Abstract
Transgenic mice are widely used in biomedical research to study gene expression, developmental biology, and gene therapy models. Bacterial artificial chromosome (BAC) transgenes direct gene expression at physiological levels with the same developmental timing and expression patterns as endogenous genes in transgenic animal models. We generated 707 transgenic founders from 86 BAC transgenes purified by three different methods. Transgenesis efficiency was the same for all BAC DNA purification methods. Polyamine microinjection buffer was essential for successful integration of intact BAC transgenes. There was no correlation between BAC size and transgenic rate, birth rate, or transgenic efficiency. A narrow DNA concentration range generated the best transgenic efficiency. High DNA concentrations reduced birth rates while very low concentrations resulted in higher birth rates and lower transgenic efficiency. Founders with complete BAC integrations were observed in all 47 BACs for which multiple markers were tested. Additional founders with BAC fragment integrations were observed for 65% of these BACs. Expression data was available for 79 BAC transgenes and expression was observed in transgenic founders from 63 BACs (80%). Consistent and reproducible success in BAC transgenesis required the combination of careful DNA purification, the use of polyamine buffer, and sensitive genotyping assays.
Keywords: Mice, Transgenic, Gene Transfer Techniques, Chromosomes, Artificial, Bacterial, BAC, Electrophoresis, Gel, Pulsed-Field, DNA, Gene Expression
Introduction
Since the first transgenic mice were generated the ability to study gene expression in whole animal models has captured the scientific imagination. The ability to introduce exogenous DNA into animal models afforded insights into numerous physiological processes that could not be gleaned from cell culture studies. Transgenic models provided insights in developmental biology, gene regulation, and the genetic basis of human disease. Initially transgenes were limited to DNA fragments of a few thousand base pairs. The public mouse genome sequence is based, in large part, on the RPCI-23 mouse genomic library constructed with the pBACe3.6 vector (Osoegawa et al. 2000). This library is an important and convenient source of BAC transgenes. Recent advances in genomic library construction based on bacterial artificial chromosome (BAC) and yeast artificial chromosome (YAC) technologies make the generation of transgenes as large as 500 kb feasible and practical.
The ideal transgene drives expression in a cell-specific, copy-dependent fashion at levels that are easily detectable or that have informative physiological effects. Small plasmid transgenes often do not meet these criteria (Giraldo and Montoliu 2001, Hammer et al. 1987, Wilson et al. 1990). Research on BAC transgenic mice has resulted in the identification of disease-causing genetic mutations (Antoch et al. 1997; Probst et al. 1998; Jones et al. 2003; Oliver et al. 2004) and the identification of gene regulatory elements (Deal et al. 2006; Dunnick et al. 2005; Krebs et al. 2003). Large genomic transgenes such as BAC, P1, and YAC clones are more likely to produce copy number dependent transgene expression, independent of position effects, that recapitulate endogenous gene expression patterns (Callow et al. 1994, Chandler et al. 2007, Giraldo and Montoliu et al. 2001). For example, the generation of transgenic models to express human APOB from plasmid transgene constructs designed with heterologous promoters and APOB cDNA produced 29 transgenic founders, of which only one expressed the transgene (Chiesa et al. 1993). Subsequent transgenic models generated with 90 kb genomic transgenes expressed APOB in 11 of 13 transgenic lines in a copy number dependent and integration site independent fashion (Callow et al. 1994). Subsequent BAC transgenic models identified distant regulatory elements that regulate physiologically relevant human APOB expression patterns (Nielsen et al. 1997).
The advent of recombineering methods to manipulate BAC DNA sequences (Court et al. 2002) further increased the usefulness of BAC transgenic mice to address questions in biomedical research. BAC transgenes with the necessary genetic instructions to direct cell-specific gene expression can be modified so that proteins or reporters are expressed in defined cell populations. Examples of cell-specific expression from BAC transgenes include physiologically relevant proteins (Sun et al. 2003), reporter proteins such as lacZ or eGFP (Lehoczky and Innis 2008; Okita et al. 2007), and exogenous proteins such as Cre recombinase (Baoan et al. 2008). BAC transgenes direct gene expression to specific cell types in tissues more precisely than many promoter mini-constructs.
The most rapid and effective method to generate BAC transgenic mice is the direct microinjection of BAC DNA into the pronucleus of fertilized mouse eggs. The large size of BAC molecules precludes the use of lentiviral- or transposase- mediated transgenesis methods (limited to 8.5-10 kb). BACs integrated into mouse embryonic stem (ES) cells can be used as a vehicle to obtain transgenic mice from germline ES cell-mouse chimeras. BAC transgenes usually include enough genomic DNA to confer endogenous gene expression patterns so that the advantages of ES cell-mediated transgenesis, such as the ability to pre-screen ES cell clones for expression, (Bronson et al. 1996), are outweighed by the additional time and technical manipulations required for recombineering of drug selection cassettes (Testa et al. 2003), ES cell culture, generation of ES cell-mouse chimeras and germline breeding. The analysis of F0 hemizygous transgenic founders derived from BAC DNA microinjection of eggs collected from mutant mouse strains can quickly identify gene function in mutant phenotype rescue experiments (Antoch et al. 1997; Probst et al. 1998). BAC transgenesis by microinjection is an effective and efficient method to produce genetically engineered mouse models.
Common questions about BAC pronuclear microinjection include 1) the best method to purify BAC DNA for microinjection, 2) the best microinjection buffer, 3) the most effective microinjection DNA concentration, and 4) whether linear or circular DNA molecule microinjection is more efficient. We provide standard methods for BAC DNA purification by commercial anion exchange kits, CsCl gradients, and size exclusion chromatography (http://www.med.umich.edu/tamc/BACDNA.html). In our experience the use of polyamine microinjection buffer and validation of BAC DNA quality by pulsed-field gel electrophoresis is essential for success, as shown by the data in this paper. We find that transgenic efficiency is independent of the purification method, the size of the BAC DNA, and the form (circular or linear) of the molecules microinjected. Genotyping with multiple markers is essential to identify transgenic founders with intact or partial BAC transgene integrations. The microinjection of BAC DNA at a concentration of 0.5 ng/ul ensures high egg survival, reasonable birth rates, and effective transgenesis.
Materials and Methods
BAC DNA Purification
Three methods were used to purify BAC DNA for microinjection. Circular BAC DNA was prepared by modified alkaline lysis and anion exchange chromatography (Camper and Saunders 2000) or by CsCl gradients (Deal et al. 2006). After isolation linear BAC DNA molecules were prepared by digestion with a rare-cutting restriction enzyme (i.e. Not I) and separated from the cloning vector backbone by size-exclusion column chromatography (Dunnick et al. 2004). Circular or linear BAC transgenes were resuspended, and diluted for microinjection in polyamine (PA) buffer that was pre-filtered through Anotop inorganic membrane filters (average pore size: 0.02 uM from Whatman, Maidstone, England) that trap and remove fine particles that plug injection needles. The filters cannot be used with DNA suspensions because the small pore size effectively traps and removes DNA molecules from solution. PA buffer (10 mM Tris-HCl, pH 7.5, 0.1 mM EDTA, 30 uM spermine, 70 uM spermidine, 100 mM NaCl) was prepared as described (Montoliu et al. 1995). Other precautions taken during BAC DNA purification included gentle mixing of BAC containing solutions (no vortex mixing), storage of BAC DNA at 4°C (no freezing temperatures), use of wide mouth pipets and pipet tips, and never allowing precipitated BAC DNA to dry completely prior to the addition of PA buffer. After resuspension in PA buffer, BAC transgenes were stored at 4°C until they were microinjected, usually three to six weeks after purification. BAC transgenes were provided by submitting laboratories. BACs that were modified, as by the insertion of reporter genes, were modified by submitting laboratories (examples: Deal et al. 2006, Dunnick et al. 2004, Dunnick et al. 2005, Krebs et al. 2003, Lehoczky et al. 2008, Sun et al. 2003).
Pulsed-field Gel Electrophoresis
Prior to microinjection BAC DNA concentration was measured by absorbance at 260 nm with a spectrophotometer (NanoDrop, Thermo Scientific, Wilmington, DE) and BAC integrity was evaluated by pulsed-field gel electrophoresis (PFGE). Aliquots of Not I -digested and - undigested BAC transgenes were sized by PFGE to test for the presence of DNA molecules of the expected size and to assess the level of DNA fragmentation. PFGE was carried out essentially as described (Saunders and Camper 2000). Agarose gels (1.2% agarose in 0.5× TBE) were run at 4°C for 17 hours at 6 V/cm, 120° angle with 1-25 second switch times on a CHEF-DR III apparatus (BioRad, Hercules, CA). Size markers used were MidRange II PFG marker (New England Biolabs, Ipswich, MA) and 2-Log DNA Ladder (New England Biolabs). After electrophoresis gels were stained with EtBr for one hour and imaged with a Kodak Gel Logic 100 Imaging System (Carestream Health, New Haven, Connecticut). Photoshop software (Adobe, San Jose, California) was used to enhance contrast of black and white images.
Animals
(C57BL/6J × SJL/J)F1, and (C57BL/6J × DBA/2J)F1 mice were obtained from the Jackson Laboratory (Bar Harbor, Maine) and are referred to as B6SJLF1, and B6D2F1. Adult males were housed one per cage, while female mice were housed five per cage. Mice were housed in static microisolator cages (Allentown Caging, Allentown, NJ). Access to water and food (LabDiet 5008, Richmond, IN) was ad libitum. Animal rooms were climate controlled to provide temperatures of 22-23° C on a 12 h light/dark cycle (lights on at 0600). All animals were housed in an AAALAC accredited facility in accordance with the National Research Council's guide for the care and use of laboratory animals.
Superovulation and egg collection
B6SJLF1 egg donors were received at 20-24 days of age. Superovulation treatments were initiated within three to six days of receipt. Egg donors were treated with 5 IU pregnant mare's serum gonadotropin (National Hormone and Peptide Program, NIDDK, Bethesda MD) in 0.1 ml phosphate buffered saline (PBS, Invitrogen, Carlsbad, CA) by intraperitoneal (IP) injection and 46-50 hours later were treated with 5 IU human chorionic gonadotropin (HCG, Sigma-Aldrich, St. Louis, MO) in 0.1 ml PBS by IP injection. Egg donors were mated with B6SJLF1 stud males immediately after HCG treatment. The following day egg donors were euthanized then oviducts were removed and rinsed in M2 medium (Sigma-Aldrich). Ampulla were collected and opened in 15 mg hyaluronidase (Sigma-Aldrich) per ml M2 to release eggs and to remove cumulus cells. Eggs were washed once in M2 and placed in BMOC-3 (Invitrogen) under mouse egg-tested mineral oil (Sigma-Aldrich) in a 37°C, 5% CO2, humidified incubator. Eggs were stored in the incubator for 30 minutes to 6 hours prior to microinjection.
Pronuclear Microinjection
DNA was injected with needles pulled from 1 mm O.D., 0.75 mm I.D. glass capillaries with internal filaments (World Precision Instruments, Sarasota FL) on a Sutter P-87 micropipette puller (Sutter Instrument Co., Novato, CA). The P-87 was configured in the following way: 4.5 mm trough filament, heat set to 415, pull set to 150, velocity set to 75, time set to 115, pressure set to 70. Microinjection needles containing transgene DNA were connected to a pneumatic microinjector (Tritech Research, Los Angeles, CA). Holding pipets (200 um O.D., 30 um I.D.) were fashioned on a microforge (Technical Products International, O'Fallon, MO). The micromanipulation workstation consisted of a motorized Nikon TE2000–S microscope (Nikon, New York) equipped with differential interference contrast optics and Narishige hanging joysticks (Narishige, New York). Fertilized eggs with visible pronuclei were selected for microinjection. Unfertilized eggs were discarded. After microinjection, eggs were washed through four 75 ul drops of BOMC-3 under mineral oil and stored in the incubator. Only intact microinjected fertilized eggs were transferred to pseudopregnant recipients.
Egg transfer
Intact microinjected eggs were transferred to the oviducts of pseudopregnant recipients prepared by mating B6D2F1 females with vasectomized B6D2F1 males. Following anesthesia with ketamine/xylazine (Zeller et al. 1998), incisions in the skin and the peritoneal wall were made over the oviduct. A serrafine clamp was applied to the ovarian fat pad and used to exteriorize the ovary, oviduct, and the tip of the uterus. A small tear was made in the bursa over the ovary. An embryo transfer pipet fashioned from a flame-pulled Pasteur pipet 180 microns in diameter at the tip was used to introduce 10-13 eggs into the infundibulum of the left oviduct. Afterwards, the reproductive tract was returned to the peritoneal cavity and the skin was closed by surgical staples. This procedure was repeated with the right oviduct. After recovering from anesthetic on a warming plate, egg recipients were housed one or two per cage.
Genotyping and Transgene Expression
Ear tags (National Band and Tag, Newport, KY) were applied to 2-week old mouse pups for identification purposes and 5–10 mm tail biopsies were collected for DNA extraction. Genomic DNA was extracted and tested for the presence of BAC transgene DNA with transgene specific PCR (Saiki et al. 1988) or Southern blot (Southern et al. 1975) assays by investigators who submitted BACs for the generation of transgenic mouse founders. Prior to microinjection PCR primers were tested and demonstrated to detect single copy genomic equivalents of BAC transgene DNA mixed with mouse tail genomic DNA (http://www.med.umich.edu/tamc/spike.html) to preclude false negative genotyping results. PCR primers specific for an endogenous mouse gene, such as beta-globin, were used to control for false negatives. PCR assays were designed with primers for the 5′ and 3′ BAC vector-genomic DNA boundaries (i.e. SP6 and T7 sequences), primers unique to the genomic DNA within the BAC, or a combination of both. Expression data was accumulated from a number of investigators over a period of years in this retrospective study of our experience with BAC transgenic mouse production. As a result, quantitative expression data with respect to copy number and relative to endogenous gene levels is unavailable beyond the assessment that expression was detected. Published data with in-depth analysis of expression with respect to detection methods, spacing of genotyping markers along BAC transgenes, recapitulation of developmental and adult expression patterns, relationship to transgene copy number, and expression relative to endogenous gene levels is available (Antoch et al. 1997, Callow et al. 1994, Chandler et al. 2007, Chiesa et al. 1993, Deal et al. 2006, Dunnick et al. 2004, Dunnick et al. 2005, Giraldo and Montoliu, 2001, Krebs et al. 2003, Lehoczky et al. 2008, Mensah-Osman et al. 2008, Nielsen et al. 1997).
Statistical Analysis
Relationships among DNA microinjection concentration, birth rate, transgenic rate and transgenic efficiencies were examined by Correlation Analysis testing in Fig. 3 and Table 2 as described (Zar, pp 377-410). In each comparison the statistical null hypothesis tested was that there was no correlation between the two factors undergoing comparison. The alternative hypothesis was that the factors correlated. The two-tailed probability was calculated for each comparison and the alternative hypothesis was accepted if the probability that the null hypothesis was true was less than 5% (P<0.05). Correlation analysis assumes that sample data is obtained from normally distributed data. Percentages form binomial instead of normal distributions. In order to meet the assumptions of a normal distribution, percentages were subjected to arcsine angular transformation to produce normally distributed data for calculations (Zar pp. 278-280).
Figure 3.
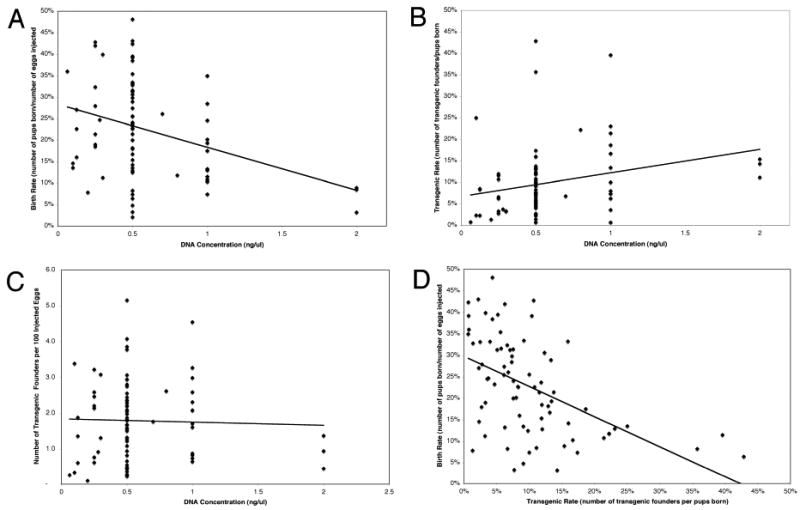
Correlations between DNA Concentration and Transgenic Efficiency.
All data were derived from microinjections of fertilized B6SJLF2 eggs with circular BAC DNA molecules purified by anion exchange chromatography and resuspended in PA buffer. 3a. Negative correlation between increasing concentrations of DNA microinjected and the proportion of live-born pups. 3b. Positive correlation between increasing concentrations of DNA microinjected and transgenic rates (proportion of transgenic pups). 3c. Absence of correlation between increasing concentrations of microinjected DNA transgenic efficiency (number of transgenic founders produced per 100 microinjected eggs). 3d. Negative correlation between birth rates and transgenic rates.
Table 2.
Correlation Between BAC DNA Concentration, Birth Rate, Transgenic Rate, and Transgenic Efficiency.
| BAC Size | BAC DNA Concentration | Birth Rate | Transgenic Ratea | Transgenic Efficiencyb | |
|---|---|---|---|---|---|
| BAC Size | - | - | n.s. | n.s. | n.s. |
| BAC DNA Concentration | - | p<0.002c | p<0.02d | n.s. | |
| Birth Rate | - | p<0.001e | n.s. | ||
| Transgenic Rate | - | n.s. |
Transgenic Rate: proportion of mouse pups born that were transgenic.
Transgenic Efficiency: Number of transgenic founders produced for every 100 microinjected eggs.
Birth rates of mouse pups declined as BAC DNA concentration increased.
Transgenic rates increased as BAC DNA concentration increased.
Birth rates decreased as transgenic rates increased.
Results
Protective Effect of Polyamine Buffer on BAC DNA
Small, sheared DNA fragments may be produced during BAC DNA purification and/or DNA storage. Microinjection of BAC DNA purified by anion exchange and resuspended in conventional TE buffer (10 mM Tris-HCl, pH 7.5, 0.25 mM EDTA) produced transgenic mice that contained BAC DNA fragments instead of intact BAC molecules. For example in one experiment 4 transgenic founders were obtained from 27 pups after 191 eggs were microinjected with BAC DNA stored in conventional buffer at 4°C for 21 days (birth rate =14%, transgenic rate = 15%, transgenic founders per 100 injected eggs = 2.1). These values are all within acceptable ranges. However, when the founders were genotyped by PCR for markers on the 3′ end of the cloning vector, the 5′ end of the cloning vector, and an internal, polymorphic genomic marker, it was found that three of four transgenic founders had only the 5′ marker and the fourth had the 5′ marker and the internal marker only. No founders had all three genotyping markers. Founders with all markers were obtained after circular BAC DNA was purified by anion exchange and resuspended in PA buffer. These observations are consistent with reports that PA buffer protects high molecular weight genomic DNA from degradation (Larin et al. 1991) and provides for the integration of intact transgenes (Schedl et al. 1993a, Schedl 1993b).
All purified BAC transgenes were tested by pulsed-field gel electrophoresis (PFGE) to determine if they were intact or sheared prior to microinjection. A representative gel analysis is shown for circular BAC 2039 (anion exchange purification, PA buffer) and linear BAC 2053 (anion exchange and column chromatography purification, PA buffer) (Fig. 1A). The presence of two strong DNA bands in BAC 2039 is indicative of intact DNA. The upper bands in Lanes 2 and 3 of the pulsed-field gel correspond to closed, circular, supercoiled BAC DNA and the lower bands correspond to relaxed open circles (Cole and Tellez 2004). Not I digestion shows the expected DNA fingerprint (Lane 4). In contrast to BAC 2039, BAC 2053 is clearly not suitable for microinjection since it contains sheared DNA instead of the expected 140 kb DNA band (Lanes 6-8). Conventional agarose gels cannot be substituted for PFGE because static field gel electrophoresis of sheared BAC DNA produces a sharp band due to lack of resolving power of conventional gels (Fig.1B). DNA smears in lanes with intact BAC DNA such as those seen in Figs. 1 and 2 are not uncommon when circular BAC molecules are run on pulsed-field gels (Cole and Tellez 2004, Sparwasser et al. 2004, Wang et al. 1995).
Figure 1.
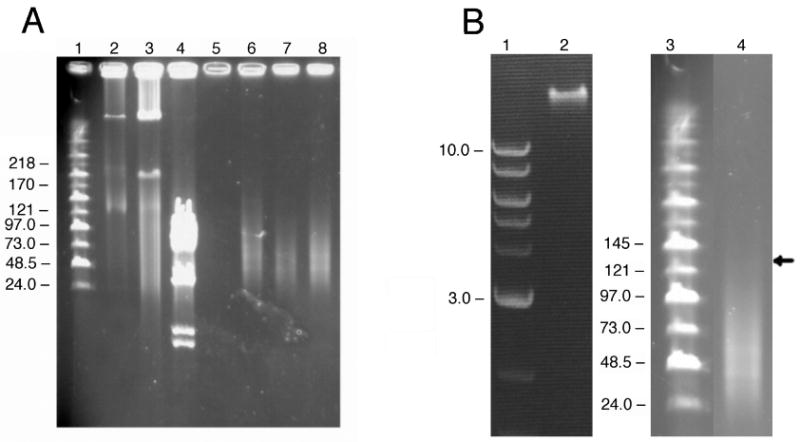
Pulsed-Field Gel Analysis of BAC DNA.
1a. Analysis of intact circular BAC DNA and sheared BAC DNA. Lane 1: Midrange PFG Marker II (New England Biolabs). Lanes 2, 3, and 4: BAC 2039 (expected size 182kb). Lane 2: Circular BAC DNA: 0.34 ug. Lane 3: Circular BAC DNA: 3.4 ug. Lane 4: Not I cut BAC DNA. Lane 5: deliberately empty. Lanes 6, 7, and 8 contain three fractions from a size exclusion column for BAC 2053. The expected size fragment is 140 kb. All three fractions contain sheared DNA smaller than 97 kb instead of intact linearized BAC DNA. 1b. Comparative analysis of sheared BAC DNA with conventional gel electrophoresis and pulsed-field gel electrophoresis. Lane 1: 2-Log DNA Ladder (New England Biolabs) separation on a conventional 0.8% agarose gel. Lane 2. Sheared BAC DNA runs as a sharp band on a conventional gel. Lane 3. Midrange PFG Marker II (New England Biolabs) separation on a pulsed-field gel. Lane 4. The same sheared BAC DNA that resolves as a sharp band on a conventional gel (Lane 2) appears as a smear upon pulsed-field gel electrophoresis. Arrow indicates expected size of BAC in Lane 4.
Figure 2.
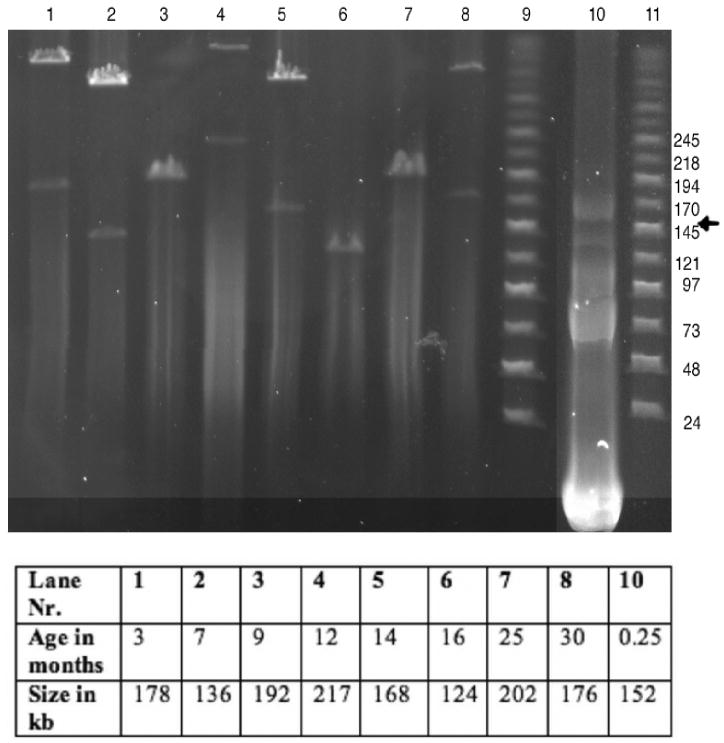
Stability of BAC DNA in Polyamine Buffer.
Circular BAC DNA was purified from bacterial cultures by modified alkaline lysis and anion exchange chromatography and resuspended in PA buffer (Lanes 1-8) or in conventional microinjection buffer (lane 10). BAC DNA preparations were stored for 1 week to 30 months at 4°C. Pulsed-field gel electrophoresis of 400 ng (Lanes 1-8) or 3,000 ng (Lane 10) of each BAC was performed. Arrow indicates expected size of BAC in Lane 10. Lane contents and time in storage are shown below the gel image. Lanes 9 and 11 contain Midrange PFG Marker II (New England Biolabs).
The average time that BAC transgenes were stored in PA buffer at 4°C before they were microinjected was 27 days with a range of 4 to 56 days. The transgenic rate for anion exchange purified BACs 1) injected 4 to 19 days after submission was 10.2% (n=12), for 2) BACs injected 20 to 39 days after submission the rate was 9.6% (n=28), and for 3) BACs injected 40 to 56 days after submission the rate was 10.4% (n=11). Unlike BACs stored in conventional microinjection buffer at 4°C, for BACs stored in PA Buffer founders with all desired markers were identified for BACs stored in PA buffer. There were no differences in transgenic rates for BACs stored in polyamine buffer up to 56 days. Pulsed field gel analysis showed that BACs stored in PA buffer at 4°C remained intact for up to 30 months (Fig. 2, Lanes 1-8). The upper DNA band corresponds to closed, circular, supercoiled BAC DNA and the lower band corresponds to relaxed open circles. BACs stored in conventional TE injection buffer and stored at 4°C degraded into sheared fragments after one week (lane 10). Survival and in vitro development of eggs microinjected with TE or PA buffers were the same (Table 1). Although polyamine buffer is chemically more complex than conventional microinjection buffer, it is not more toxic to fertilized eggs and protects BAC DNA from degradation. All BAC DNA transgenes described below were resuspended in polyamine buffer to improve transgenic mouse yields.
Table 1.
Absence of Polyamine Injection Buffer Toxicity
| Treatment | Intact Eggs/Injected Eggsa | 2-Cell Eggs/Intact Eggsb | Blastocysts/Intact Eggsc |
|---|---|---|---|
| Trial One | |||
| None | 20/ - | 20/20 | 18/20 |
| TE Injection | 14/20 | 13/14 | 8/14 |
| PA Injection | 16/20 | 15/16 | 8/15 |
| Trial Two | |||
| None | 20/ - | 20/20 | 13/20 |
| TE Injection | 15/20 | 15/15 | 11/15 |
| PA Injection | 13/20 | 13/13 | 12/13 |
| Trial Three | |||
| None | 20/ - | 19/20 | 15/19 |
| TE Injection | 20/20 | 19/20 | 12/19 |
| PA Injection | 16/20 | 15/16 | 12/15 |
Number of fertilized eggs that survived pronuclear microinjection without lysis.
Number of surviving eggs that divided to 2-cell eggs.
Number of 2-cell eggs that developed to blastocysts in cell culture.
Effect of BAC Size and DNA Concentration on Transgenesis
Intact circular BAC transgenes were purified by anion exchange and resuspended in polyamine buffer prior to microinjection into fertilized B6SJF2 mouse eggs. Effects of BAC size and DNA concentration on birth rates, proportion of transgenic founders born and transgenic efficiency (transgenic founders produced per 100 microinjected eggs) were analyzed by Correlation Analysis. BAC size did not influence birth rates, transgenic rates, or transgenic efficiency (Table 2). BAC DNA concentration was inversely correlated with birth rates (Fig. 3a) and positively correlated with transgenic rates (Fig. 3b) and did not correlate with transgenic efficiency (Fig. 3c). In addition, an inverse correlation was observed between birth rates and transgenic rates (Fig. 3d, Table 2). Since there was no correlation between DNA concentration and transgenic efficiency, BAC DNA was routinely microinjected at a concentration of 0.5 ng/ul. At this concentration birth rates are higher than at 1 ng/ul, which compensates for a slightly lower transgenic rate. The higher birth rates allow for successful transgenic production when other factors (genetic background, impure DNA samples) cause lower than average birth rates. Circular BAC transgenes purified by anion exchange chromatography, resuspended in PA buffer, and microinjected at 0.5 ng/ul into B6SJLF2 fertilized eggs resulted in 1) average birth rate of 24% ± standard deviation of 0.1%, 2) average transgenic rate of 9.4% ± 0.1%, 3) average transgenic efficiency of 1.9 ± 1.1, and 4) and average yield of 7 ± 4.2 transgenic founders per BAC transgene, (n=46).
Effect of Purification Method on Transgenic Efficiency
Transgenic efficiency for BAC transgenes purified by ion exchange, CsCl gradient, and size exclusion chromatography methods was compared. The effect of the DNA purification method was isolated by examining data from microinjections when the same injection buffer, DNA concentration, and genetic background were used. Results from BACs resuspended in polyamine buffer, microinjected at 0.5 ng/ul into fertilized B6SJLF2 eggs are summarized in Table 8. There is a clear trend to lower transgenic rates in the CsCl method; however, the CsCl transgenic efficiency was the same as other purification methods. Statistical analysis (ANOVA) showed no significant differences in birth rates, transgenic rates, or transgenic efficiency that could be attributed to DNA purification methods.
Table 8.
Effect of BAC DNA Purification Method on Transgenesis Efficiency
| Purification Method | Number of BACs | Birth Ratea | Transgenic Rateb | Transgenic Efficiencyc |
|---|---|---|---|---|
| Ion Exchange | 22 | 21.8 ± 11.0% | 11.1 ± 8.7% | 2.0 ± 1.1 |
| CsCl Gradient | 12 | 28.2 ± 6.9% | 6.7 ± 2.4% | 1.9 ± 0.9 |
| Size Exclusion | 7 | 26.7 ± 15.0% | 11.4 ± 12.8% | 1.9 ± 1.3 |
Average percentage of pups born from transferred eggs +/- standard deviation.
Average percentage of transgenic founders born +/- standard deviation.
Average number of transgenic founders born per 100 microinjected eggs +/- standard deviation.
Effect of Linear or Circular BAC DNA on Transgenic Efficiency
Transgenesis efficiency for linearized or circular BAC transgene DNA was compared. All transgenes were purified by ion exchange chromatography, resuspended in PA buffer, and microinjected at 0.5 ng/ul into fertilized B6SJLF2 eggs. Linear BAC transgenes were prepared by restriction enzyme digestion and size exclusion chromatography was used to isolate the microinjection fragment as described (Dunnick et al., 2004). Three linear and four circular BAC transgenes were compared (Table 9). Although statistical analysis found no difference between linear and circular BAC transgenes, the results show a tendency for higher transgenic efficiency in linear BAC DNA molecules (overall 2.53 founders produced for every 100 microinjected eggs compared to 1.48 founders per 100 eggs for circular BAC DNA).
Table 9.
Effect of BAC DNA Physical Form on Transgenesis Efficiency
| Physical Form | Number of BACs | Birth Ratea | Transgenic Rateb | Transgenic Efficiencyc |
|---|---|---|---|---|
| Linear | 3 | 18.7 ± 14.5% | 19.6 ± 17.0% | 2.53 ± 1.4 |
| Circular | 4 | 32.8 ± 14.0% | 5.3 ± 4.4% | 1.48 ± 1.1 |
Average percentage of pups born from transferred eggs +/- standard error.
Average percentage of transgenic founders born +/- standard error.
Average number of transgenic founders born per 100 microinjected eggs +/- standard error
BAC Transgene Integration and Frequency of Expression
Two or more unique genetic markers were available to identify transgenic founder mice for 47 BACs. Animals positive for all markers were considered to carry complete and intact BAC DNA molecules. The absence of one or more markers indicated that part of the BAC was missing from the chromosomal integration site. Results for 372 transgenic founders generated from 47 BAC transgenes showed that 244 (66%) were positive for all markers (Tables 4, 5, 6). Transgenic mice that had all of the genotyping markers were generated for every BAC transgene. For 17 BACs all of the founders were positive for all markers. However, not all of the mice generated from the other 30 BACs were positive for all genotyping markers. The likelihood of transgenic founders with partial BAC integrations was the same for ion exchange purified and CsCl purified transgenes (36%). Comparison with size exclusion purified BAC transgenes cannot be made since no founders were genotyped with more than one marker. Transgenic mice expressed genes from 63 of the 79 BAC transgenes for which expression data was available, although in two BACs, only mRNA expression was detected. No expression was observed from 16 BAC transgenes. Transgenic mouse founders that were identified with two or more genotyping markers were more than twice as likely to express the transgene as founders that were identified with a single genotyping assay. Overall 80% of the BACs resulted in transgenic mice that expressed the transgene, including those founders that were genotyped with a single marker.
Table 4.
Ion Exchange Purified DNA. BAC Transgene Integration Analyzed with Two or Three Genetic Markers.
| BAC Transgene | Size (kb) | Number of Foundersa | SP6 Markerb | T7 Markerc | Other Markerd | Expression Positive?e |
|---|---|---|---|---|---|---|
| 1160 | 70 | 4 | + | + | n.a. | no |
| 5 | - | + | n.a. | |||
| 1166 | 200 | 4 | + | + | n.a. | yes |
| 2 | - | + | n.a. | |||
| 1423 | 189 | 3 | + | + | n.a. | yes |
| 2 | + | - | n.a. | |||
| 2 | - | + | n.a. | |||
| 1634 | 240 | 8 | + | + | n.a. | yes |
| 1638 | 160 | 8 | + | + | n.a. | yes |
| 1 | + | - | n.a. | |||
| 2 | - | + | n.a. | |||
| 1740 | 195 | 8 | + | + | n.a. | yes |
| 1742 | 230 | 6 | + | + | n.a. | no |
| 1776 | 176 | 4 | + | + | n.a. | yes |
| 1 | - | + | n.a. | |||
| 1926 | 6 | + | + | n.a. | no | |
| 1 | + | - | n.a. | |||
| 2 | - | + | n.a. | |||
| 1962 | 4 | + | + | n.a. | no | |
| 1 | + | - | n.a. | |||
| 1951 | 6 | + | + | + | yes | |
| 4 | + | + | - | |||
| 1 | - | + | - | |||
| 1599 | 191 | 10 | n.a. | n.a. | +,+ | yes |
| 1673 | 196 | 6 | n.a. | n.a. | +,+ | yes |
| 1762 | 188 | 6 | n.a. | n.a. | +,+ | no |
| 2 | n.a. | n.a. | +,- | |||
| 1763 | 203 | 4 | n.a. | n.a. | +,+ | yes |
| 2 | n.a. | n.a. | -,+ | |||
| 1 | - | n.a. | +,- | |||
| 1764 | 246 | 4 | n.a. | n.a. | +,+ | yes |
| 4 | n.a. | n.a. | -,+ | |||
| 1 | n.a. | n.a. | +,- | |||
| 1816 | 160 | 3 | n.a. | n.a. | +,+ | yes |
| 1826 | 117.5 | 7 | n.a. | n.a. | +,+ | yes |
| 1 | n.a. | n.a. | -,+ | |||
| 1827 | 125 | 4 | n.a. | n.a. | +,+ | yes |
| 1 | n.a. | n.a. | +,- | |||
| 1838 | 190 | 4 | n.a. | n.a. | +,+ | yes |
| 2 | n.a. | n.a. | -,+ | |||
| 1 | n.a. | n.a. | +,- | |||
| 1911 | 115 | 7 | n.a. | n.a. | +,+ | yes |
| 5 | n.a. | n.a. | +,- | |||
| 7 | n.a. | n.a. | +,- | |||
| 1967 | 135 | 4 | n.a. | n.a. | +,+ | n.a. |
| 2 | n.a. | n.a. | +,- | |||
| 2 | n.a. | n.a. | -,+ | |||
| 1968 | 135 | 5 | n.a. | n.a. | +,+ | n.a. |
Number of transgene integration positive founder mice.
Result of PCR assay to detect SP6 – genomic boundary in BAC transgene.
Result of PCR assay to detect T7 – genomic boundary in BAC transgene.
Result of PCR assay to detect other markers in BAC transgene or cloning vector.
Outcome of transgene expression analyses.
n.a. Data are not available.
Table 5.
Ion Exchange BAC Transgene DNA Purification and Expression: Transgenic Founders Identified with Two to Nine Markers Internal to BAC.
| BAC Transgene | Size (kb) | Number of Foundersa | Genetic Markers | Positive Markers | Expression Positive?b |
|---|---|---|---|---|---|
| 1584 | 192 | 2 | 6 | 6 | yes |
| 4 | 6 | <6 | |||
| 1585 | 196 | 8 | 6 | 6 | yes |
| 1586 | 196 | 12 | 6 | 6 | yes |
| 1596 | 92 | 3 | 6 | 6 | yes |
| 1 | 6 | 4 | |||
| 1643 | 264 | 1 | 9 | 9 | yes |
| 1 | 9 | 7 | |||
| 2 | 9 | 5 | |||
| 1665 | 200 | 2 | 5 | 5 | yes |
| 1 | 5 | 3 | |||
| 1 | 5 | 1 | |||
| 1672 | 180 | 10 | 6 | 6 | yes |
| 2 | 6 | 2 | |||
| 1674 | 190 | 6 | 6 | 6 | yes |
| 1753 | 190 | 11 | 6 | 6 | yes |
| 1861 | 190 | 3 | 6 | 6 | n.a. |
Number of transgene integration positive founder mice.
Outcome of transgene expression analyses.
n.a. Data are not available.
Table 6.
CsCl Purified BAC DNA Transgene Integration. Two or More Genotyping Markers for Integration Show Evidence of Fragmented BAC Molecules.
| BAC Transgene | Size (kb) | Number of Foundersa | SP6 Assayb | T7 Assayc | Internal Assayd | Expression Positive?e |
|---|---|---|---|---|---|---|
| 1417 | 219 | 6 | + | + | + | yes |
| 5 | + | + | - | |||
| 3 | + | - | - | |||
| 1537 | 76 | 9 | + | + | + | yes |
| 6 | + | - | + | |||
| 1557 | 225 | 3 | + | + | + | yes |
| 2 | - | - | + | |||
| 2 | - | + | - | |||
| 1598 | 225 | 5 | + | + | + | yes |
| 1732 | 240 | 3 | + | + | + | yes |
| 3 | + | + | - | |||
| 5 | - | - | + | |||
| 2 | + | - | + | |||
| 1 | + | - | - | |||
| 1733 | 175 | 5 | + | + | + | yes |
| 2 | - | - | + | |||
| 1652 | 192 | 4 | n.a. | n.a. | +,+ | n.a. |
| 1653 | 174 | 9 | n.a. | n.a. | +,+ | n.a. |
| 1 | n.a. | n.a. | +,- | |||
| 1 | n.a. | n.a. | -,+ | |||
| 1669 | 204 | 3 | n.a. | n.a. | +,+ | n.a. |
| 2 | n.a. | n.a. | +,- | |||
| 1928 | 232 | 7 | + | + | + | yes |
| 11 | + | - | - | |||
| 3 | - | + | - | |||
| 1 | - | - | + | |||
| 5 | + | + | - | |||
| 1942 | 232 | 7 | + | + | + | yes |
| 5 | + | + | - | |||
| 1 | - | + | - | |||
| 1697 | 300 | 3 | n.a. | n.a. | + | yes |
| 1698 | 350 | 6 | n.a. | n.a. | + | yes |
| 1904 | 200 | 8 | n.a. | n.a. | + | yes |
Number of transgene integration positive founder mice.
Result of PCR assay to detect SP6 – genomic boundary in BAC transgene.
Result of PCR assay to detect T7 – genomic boundary in BAC transgene.
Result of PCR assay to detect internal sequences unique to BAC transgene.
Outcome of transgene expression analyses.
n.a. Data are not available.
Discussion
BAC DNA transgenesis requires the purification of intact BAC DNA. Birth rates and transgenic rates are dependent on carefully calibrated concentrations of microinjected DNA (Fig. 3). The use of polyamine microinjection buffer protects BAC DNA from fragmentation and does not interfere with in vitro development of microinjected eggs (Figs. 1 and 2, Table 1). Transgenesis efficiency is independent of the BAC size, BAC DNA purification method, and form (linear or circular) of BAC molecules that are microinjected (Table 2). Transgenic founders that express genes contained in BACs are generated for the majority of BAC transgenes. Genes in some BACs were not expressed, perhaps because of missing regulatory elements in genes that are too large to be contained in a single BAC or the absence of a complete genetic context. Multiple genotyping markers show that it is not unusual to generate transgenic founders that contain intact BAC molecules and founders that contain BAC fragments from the same BAC transgene (Tables 4-6). Complete BACs are more likely to provide physiologically relevant expression patterns and copy dependent expression levels while the examination of fragmented BACs may be useful to identify regulatory DNA sequences (Deal et al. 2006, Dunnick et al. 2004, Dunnick et al. 2005, Krebs et al. 2003, Lehoczky et al. 2008, Sun et al. 2003). An important key to success in BAC transgenesis is to control microinjection DNA concentrations so that the number of pups born and the number of transgenic founders generated are balanced.
Identification of a BAC DNA concentration that provides reasonable post-microinjection survival rates, birth rates, and transgenic rates is crucial. Eggs injected with BAC DNA molecules are more sensitive to the toxicity of high DNA concentrations than those injected with high concentrations of small plasmid transgenes. For example, when BAC 1282 was microinjected at 1 ng/ul we observed an extremely low birth rate (10 pups born from 443 microinjected eggs). Upon 10 fold dilution of the injection concentration to 0.1 ng/ul the birth rate improved to 107 pups from 730 injected eggs, but only two transgenic founders were identified. An ideal concentration would balance birth rates and transgenic rates more evenly. Compared to small plasmid transgenes, which are microinjected at 1-2 ng/ul (Brinster et al. 1985), the concentration range for BACs is narrower and can vary among BAC DNA preparations. The molarity of small plasmid transgene molecules results in the injection of a few hundred transgene molecules per mouse egg. During BAC microinjection 10 to 100 fold fewer DNA molecules are introduced per egg. The percentage of transgenic pups born from the microinjection of large DNA transgenes is comparable to the percentage obtained from small DNA microinjection (Schedl et al. 1993b; Callow et al. 1994; Antoch et al. 1997; Probst et al. 1998). As many as 1000 copies of small transgenes can integrate into a single chromosomal site in transgenic mice (Lo 1986). In BAC transgenic mice, the majority of BAC transgene concatemers contain fewer than a dozen copies (Camper and Saunders 2000; Giraldo and Montoliu 2001), although integration of a 76-copy BAC concatemer has been reported (Chandler et al. 2007). The lower copy numbers typically observed in BAC transgenesis most likely reflects the smaller number of DNA molecules microinjected into each fertilized egg. Estimates of a few hundred plasmid molecules microinjected per fertilized mouse egg (Brinster et al. 1985) are consistent with the microinjection of a few tens of BAC molecules per egg. Even though fewer molecules are microinjected, BAC transgenic efficiency is comparable to that of smaller plasmid transgenes.
High BAC DNA concentrations result in viscous DNA solutions that can be observed as they flow out of injection needles and mix with the media in the microinjection chamber. Such sticky DNA preparations trap nucleoli upon pronuclear injection and pull them out of the egg as the microinjection needle is removed. This results in egg lysis, which reduces the number of viable pups that can be obtained. We use a concentration of 0.5 ng/ul is for the initial microinjection session. This is adjusted downward to a lower concentration if one of the following conditions is noted: high viscosity of the microinjection solution causing difficulty with the injections (“sticky” needles); an extraordinarily high rate of lysis of injected eggs during the injection session (less than 70% survival); a low 2-cell embryo rate for injected eggs allowed to culture overnight (less than 70%), or an unusually low birth rate for pups (less than 10%). If a high birth rate, accompanied by a low transgenic rate is the result, then the DNA is reinjected at a higher concentration. On those occasions when BAC transgenic mice are not produced in the first microinjection sessions, the most practical solution is to purify and microinject a new BAC DNA preparation.
Examination of transgenic efficiency (the number of transgenic founders produced per microinjected eggs) facilitates comparisons between technical methods. In this context the purification methods of ion exchange, column chromatography, or CsCl gradients are equivalent (Table 8). Differences in transgenic efficiency between linear and circular BAC DNA fragments were not observed (reviewed in Camper and Saunders 2000), unlike those differences reported for linear and supercoiled small plasmid transgenes (Brinster et al. 1985). Systematic interference with expression by the plasmid backbone of the BAC cloning vector was not observed, although this was reported for plasmid transgenes (Hammer et al. 1987; Kjer-Nielsen et al. 1992; Townes et al. 1985).
In transgenesis with circular BAC DNA it is sometimes assumed that random breaks will occur in the BAC, interrupt the gene of interest (particularly if the gene occupies most of the BAC), and preclude gene expression. This intuitive assumption about transgene DNA integration is incongruent with existing evidence and models for exogenous DNA insertion into chromosomes in fertilized eggs. Integration of experimentally introduced DNA in eggs occurs after a process of homologous recombination between transgene molecules to form a tandem array (Luciw et al. 1983; Wagner et al. 1996; reviewed by Bishop 1996). The process of recombination resolves DNA fragments into intact molecules (Fiorenza et al. 2001). Some research groups have taken advantage of this process to build larger transgenes from smaller DNA molecules, including plasmid (Keegan et al. 1994), cosmid (Tacken et al. 2001) and P1 clones (Wagner et al. 1996). There is no a priori advantage for the use of linear BAC DNA since both circular BACs and linear fragments mediate transgenesis efficiently with the same expression outcomes.
Either circular or linearized BAC DNA can be used for pronuclear microinjection. Thus, circular BAC DNA most commonly is microinjected since it is simpler to purify. Transgenic efficiency is the same among BAC DNA molecules ranging in size from 61 to 303 kb. Circular BAC DNA is conveniently isolated from bacterial cultures with available commercial kits. Careful DNA preparation and the use of polyamine microinjection buffer protect circular BACs from shearing. The isolation of linearized BACs free of the cloning vector backbone or the isolation of subfragments requires additional manipulations that reduce DNA yields and risk DNA shearing. Unless there is a cogent reason for BAC DNA fragment purification, such as the analysis of gene regulatory regions or the exclusion of genes from multigenic BACs, the preparation of transgenic mice with circular BAC DNA molecules is warranted.
The desired outcome of BAC transgenesis is the expression of genes contained within BACs. Because of their size, it is advantageous to genotype BAC transgenic mice with multiple markers across the BAC. Transgenic mouse lines positive for two or more markers contained in the BAC were more than twice as likely to express the gene of interest as lines positive for only one marker (Tables 3-7). Two-thirds of BACs for which multiple markers were available produced transgenic lines that contained BAC fragments in addition to lines that contained intact BACs (Tables 4-6). Genotyping transgenic founders with multiple markers is essential for the identification of genes and regulatory elements that confer gene expression in the founders. Overall 80% of BACs produced transgenic lines that expressed the desired genes. Despite the need to differentiate between intact and incomplete BAC integrations, the analysis of partial BAC transgenes can be advantageous for the identification of genes in positional cloning projects (Antoch et al. 1997; Jones et al. 2003) or the identification of gene regulatory elements (Deal et al. 2006).
Table 3.
Ion Exchange BAC DNA Purification and Expression: Transgenic Founders Identified with One Genetic Marker Internal to BAC Transgene.
| BAC Transgene | Size (kb) | Number of Foundersa | Expression Positive?b |
|---|---|---|---|
| 1164 | 150 | 16 | no |
| 1165 | 150 | 11 | no |
| 1233 | 200 | 13 | no |
| 1242 | 200 | 14 | yes |
| 1243 | 200 | 19 | yes |
| 1246 | 108 | 9 | yes |
| 1259 | 130 | 3 | yes |
| 1260 | 130 | 12 | yes |
| 1282 | 130 | 4 | yes |
| 1302 | 250 | 2 | no |
| 1327 | 120 | 8 | yes |
| 1375 | 130 | 5 | yes |
| 1654 | 190 | 16 | yes |
| 1679 | 303 | 10 | yesc |
| 1711 | 134 | 4 | yes |
| 1741 | 196 | 8 | yes |
| 1745 | 188 | 3 | yesc |
| 1748 | 197 | 3 | yes |
| 1770 | 200 | 7 | no |
| 1789 | 197 | 6 | no |
| 1824 | 194 | 18 | yes |
| 1858 | 188 | 8 | yes |
| 1902 | 190 | 8 | yes |
| 1921 | 222 | 5 | yes |
| 1952 | 85 | 4 | n.a. |
Number of transgene integration positive founder mice.
Outcome of transgene expression analyses.
RNA transcript only was detected.
Table 7.
Size Exclusion BAC DNA Purification and Expression: Transgenic Founders Identified with a Single Genetic Marker Unique to the BAC Transgene.
| BAC Transgene | Size (kb) | Number of Foundersa | Expression Positive?b |
|---|---|---|---|
| 1444 | 230 | 14 | yes |
| 1472 | 80 | 4 | no |
| 1475 | 80 | 4 | yes |
| 1481 | 230 | 8 | yes |
| 1487 | 50 | 4 | yes |
| 1756 | 217 | 4 | no |
| 1788 | 100 | 4 | no |
| 1796 | 195 | 10 | yes |
| 1812 | 97 | 16c | no |
| 1830 | 230 | 3 | yes |
| 1839 | 215 | 12 | yes |
| 1842 | 200 | 7c | yes |
| 1972 | 230 | 7c | yes |
| 1980 | 142 | 5d | no |
Number of transgene integration positive founder mice.
Outcome of transgene expression analyses.
Two markers internal to the BAC transgene were used for genotyping.
Five markers internal to the BAC transgene were used for genotyping.
Production of transgenic mice with large transgenes is more difficult due to stringent requirements for highly purified intact DNA molecules. Multiple genotyping markers across BAC transgenes are essential to verify that intact BAC molecules integrate in transgenic founders. Compared to plasmid DNA molecules the DNA concentration for effective transgenesis falls into a more narrow range for BACs than for plasmid transgenes. Similarly, the optimum BAC DNA transgenesis conditions that provide good post-injection survival rates, reasonable birth rates, and effective transgenic rates are more difficult to establish than with plasmid DNA transgenes.
Acknowledgments
We thank Tina Jones and Corey Ziebell for their management of the transgenic production mouse colonies and we thank Susan Allen for her editorial assistance. The Transgenic Animal Model Core of the University of Michigan's Biomedical Research Core Facilities is supported by the University of Michigan Cancer Center (NIH CA046592), the University of Michigan Rheumatic Diseases Center Core (NIH AR048310), the University of Michigan Gastrointestinal Peptide Research Center (NIH DK034933), and the Nathan Shock Center for the Biology of Aging (NIH AG013283).
References
- Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JO. Chromosomal insertion of foreign DNA. Reprod Nutr Dev. 1996;36:607–618. [PubMed] [Google Scholar]
- Baoan J, Song J, Tsou L, Bi Y, Gaiser S, Mortensen R, Logsdon C. Robust acinar cell transgene expression of CreErT via BAC recombineering. Genesis. 2008;35:390–395. doi: 10.1002/dvg.20411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Chen HY, Trumbauer ME, Yagle MK, Palmiter RD. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc Natl Acad Sci USA. 1985;82:4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson SK, Plaehn EG, Kluckman KD, Hagaman JR, Maeda N, Smithies O. Single-copy transgenic mice with chosen-site integration. Proc Natl Acad Sci USA. 1996;93:9067–9072. doi: 10.1073/pnas.93.17.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow MJ, Stoltzfus LJ, Lawn RM, Rubin EM. Expression of human apolipoprotein B and assembly of lipoprotein(a) in transgenic mice. Proc Natl Acad Sci USA. 1994;91:2130–2134. doi: 10.1073/pnas.91.6.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camper SA, Saunders TL. Transgenic rescue of mutant phenotypes using large DNA fragments. In: Accili D, editor. Genetic manipulation of receptor expression and function. Wiley-Liss; New York: 2000. pp. 1–22. [Google Scholar]
- Chandler KJ, Chandler RL, Broeckelmann EM, Hou Y, Southard-Smith EM, Mortlock DP. Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mamm Genome. 2007;18:693–708. doi: 10.1007/s00335-007-9056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiesa G, Johnson DF, Yao Z, Innerarity TL, Mahley RW, Young SG, Hammer RH, Hobbs HH. Expression of human apolipoprotein B100 in transgenic mice. Editing of human apolipoprotein B100 mRNA. J Biol Chem. 1993;268:23747–23750. [PubMed] [Google Scholar]
- Cole KD, Tellez CM. Separation of large circular DNA by electrophoresis in agarose gels. Biotechnol Prog. 2002;18:82–87. doi: 10.1021/bp010135o. [DOI] [PubMed] [Google Scholar]
- Court DL, Sawitzke JA, Thomason LC. Genetic Engineering using Homologous Recombination. Annu Rev Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- Deal KK, Cantrell VA, Chandler RL, Saunders TL, Mortlock DP, Southard-Smith EM. Distant regulatory elements in a Sox10-betaGEO BAC transgene are required for expression of Sox10 in the enteric nervous system and other neural crest-derived tissues. Dev Dyn. 2006;235:1413–1432. doi: 10.1002/dvdy.20769. [DOI] [PubMed] [Google Scholar]
- Dunnick WA, Shi J, Graves KA, Collins JT. Germline transcription and switch recombination of a transgene containing the entire H chain constant region locus: Effect of a mutation in a STAT6 binding site in the gamma1 promoter. J Immun. 2004;173:5531–5539. doi: 10.4049/jimmunol.173.9.5531. [DOI] [PubMed] [Google Scholar]
- Dunnick WA, Shi J, Graves KA, Collins JT. The 3′ end of the heavy chain constant region locus enhances germline transcription and switch recombination of the four gamma genes. J Exp Med. 2005;201:1459–1466. doi: 10.1084/jem.20041988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorenza MT, Bevilacqua A, Bevilacqua S, Mangia F. Growing dictyate oocytes, but not early preimplantation embryos, of the mouse display high levels of DNA homologous recombination by single-strand annealing and lack DNA nonhomologous end joining. Dev Biol. 2001;233:214–224. doi: 10.1006/dbio.2001.0199. [DOI] [PubMed] [Google Scholar]
- Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 2001;10:83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- Hammer RE, Krumlauf R, Camper SA, Brinster RL, Tilghman SM. Diversity of alpha-fetoprotein gene expression in mice is generated by a combination of separate enhancer elements. Science. 1987;235:53–58. doi: 10.1126/science.2432657. [DOI] [PubMed] [Google Scholar]
- Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z, Davies E, Hajnocyzky, Saunders TL, Van Keuren ML, Fernandes-Alnemri T, Meisler MH, Alnemri ES. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature. 2003;425:721–727. doi: 10.1038/nature02052. [DOI] [PubMed] [Google Scholar]
- Krebs CJ, Larkins LK, Price R, Tullis KM, Miller RD, Robins DM. Regulator of sex-limitation (Rsl) encodes a pair of KRAB zinc-finger genes that control sexually dimorphic liver gene expression. Genes Dev. 2003;17:2664–2674. doi: 10.1101/gad.1135703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan CE, Karolyi IJ, Burrows H, Camper SA, Seasholtz AF. Homologous recombination in fertilized mouse eggs and assessment of heterologous LCR function. Transgenics. 1994;1:439–449. [Google Scholar]
- Kjer-Nielsen L, Holmberg K, Perera JD, McCluskey J. Impaired expression of chimaeric major histocompatibility complex transgenes associated with plasmid sequences. Transgenic Res. 1992;1:182–187. doi: 10.1007/BF02522537. [DOI] [PubMed] [Google Scholar]
- Larin Z, Monaco AP, Lehrach H. Yeast artificial chromosome libraries containing large inserts from mouse and human DNA. Proc Natl Acad Sci USA. 1991;88:4123–4127. doi: 10.1073/pnas.88.10.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehoczky JA, Innis JW. BAC transgenic analysis reveals enhancers sufficient for Hoxa13 and neighborhood gene expression in mouse embryonic distal limbs and genital bud. Evol Dev. 2008;10:421–432. doi: 10.1111/j.1525-142X.2008.00253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CW. Localization of low abundance DNA sequences in tissue sections by in situ hybridization. J Cell Sci. 1986;81:143–162. doi: 10.1242/jcs.81.1.143. [DOI] [PubMed] [Google Scholar]
- Luciw PA, Bishop JM, Varmus HE, Capecchi MR. Location and function of retroviral and SV40 sequences that enhance biochemical transformation after microinjection of DNA. Cell. 1983;33:705–716. doi: 10.1016/0092-8674(83)90013-2. [DOI] [PubMed] [Google Scholar]
- Montoliu L, Bock CT, Schutz G, Zentgraf H. Visualization of large DNA molecules by electron microscopy with polyamines: application to the analysis of yeast endogenous and artificial chromosomes. J Mol Biol. 1995;246:486–492. doi: 10.1006/jmbi.1994.0100. [DOI] [PubMed] [Google Scholar]
- Mensah-Osman E, Labut E, Zavros Y, El-Zaatari M, Law DJ, Merchant JL. Regulated expression of the human gastrin gene in mice. Regul Pept. 2008;151:115–122. doi: 10.1016/j.regpep.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen LB, McCormick SP, Pierotti V, Tam C, Gunn MD, Shizuya H, Young SG. Human apolipoprotein B transgenic mice generated with 207- and 145-kilobase pair bacterial artificial chromosomes. Evidence that a distant 5′-element confers appropriate transgene expression in the intestine. J Biol Chem. 1997;272:29752–29758. doi: 10.1074/jbc.272.47.29752. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Oliver ER, Saunders TL, Tarlé SA, Glaser T. Ribosomal protein L24 defect in Belly spot and tail (Bst), a mouse Minute. Development. 2004;131:3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa K, Tateno M, Woon PY, Frengen E, Mammoser AG, Catanese JJ, Hayashizaki Y, de Jong PJ. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 2000;10:116–128. [PMC free article] [PubMed] [Google Scholar]
- Probst FJ, Fridell RA, Raphael Y, Saunders TL, Wang A, Liang Y, Morell RJ, Touchman JW, Lyons RH, Noben-Trauth K, Friedman TB, Camper SA. Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science. 1998;280:1444–1447. doi: 10.1126/science.280.5368.1444. [DOI] [PubMed] [Google Scholar]
- Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Schedl A, Larin Z, Montoliu L, Thies E, Kelsey G, Lehrach H, Schütz G. A method for the generation of YAC transgenic mice by pronuclear microinjection. Nucleic Acids Res. 1993a;21:4783–4787. doi: 10.1093/nar/21.20.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl A, Montoliu L, Kelsey G, Schutz G. A yeast artificial chromosome covering the tyrosinase region confers copy number dependent expression in transgenic mice. Nature. 1993b;362:258–261. doi: 10.1038/362258a0. [DOI] [PubMed] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sparwasser T, Gong S, Li JY, Eberl G. General method for the modification of different BAC types and the rapid generation of BAC transgenic mice. Genesis. 2004;38:39–50. doi: 10.1002/gene.10249. [DOI] [PubMed] [Google Scholar]
- Sun H, Yang TL, Yang A, Wang X, Ginsburg D. The murine platelet and plasma factor V pools are biosynthetically distinct and sufficient for minimal hemostasis. Blood. 2003;102:2856–2861. doi: 10.1182/blood-2003-04-1225. [DOI] [PubMed] [Google Scholar]
- Tacken PJ, van der Zee A, Beumer TL, Florijn RJ, Gijpels MJ, Havekes LM, Frants RR, van Dijk KW, Hofker MH. Effective generation of very low density lipoprotein receptor transgenic mice by overlapping genomic DNA fragments: high testis expression and disturbed spermatogenesis. Transgenic Res. 2001;10:211–221. doi: 10.1023/a:1016682520887. [DOI] [PubMed] [Google Scholar]
- Testa G, Zhang Y, Vintersten K, Benes V, Pijnappel WW, Chambers I, Smith AJ, Smith AG, Stewart AF. Engineering the mouse genome with bacterial artificial chromosomes to create multipurpose alleles. Nat Biotechnol. 2003;21:443–447. doi: 10.1038/nbt804. [DOI] [PubMed] [Google Scholar]
- Townes TM, Lingrel JB, Chen HY, Brinster RL, Palmiter RD. Erythroid-specific expression of human beta-globin genes in transgenic mice. EMBO J. 1985;4:1715–1723. doi: 10.1002/j.1460-2075.1985.tb03841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner SD, Gross G, Cook GP, Davies SL, Neuberger MS. Antibody expression from the core region of the human IgH locus reconstructed in transgenic mice using bacteriophage P1 clones. Genomics. 1996;35:405–414. doi: 10.1006/geno.1996.0379. [DOI] [PubMed] [Google Scholar]
- Wang M, Lai E. Pulsed field separation of large supercoiled and open-circular DNAs and its application to bacterial artificial chromosome cloning. Electrophoresis. 1995;16:1–7. doi: 10.1002/elps.1150160102. [DOI] [PubMed] [Google Scholar]
- Wilson C, Bellen HJ, Gehring WJ. Position effects on eukaryotic gene expression. Ann Rev Cell Biol. 1990;6:679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
- Zar JH. Biostatistical analysis. 4th. Prentice Hall; Upper Saddle River, New Jersey: 1998. [Google Scholar]
- Zeller W, Meier G, Burki K, Panoussis B. Adverse Effects of Tribromoethanol as used in the production of transgenic mice. Lab Animals. 1998;32:407–413. doi: 10.1258/002367798780599811. [DOI] [PubMed] [Google Scholar]


