling AC. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer SE, Wienholds E, Plasterk RH. Regulated transposition of a fish transposon in the mouse germ line. Proc Natl Acad Sci USA. 2001;98:6759–6764. doi: 10.1073/pnas.121569298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CM, Dupuy AJ, Fritz S, Roberg-Perez KJ, Fletcher CF, Largaespada DA. Transposon mutagenesis of the mouse germline. Genetics. 2003;165:243–256. doi: 10.1093/genetics/165.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Ivics Z, Izsvak Z, Bradley A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc Natl Acad Sci USA. 1998;95:10769–10773. doi: 10.1073/pnas.95.18.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner HK, Belachew A, Burgess D, Harding S, Ralston M, Ralston E. Distribution of unlinked receptor sites for transposed Ac elements from the bz-m2(Ac) allele in maize. Genetics. 1994;136:261–279. doi: 10.1093/genetics/136.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Greenblatt IM, Dellaporta SL. Transposition of Ac from the P locus of maize into unreplicated chromosomal sites. Genetics. 1987;117:109–116. doi: 10.1093/genetics/117.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft I, Dean C. Transposition pattern of the maize element Ds in Arabidopsis thaliana. Genetics. 1993;134:1221–1229. doi: 10.1093/genetics/134.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida C, Onouchi H, Koizumi J, Hamada S, Semiarti E, Torikai S, Machida Y. Characterization of the transposition pattern of the Ac element in Arabidopsis thaliana using endonuclease I-SceI. Proc Natl Acad Sci USA. 1997;94:8675–8680. doi: 10.1073/pnas.94.16.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner HK, Keller J, Harper E, Ralston E. Variable patterns of transposition of the maize element activator in tobacco. Plant Cell. 1991;3:473–482. doi: 10.1105/tpc.3.5.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzile F, Yoder JI. Pattern of somatic transposition in a high copy Ac tomato line. Plant J. 1992;2:173–179. [PubMed] [Google Scholar]
- Osborne BI, Corr CA, Prince JP, Hehl R, Tanksley SD, McCormick S, Baker B. Ac transposition from a T-DNA can generate linked and unlinked clusters of insertions in the tomato genome. Genetics. 1991;129:833–844. doi: 10.1093/genetics/129.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S, Larondelle Y, Rossberg M, Furtek D, Theres K. Transgenic tomato lines containing Ds elements at defined genomic positions as tools for targeted transposon tagging. Mol Gen Genet. 1994;243:666–673. doi: 10.1007/BF00279576. [DOI] [PubMed] [Google Scholar]
- Kunze R, Weil CF. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editor. Washington, DC: ASM Press; 2002. The hAT and CACTA superfamilies of plant transposons; pp. 565–610. [Google Scholar]
- Keng VW, Yae K, Hayakawa T, Mizuno S, Uno Y, Yusa K, Kokubu C, Kinoshita T, Akagi K, Jenkins NA, Copeland NG, Horie K, Takeda J. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat Methods. 2005;2:763–769. doi: 10.1038/nmeth795. [DOI] [PubMed] [Google Scholar]
- Hansen GM, Markesich DC, Burnett MB, Zhu Q, Dionne KM, Richter LJ, Finnell RH, Sands AT, Zambrowicz BP, Abuin A. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res. 2008;18:1670–1679. doi: 10.1101/gr.078352.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubu C, Horie K, Abe K, Ikeda R, Mizuno S, Uno Y, Ogiwara S, Ohtsuka M, Isotani A, Okabe M, Imai K, Takeda J. A transposon-based chromosomal engineering method to survey a large cis-regulatory landscape in mice. Nat Genet. 2009;41:946–952. doi: 10.1038/ng.397. [DOI] [PubMed] [Google Scholar]
- Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Fritz S, Largaespada DA. Transposition and gene disruption in the male germline of the mouse. Genesis. 2001;30:82–88. doi: 10.1002/gene.1037. [DOI] [PubMed] [Google Scholar]
- Roberg-Perez K, Carlson CM, Largaespada DA. MTID: a database of Sleeping Beauty transposon insertions in mice. Nucleic Acids Res. 2003;31:78–81. doi: 10.1093/nar/gkg045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM, Wilber A, Carlson CM, Lobitz PD, Clark KJ, Hackett PB, McIvor RS, Largaespada DA. Conditional gene expression in the mouse using a Sleeping Beauty gene-trap transposon. BMC Biotechnol. 2006;6:30. doi: 10.1186/1472-6750-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie K, Kuroiwa A, Ikawa M, Okabe M, Kondoh G, Matsuda Y, Takeda J. Efficient chromosomal transposition of a Tc1/mariner- like transposon Sleeping Beauty in mice. Proc Natl Acad Sci USA. 2001;98:9191–9196. doi: 10.1073/pnas.161071798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada K, Ishishita S, Tosaka K, Takahashi RI, Ueda M, Keng VW, Horie K, Takeda J. Transposon-tagged mutagenesis in the rat. Nat Methods. 2007;4:131–3. doi: 10.1038/nmeth1002. [DOI] [PubMed] [Google Scholar]
- Lu B, Geurts AM, Poirier C, Petit DC, Harrison W, Overbeek PA, Bishop CE. Generation of rat mutants using a coat color-tagged Sleeping Beauty transposon system. Mamm Genome. 2007;18:338–346. doi: 10.1007/s00335-007-9025-5. [DOI] [PubMed] [Google Scholar]
- Horie K, Yusa K, Yae K, Odajima J, Fischer SE, Keng VW, Hayakawa T, Mizuno S, Kondoh G, Ijiri T, Matsuda Y, Plasterk RH, Takeda J. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol Cell Biol. 2003;23:9189–9207. doi: 10.1128/MCB.23.24.9189-9207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford WL, Cohn JB, Cordes SP. Gene-trap mutagenesis: past, present and beyond. Nat Rev Genet. 2001;2:756–768. doi: 10.1038/35093548. [DOI] [PubMed] [Google Scholar]
- Izsvák Z, Frohlich J, Grabundzija I, Shirley JR, Powell HM, Chapman KM, Ivics Z, Hamra FK. Generating knockout rats by transposon mutagenesis in spermatogonial stem cells. Nat Methods. 2010;7:443–445. doi: 10.1038/nmeth.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yae K, Keng VW, Koike M, Yusa K, Kouno M, Uno Y, Kondoh G, Gotow T, Uchiyama Y, Horie K, Takeda J. Sleeping beauty transposon-based phenotypic analysis of mice: lack of Arpc3 results in defective trophoblast outgrowth. Mol Cell Biol. 2006;26:6185–6196. doi: 10.1128/MCB.00018-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM, Collier LS, Geurts JL, Oseth LL, Bell ML, Mu D, Lucito R, Godbout SA, Green LE, Lowe SW, Hirsch BA, Leinwand LA, Largaespada DA. Gene mutations and genomic rearrangements in the mouse as a result of transposon mobilization from chromosomal concatemers. PLoS Genet. 2006;2:e156. doi: 10.1371/journal.pgen.0020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray YH. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000;16:461–468. doi: 10.1016/S0168-9525(00)02104-1. [DOI] [PubMed] [Google Scholar]
- Urasaki A, Asakawa K, Kawakami K. Efficient transposition of the Tol2 transposable element from a single-copy donor in zebrafish. Proc Natl Acad Sci USA. 2008;105:19827–19832. doi: 10.1073/pnas.0810380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold AM, Balciunas D, Sivasubbu S, Clark KJ, Bedell VM, Westcot SE, Myers SR, Moulder GL, Thomas MJ, Ekker SC. Nicotine response genetics in the zebrafish. Proc Natl Acad Sci USA. 2009;106:18662–18667. doi: 10.1073/pnas.0908247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy AJ, Rogers LM, Kim J, Nannapaneni K, Starr TK, Liu P, Largaespada DA, Scheetz TE, Jenkins NA, Copeland NG. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69:8150–8156. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy AJ. Transposon-based screens for cancer gene discovery in mouse models. Semin Cancer Biol. 2010;20:361–8. doi: 10.1016/j.semcancer.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA. Harnessing transposons for cancer gene discovery. Nat Rev Cancer. 2010;10:696–706. doi: 10.1038/nrc2916. [DOI] [PubMed] [Google Scholar]
- Collier LS, Adams DJ, Hackett CS, Bendzick LE, Akagi K, Davies MN, Diers MD, Rodriguez FJ, Bender AM, Tieu C, Matise I, Dupuy AJ, Copeland NG, Jenkins NA, Hodgson JG, Weiss WA, Jenkins RB, Largaespada DA. Whole-body sleeping beauty mutagenesis can cause penetrant leukemia/lymphoma and rare high-grade glioma without associated embryonic lethality. Cancer Res. 2009;69:8429–8437. doi: 10.1158/0008-5472.CAN-09-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy AJ, Jenkins NA, Copeland NG. Sleeping beauty: a novel cancer gene discovery tool. Hum Mol Genet. 2006;15(Spec No 1):R75–79. doi: 10.1093/hmg/ddl061. [DOI] [PubMed] [Google Scholar]
- Keng VW, Villanueva A, Chiang DY, Dupuy AJ, Ryan BJ, Matise I, Silverstein KA, Sarver A, Starr TK, Akagi K, Tessarollo L, Collier LS, Powers S, Lowe SW, Jenkins NA, Copeland NG, Llovet JM, Largaespada DA. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27:264–74. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, Gupta M, O'Sullivan MG, Matise I, Dupuy AJ, Collier LS, Powers S, Oberg AL, Asmann YW, Thibodeau SN, Tessarollo L, Copeland NG, Jenkins NA, Cormier RT, Largaespada DA. A Transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–50. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier LS, Largaespada DA. Hopping around the tumor genome: transposons for cancer gene discovery. Cancer Res. 2005;65:9607–9610. doi: 10.1158/0008-5472.CAN-05-3085. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Conspiracy of silence among repeated transgenes. Bioessays. 1998;20:532–535. doi: 10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Kaufman CD, Izsvak Z, Katzer A, Ivics Z. Frog Prince transposon-based RNAi vectors mediate efficient gene knockdown in human cells. J RNAi Gene Silenc. 2005;1:97–104. [PMC free article] [PubMed] [Google Scholar]
- Tamhane M, Akkina R. Stable gene transfer of CCR5 and CXCR4 siRNAs by sleeping beauty transposon system to confer HIV-1 resistance. AIDS Res Ther. 2008;5:16. doi: 10.1186/1742-6405-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban TI, Apati A, Nemeth A, Varga N, Krizsik V, Schamberger A, Szebenyi K, Erdei Z, Varady G, Karaszi E, Homolya L, Német K, Gócza E, Miskey C, Mátés L, Ivics Z, Izsvák Z, Sarkadi B. Applying a "double-feature" promoter to identify cardiomyocytes differentiated from human embryonic stem cells following transposon-based gene delivery. Stem Cells. 2009;27:1077–1087. doi: 10.1002/stem.45. [DOI] [PubMed] [Google Scholar]
- Lacoste A, Berenshteyn F, Brivanlou AH. An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell. 2009;5:332–342. doi: 10.1016/j.stem.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Kahlig KM, Saridey SK, Kaja A, Daniels MA, George AL Jr, Wilson MH. Multiplexed transposon-mediated stable gene transfer in human cells. Proc Natl Acad Sci USA. 2010;107:1343–1348. doi: 10.1073/pnas.0910383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- VandenDriessche T, Ivics Z, Izsvak Z, Chuah MK. Emerging potential of transposons for gene therapy and generation of induced pluripotent stem cells. Blood. 2009;114:1461–1468. doi: 10.1182/blood-2009-04-210427. [DOI] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R, Fain-Thornton J, Craig NL. piggyBac can bypass DNA synthesis during cut and paste transposition. Embo J. 2008;27:1097–1109. doi: 10.1038/emboj.2008.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz E, van Luenen HG, Schaerringer B, Plasterk RH, Driever W. Transposition of the nematode Caenorhabditis elegans Tc3 element in the zebrafish Danio rerio. Curr Biol. 1998;8:82–88. doi: 10.1016/S0960-9822(98)70038-7. [DOI] [PubMed] [Google Scholar]
- Fadool JM, Hartl DL, Dowling JE. Transposition of the mariner element from Drosophila mauritiana in zebrafish. Proc Natl Acad Sci USA. 1998;95:5182–5186. doi: 10.1073/pnas.95.9.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene 'knockdown' in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Sinzelle L, Vallin J, Coen L, Chesneau A, Pasquier DD, Pollet N, Demeneix B, Mazabraud A. Generation of trangenic Xenopus laevis using the Sleeping Beauty transposon system. Transgenic Res. 2006;15:751–60. doi: 10.1007/s11248-006-9014-6. [DOI] [PubMed] [Google Scholar]
- Hamlet MR, Yergeau DA, Kuliyev E, Takeda M, Taira M, Kawakami K, Mead PE. Tol2 transposon-mediated transgenesis in Xenopus tropicalis. Genesis. 2006;44:438–445. doi: 10.1002/dvg.20234. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Clark K, Carlson CM, Fritz S, Davidson AE, Markley KM, Finley K, Fletcher CF, Ekker SC, Hackett PB, Horn S, Largaespada DA. Mammalian germ-line transgenesis by transposition. Proc Natl Acad Sci USA. 2002;99:4495–4499. doi: 10.1073/pnas.062630599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilber A, Frandsen JL, Geurts JL, Largaespada DA, Hackett PB, McIvor RS. RNA as a source of transposase for sleeping beauty-mediated gene insertion and expression in somatic cells and tissues. Mol Ther. 2006;13:625–630. doi: 10.1016/j.ymthe.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Carlson CM, Frandsen JL, Kirchhof N, McIvor RS, Largaespada DA. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc Natl Acad Sci USA. 2005;102:17059–17064. doi: 10.1073/pnas.0502974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyama K, Kawakami K, Yagita K. A simple and highly efficient transgenesis method in mice with the Tol2 transposon system and cytoplasmic microinjection. Genomics. 2010;95:306–311. doi: 10.1016/j.ygeno.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Suster ML, Sumiyama K, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish and mice. BMC Genomics. 2009;10:477. doi: 10.1186/1471-2164-10-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahner D, Stuhlmann H, Stewart CL, Harbers K, Lohler J, Simon I, Jaenisch R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison BS, Yant SR, Mikkelsen JG, Kay MA. Postintegrative gene silencing within the Sleeping Beauty transposition system. Mol Cell Biol. 2007;27:8824–8833. doi: 10.1128/MCB.00498-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant SR, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay MA. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- Ohlfest JR, Frandsen JL, Fritz S, Lobitz PD, Perkinson SG, Clark KJ, Nelsestuen G, Key NS, McIvor RS, Hackett PB, Largaespada DA. Phenotypic correction and long-term expression of factor VIII in hemophilic mice by immunotolerization and nonviral gene transfer using the Sleeping Beauty transposon system. Blood. 2005;105:2691–2698. doi: 10.1182/blood-2004-09-3496. [DOI] [PubMed] [Google Scholar]
- Aronovich EL, Bell JB, Khan SA, Belur LR, Gunther R, Koniar B, Schachern PA, Parker JB, Carlson CS, Whitley CB, McIvor RS, Gupta P, Hackett PB. Systemic correction of storage disease in MPS I NOD/SCID mice using the sleeping beauty transposon system. Mol Ther. 2009;17:1136–1144. doi: 10.1038/mt.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kren BT, Unger GM, Sjeklocha L, Trossen AA, Korman V, Diethelm-Okita BM, Reding MT, Steer CJ. Nanocapsule-delivered Sleeping Beauty mediates therapeutic Factor VIII expression in liver sinusoidal endothelial cells of hemophilia A mice. J Clin Invest. 2009;119:2086–2099. doi: 10.1172/JCI34332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Mah C, Fletcher BS. Sustained FVIII expression and phenotypic correction of hemophilia a in neonatal mice using an endothelial-targeted Sleeping Beauty transposon. Mol Ther. 2006;13:1006–1015. doi: 10.1016/j.ymthe.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Belur LR, Frandsen JL, Dupuy AJ, Ingbar DH, Largaespada DA, Hackett PB, Scott McIvor R. Gene insertion and long-term expression in lung mediated by the Sleeping Beauty transposon system. Mol Ther. 2003;8:501–507. doi: 10.1016/S1525-0016(03)00211-9. [DOI] [PubMed] [Google Scholar]
- Ohlfest JR, Demorest ZL, Motooka Y, Vengco I, Oh S, Chen E, Scappaticci FA, Saplis RJ, Ekker SC, Low WC, Freese AB, Largaespada DA. Combinatorial antiangiogenic gene therapy by nonviral gene transfer using the sleeping beauty transposon causes tumor regression and improves survival in mice bearing intracranial human glioblastoma. Mol Ther. 2005;12:778–788. doi: 10.1016/j.ymthe.2005.07.689. [DOI] [PubMed] [Google Scholar]
- Xue X, Huang X, Nodland SE, Mates L, Ma L, Izsvak Z, Ivics Z, LeBien TW, McIvor RS, Wagner JE, Zhou X. Stable gene transfer and expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells by a hyperactive Sleeping Beauty transposon system. Blood. 2009;114:1319–1330. doi: 10.1182/blood-2009-03-210005. [DOI] [PubMed] [Google Scholar]
- Sinn PL, Sauter SL, McCray PB Jr. Gene therapy progress and prospects: development of improved lentiviral and retroviral vectors--design, biosafety and production. Gene Ther. 2005;12:1089–1098. doi: 10.1038/sj.gt.3302570. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, Sorensen R, Forster A, Fraser P, Cohen JI, de Saint Basile G, Alexander I, Wintergerst U, Frebourg T, Aurias A, Stoppa-Lyonnet D, Romana S, Radford-Weiss I, Gross F, Valensi F, Delabesse E, Macintyre E, Sigaux F, Soulier J, Leiva LE, Wissler M, Prinz C, Rabbitts TH, Le Deist F, Fischer A, Cavazzana-Calvo M. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Baum C, von Kalle C, Staal FJ, Li Z, Fehse B, Schmidt M, Weerkamp F, Karlsson S, Wagemaker G, Williams DA. Chance or necessity? Insertional mutagenesis in gene therapy and its consequences. Mol Ther. 2004;9:5–13. doi: 10.1016/j.ymthe.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, Clappier E, Caccavelli L, Delabesse E, Beldjord K, Asnafi V, MacIntyre E, Dal Cortivo L, Radford I, Brousse N, Sigaux F, Moshous D, Hauer J, Borkhardt A, Belohradsky BH, Wintergerst U, Velez MC, Leiva L, Sorensen R, Wulffraat N, Blanche S, Bushman FD, Fischer A, Cavazzana-Calvo M. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrasher AJ, Gaspar HB, Baum C, Modlich U, Schambach A, Candotti F, Otsu M, Sorrentino B, Scobie L, Cameron E, Blyth K, Neil J, Abina SH, Cavazzana-Calvo M, Fischer A. Gene therapy: X-SCID transgene leukaemogenicity. Nature. 2006;443:E5–6. doi: 10.1038/nature05219. discussion E6-7. [DOI] [PubMed] [Google Scholar]
- Hackett PB, Largaespada DA, Cooper LJ. A transposon and transposase systemfor human application. Mol Ther. 2010;18:674–683. doi: 10.1038/mt.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walisko O, Schorn A, Rolfs F, Devaraj A, Miskey C, Izsvak Z, Ivics Z. Transcriptional activities of the Sleeping Beauty transposon and shielding its genetic cargo with insulators. Mol Ther. 2008;16:359–369. doi: 10.1038/sj.mt.6300366. [DOI] [PubMed] [Google Scholar]
- Moldt B, Yant SR, Andersen PR, Kay MA, Mikkelsen JG. Cis-acting gene regulatory activities in the terminal regions of sleeping beauty DNA transposon-based vectors. Hum Gene Ther. 2007;18:1193–1204. doi: 10.1089/hum.2007.099. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Katzer A, Stuwe EE, Fiedler D, Knespel S, Izsvak Z. Targeted Sleeping Beauty transposition in human cells. Mol Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Izsvak Z. Transposons for gene therapy! Curr Gene Ther. 2006;6:593–607. doi: 10.2174/156652306778520647. [DOI] [PubMed] [Google Scholar]
- Fernando S, Fletcher BS. Sleeping beauty transposon-mediated nonviral gene therapy. BioDrugs. 2006;20:219–229. doi: 10.2165/00063030-200620040-00003. [DOI] [PubMed] [Google Scholar]
- Ehrhardt A, Engler JA, Xu H, Cherry AM, Kay MA. Molecular analysis of chromosomal rearrangements in mammalian cells after phiC31-mediated integration. Hum Gene Ther. 2006;17:1077–1094. doi: 10.1089/hum.2006.17.1077. [DOI] [PubMed] [Google Scholar]
- Liu J, Jeppesen I, Nielsen K, Jensen TG. Phi c31 integrase induces chromosomal aberrations in primary human fibroblasts. Gene Ther. 2006;13:1188–1190. doi: 10.1038/sj.gt.3302789. [DOI] [PubMed] [Google Scholar]
- Cadinanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausl MA, Zhang W, Muther N, Rauschhuber C, Franck HG, Merricks EP, Nichols TC, Kay MA, Ehrhardt A. Hyperactive sleeping beauty transposase enables persistent phenotypic correction in mice and a canine model for hemophilia B. Mol Ther. 2010;18:1896–1906. doi: 10.1038/mt.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Urda S, Thyagarajan B, Keene DR, Lin Q, Fang M, Calos MP, Khavari PA. Stable nonviral genetic correction of inherited human skin disease. Nat Med. 2002;8:1166–1170. doi: 10.1038/nm766. [DOI] [PubMed] [Google Scholar]
- Montini E, Held PK, Noll M, Morcinek N, Al-Dhalimy M, Finegold M, Yant SR, Kay MA, Grompe M. In vivo correction of murine tyrosinemia type I by DNA-mediated transposition. Mol Ther. 2002;6:759–769. doi: 10.1006/mthe.2002.0812. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Kren BT, Wong PY, Low WC, Steer CJ. Sleeping Beauty-mediated down-regulation of huntingtin expression by RNA interference. Biochem Biophys Res Commun. 2005;329:646–652. doi: 10.1016/j.bbrc.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Zhu J, Kren BT, Park CW, Bilgim R, Wong PY, Steer CJ. Erythroid-specific expression of beta-globin by the sleeping beauty transposon for Sickle cell disease. Biochemistry. 2007;46:6844–6858. doi: 10.1021/bi6024484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronovich EL, Bell JB, Belur LR, Gunther R, Koniar B, Erickson DC, Schachern PA, Matise I, McIvor RS, Whitley CB, Hackett PB. Prolonged expression of a lysosomal enzyme in mouse liver after Sleeping Beauty transposon-mediated gene delivery: implications for non-viral gene therapy of mucopolysaccharidoses. J Gene Med. 2007;9:403–415. doi: 10.1002/jgm.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng PD, Cohen CJ, Yang S, Hsu C, Jones S, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA. Efficient nonviral Sleeping Beauty transposon-based TCR gene transfer to peripheral blood lymphocytes confers antigen-specific antitumor reactivity. Gene Ther. 2009;16:1042–1049. doi: 10.1038/gt.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CX, Shi D, Wu WJ, Ding YF, Feng DM, Lu B, Chen HM, Yao JH, Shen Q, Lu DR, Xue JL. Insulin expression in livers of diabetic mice mediated by hydrodynamics-based administration. World J Gastroenterol. 2004;10:567–572. doi: 10.3748/wjg.v10.i4.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Sanz S, Heggestad AD, Antharam V, Notterpek L, Fletcher BS. Endothelial targeting of the Sleeping Beauty transposon within lung. Mol Ther. 2004;10:97–105. doi: 10.1016/j.ymthe.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Izsvák Z, Ivics Z. Sleeping beauty transposition: biology and applications for molecular therapy. Mol Ther. 2004;9:147–156. doi: 10.1016/j.ymthe.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Hackett PB, Ekker SC, Largaespada DA, McIvor RS. Sleeping beauty transposon-mediated gene therapy for prolonged expression. Adv Genet. 2005;54:189–232. doi: 10.1016/S0065-2660(05)54009-4. full_text. [DOI] [PubMed] [Google Scholar]
- Essner JJ, McIvor RS, Hackett PB. Awakening gene therapy with Sleeping Beauty transposons. Curr Opin Pharmacol. 2005;5:513–519. doi: 10.1016/j.coph.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Belay E, Matrai J, Acosta-Sanchez A, Ma L, Quattrocelli M, Mates L, Sancho-Bru P, Geraerts M, Yan B, Vermeesch J, Rincón MY, Samara-Kuko E, Ivics Z, Verfaillie C, Sampaolesi M, Izsvák Z, Vandendriessche T, Chuah MK. Novel hyperactive transposons for genetic modification of induced pluripotent and adult stem cells: a nonviral paradigm for coaxed differentiation. Stem Cells. 2010;28:1760–1771. doi: 10.1002/stem.501. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. A fresh look at iPS cells. Cell. 2009;137:13–17. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]
- Williams DA. Sleeping beauty vector system moves toward human trials in the United States. Mol Ther. 2008;16:1515–1516. doi: 10.1038/mt.2008.169. [DOI] [PubMed] [Google Scholar]
- Bonini C, Grez M, Traversari C, Ciceri F, Marktel S, Ferrari G, Dinauer M, Sadat M, Aiuti A, Deola S, Radrizzani M, Hagenbeek A, Apperley J, Ebeling S, Martens A, Kolb HJ, Weber M, Lotti F, Grande A, Weissinger E, Bueren JA, Lamana M, Falkenburg JH, Heemskerk MH, Austin T, Kornblau S, Marini F, Benati C, Magnani Z, Cazzaniga S, Toma S, Gallo-Stampino C, Introna M, Slavin S, Greenberg PD, Bregni M, Mavilio F, Bordignon C. Safety of retroviral gene marking with a truncated NGF receptor. Nat Med. 2003;9:367–369. doi: 10.1038/nm0403-367. [DOI] [PubMed] [Google Scholar]
- Singh H, Manuri PR, Olivares S, Dara N, Dawson MJ, Huls H, Hackett PB, Kohn DB, Shpall EJ, Champlin RE, Cooper LJ. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;68:2961–2971. doi: 10.1158/0008-5472.CAN-07-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Guo H, Kang J, Choi S, Zhou TC, Tammana S, Lees CJ, Li ZZ, Milone M, Levine BL, Tolar J, June CH, Scott McIvor R, Wagner JE, Blazar BR, Zhou X. Sleeping Beauty transposon-mediated engineering of human primary T cells for therapy of CD19+ lymphoid malignancies. Mol Ther. 2008;16:580–589. doi: 10.1038/sj.mt.6300404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pule MA, Savoldo B, Myers GD, Rossig C, Russell HV, Dotti G, Huls MH, Liu E, Gee AP, Mei Z, Yvon E, Weiss HL, Liu H, Rooney CM, Heslop HE, Brenner MK. Virus-specific T cells engineered to coexpress tumor-specific receptors: persistence and antitumor activity in individuals with neuroblastoma. Nat Med. 2008;14:1264–1270. doi: 10.1038/nm.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BG, Jensen MC, Wang J, Chen EY, Wood BL, Greisman HA, Qian X, James SE, Raubitschek A, Forman SJ, Gopal AK, Pagel JM, Lindgren CG, Greenberg PD, Riddell SR, Press OW. Adoptive immunotherapy for indolent non-Hodgkin lymphoma and mantle cell lymphoma using genetically modified autologous CD20-specific T cells. Blood. 2008;112:2261–2271. doi: 10.1182/blood-2007-12-128843. [DOI] [PMC free article] [PubMed] [Google Scholar]