1. Structure
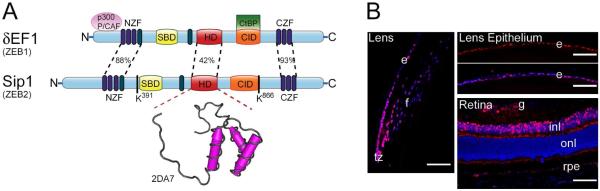
Smad Interacting Protein 1 (Sip1, also known as ZEB2, ZFHX1B, SMADIP1, FLJ42816, KIAA0569; accession #NP_055610/human and #NP_056568/mouse) is a 1,214-amino acid, 140 kDa protein which is a member of the ZEB family of two-handed zinc finger/homeodomain proteins. Sip1 is highly conserved in vertebrates, sharing 92% identity between humans and Xenopus, while it is 69% similar to its closest relative, δ-Crystallin Enhancer Binding Factor 1 (δEF1, also known as ZEB1, ZFHX1A, TCF8, BZP, AREB6, FECD6, NIL2A, ZFHEP, MGC133261). ZEB transcription factors are characterized by two separate clusters of zinc fingers, four at the N-terminus (three CCHH and one CCHC) and three at the C-terminus (all CCHH), (Fig. 1A), both of which must bind for transcriptional regulation. The homeodomain-like region, located near the center of the protein, folds into a typical structure with three alpha helices (PDB: 2DA7), although it seems to participate mainly in protein-protein interactions. A Smad binding domain and additional co-repressor (e.g. CtBP) and co-activator (e.g. p300 and P/CAF) binding domains are also found in both Sip1 and δEF1 (Verschueren et al., 1999; Vandewalle et al., 2009). Sip1 is SUMOylated at Lys-391 and Lys-866 which reduces its potency as a transcriptional repressor.
Fig. 1.
(A) Schematic diagram of the ZEB family transcription factors, Sip1 and δEF1, which are characterized by the presence of two zinc-finger hands at the N- and C-terminus (NZF and CZF, respectively) and a central homeodomain-like region (HD). The protein crystal structure for the homeodomain of Sip1 has the expected three alpha helix configuration (PDB 2DA7). The ZEB proteins also contain a Smad binding domain (SBD) and a CtBP interacting domain (CID). The co-activators p300 and P/CAF can also bind to the proteins in the N-terminal region. The two Sip1 SUMOlation sites, K391 and K866, are also shown. (B) Immunostaining with an anti-Sip1 antibody shows that Sip1 protein is found in the neural retina as well as in the lens epithelium and transitional zone The cornea showed a high level of background staining and was not included. Red: Sip1 (Santa Cruz); Blue: Nuclear/DNA; scale bars = 77μm. Abbreviations: e, lens epithelium; tz, lens transition zone; f, lens fiber cells; g, ganglion cells; inl, inner nuclear layer; onl, outer nuclear layer; rpe, retinal pigmented epithelium.
2. Function
Sip1 is expressed by numerous tissues during embryonic development, including the neural crest, neuroepithelium and limb buds. Later Sip1 expression regulates the morphogenesis of the neural crest derived craniofacial mesenchyme and parasympathetic ganglia. Complete loss of Sip1 function in mice leads to death by E9.5 due to a lack of neural tube closure (Vandewalle et al., 2009). In the eye, Sip1 is highly expressed in the lens epithelium and the neural retina (Fig. 1B). Deletion of the Sip1 gene from the lens placode causes failure of lens vesicle separation, small lens size, and deficient crystallin expression (Yoshimoto et al., 2005).
The zinc finger hands of ZEB transcription factors, including Sip1, bind to the consensus sequence, 5′-CACCT(G), which overlaps with the consensus for the bHLH DNA-binding proteins leading to the proposal that ZEB proteins compete for these binding sites to regulate transcription (Verschueren et al., 1999). In Xenopus, over-expression of Sip1 blocks the expression of the activin-dependent gene Brachyury (Xbra) by direct binding to the Xbra promoter, thus disrupting posterior mesoderm development. Sip1 can also interact with the receptor regulated Smads 1, 2, 3, 5 and 8 via the Smad interacting domain suggesting that Sip1 crosstalks with both Transforming Growth Factor Beta (TGF-β) and the Bone Morphogenetic Protein (BMP) pathways. Verschueren et al. (1999) showed that Sip1 binds tightly to activated Smads in the absence of other co-factors, while δEF1 requires p300, a co-activator, to be present for SMAD interaction. Differential co-activator and co-repressor binding to Sip1 versus δEF1 may result in large functional consequences, although this is controversial. Notably, Sip1 binds Smad3 with higher affinity than Smad2, indicating that Sip1 may contribute to functional differences between the R-Smads in vivo. Sip1 can regulate Smad-mediated transcription even when ZEB binding sites are absent from a target gene, but the presence of ZEB binding sites appears to concentrate the SMAD-ZEB complex at gene promoters more efficiently (Vandewalle et al., 2009). For example, Sip1 binds to and activates the Foxe3 promoter in vitro and Foxe3 promoter activity was further enhanced by the binding of Smad 8 to the Smad binding domain of Sip1 (Yoshimoto et al., 2005). These observations suggest an important role for Sip1 in fine tuning the transcriptional consequences of TGFβ and BMP signaling.
Much of the literature regarding Sip1 focuses on its role in driving the transition from epithelial to mesenchymal phenotypes (EMT), particularly during cancer progression. Sip1 and/or the SMAD-Sip1 complex can repress epithelial specific cell gene expression (claudins, ZO-3, connexins, E-cadherin, plakophilin 2, desmoplakin, and crumbs3) and activate mesenchymal gene expression (vitronectin, vimentin, N-cadherin, and MMPs) in numerous cell types (Vandewalle et al., 2009). Interestingly, multiple microRNAs which repress Sip1 translation are also downregulated by Sip1 providing a potential feedback loop leading to the upregulation of Sip1 expression during EMT. While the list of target genes Sip1 can regulate, via SMAD pathways or directly, is growing rapidly, the full complexity of ZEB family function remains elusive.
3. Disease involvement
A diversity of heterozygous mutations in the Sip1 gene result in Mowat-Wilson Syndrome (MWS; MIM 235730) which typically presents as Hirschsprung’s disease/severe constipation, mental retardation, microcephaly, short stature, and a distinctive facial appearance (Zweier et al., 2005). Ocular abnormalities, including microphthalmia, cataract, iris coloboma and Axenfeld anomaly, were seen in 14.3% of MWS patients consistent with the high levels of Sip1 expressed in the developing eye and the ocular phenotype of conditional knockout mice lacking Sip1 in the lens (Yoshimoto et al., 2005; Zweier et al., 2005). While there is no apparent genotype-phenotype correlation between the presence and severity of either Hirschsprung’s disease or ocular abnormalities in MWS patients, Sip1 haploinsufficiency correlates with a more prominent facial phenotype. The diverse phenotypic abnormalities of MWS patients indicate that Sip1 functions in multiple developmental pathways.
Sip1 levels are elevated in multiple types of cancer, including breast, ovarian, gastric, pancreatic, and oral squamous cell carcinoma. Further, Sip1 appears to regulate the expression of multiple genes involved in EMT (see above, Vandewalle et al., 2009). This, along with the phenotypic spectrum of Sip1 mutant animals, suggests that Sip1 controls whether a cell is more “epithelial” or “mesenchymal” in normal tissues while its pathological upregulation can re-engineer epithelial cells to be more mesenchymal leading to cancer progression or pathological fibrosis.
4. Future Studies
While advances have been made in uncovering the role of Sip1 in disease (e.g. cancer, Mowat-Wilson syndrome), the diverse function of this transcription factor in different cell types needs further investigation. For instance, Sip1 is present in the normal lens and retina (see Fig. 1B). Further, numerous genes functionally important for the eye have been reported to be ZEB target genes including E-cadherin, N-cadherin, vimentin and crystallins. Conditionally deleting Sip1 in the early stages of lens formation demonstrates that it is necessary for normal eye development (Yoshimoto et al., 2005); however, it is unclear whether this gene is also involved in the later differentiation of ocular tissues or how this gene participates in the known transcription factor networks functioning during eye development. Overall though, it can be anticipated that Sip1 function in the eye is different from that seen in cancer, particularly since appreciable amounts of Sip1 are found in normal lens epithelial cells (Fig. 1B) that co-express E-cadherin (reported to be repressed by Sip1), and vimentin (reported to be activated by Sip1). However, the known functions of Sip1 during EMT associated with organ development and cancer may be applicable to ocular pathologies such as posterior capsular opacification, proliferative vitreoretinopathy, and corneal scarring. While mechanistically complex, the ZEB family has already placed itself at the center of normal eye development and EMT and warrants further investigation.
Acknowledgements
This work was supported by the National Eye Institute (EY12221).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Vandewalle C, et al. The role of the ZEB family of transcription factors in development and disease. Cell. Mol. Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschueren K, et al. Sip1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J. Biol. Chem. 1999;274:20489–20498. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- Yoshimoto A, et al. Regulation of ocular lens development by Smad-interacting protein 1 involving Foxe3 activation. Development. 2005;132:4437–4448. doi: 10.1242/dev.02022. [DOI] [PubMed] [Google Scholar]
- Zweier C, et al. Clinical and mutational spectrum of Mowat-Wilson Syndrome. Eur. J. Med. Genet. 2005;48:97–111. doi: 10.1016/j.ejmg.2005.01.003. [DOI] [PubMed] [Google Scholar]