Abstract
We and others have previously reported linkage to schizophrenia on chromosome 10q25-q26 but, to date, a susceptibility gene in the region has not been identified. We examined data from 3606 SNPs mapping to 10q25-q26 that had been typed in a genome-wide association study (GWAS) of schizophrenia (479 UK cases/2937 controls). SNPs with p<0.01 (n=40) were genotyped in an additional 163 UK cases and those markers that remained nominally significant at p<0.01 (n=22) were genotyped in replication samples from Ireland, Germany and Bulgaria consisting of a total of 1664 cases with schizophrenia and 3541 controls. Only one SNP, rs17101921, was nominally significant after meta-analyses across the replication samples and this was genotyped in an additional six samples from the US/Australia, Germany, China, Japan, Israel and Sweden (n= 5142 cases/ 6561 controls). Across all replication samples, the allele at rs17101921 that was associated in the GWAS showed evidence for association independent of the original data (OR 1.17 (95% CI 1.06-1.29), p=0.0009). The SNP maps 85kb from the nearest gene encoding fibroblast growth factor receptor 2 (FGFR2) making this a potential susceptibility gene for schizophrenia.
Keywords: FGFR2, schizophrenia, Genome Wide Association Study
Introduction
Schizophrenia is a severe and debilitating psychiatric disorder with a lifetime risk of approximately 1%. Symptoms include hallucinations and delusions, disorganised behaviour, reduced drive and altered emotional reactivity1. Pathophysiology is largely unknown, but genes are known to make a major contribution to risk and heritability is greater than 0.82. Epidemiological studies suggest that disease predisposition is likely to be the result of multiple genes of modest to small effect3; a conclusion that is supported by meta-analysis of linkage studies4, by positional cloning studies5-8, and now by genome wide association analysis9. Molecular genetic studies also suggest a proportion of cases are associated with sub-microscopic chromosomal abnormalities often referred to as copy number variations (CNVs)10-14.
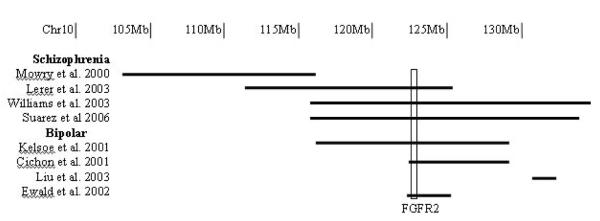
We previously undertook a genome wide linkage study of schizophrenia based on 353 affected sibling pairs from the UK, Sweden and the US. Our strongest finding was a genome wide significant LOD score of 3.87 at chromosome 10q25.3-q26.3 (116-135Mb)15. Six other independent studies of schizophrenia or bipolar disorder (BD) have also reported linkage to chromosome 10q25-q2616-21, (figure 1). Levinson et al,22 reported a genome scan of schizophrenia using 48 pedigrees from Australia and the US. One of the most significant findings was at D10s1239 (103Mb), with a non-parametric linkage score of 2.02. Mowry and colleagues16 reported a follow up of the more promising regions from that study based upon greater marker density and an extended sample. They obtained supportive evidence for linkage to 10q, although not at genome wide significant levels. Lerer et al,19 also reported data from a genome scan of schizophrenia in 21 large families (155 affected individuals) from Israel. They found suggestive evidence for linkage to a region spanning D10s543 (112Mb) to D10s587 (125Mb). More recently, genome wide linkage analysis of 409 schizophrenia pedigrees of European ancestry and African American ancestry21, showed suggestive evidence for linkage at 10q25.3-q26.3, with a LOD-1 region of 116-134Mb.
Figure 1.
Linkage findings at chr10q25-q26. Mb; Megabase.
There are also three reports of linkage to this region in BD, a disorder that may share some genetic liability with schizophrenia23. First, Kelsoe et al,17 found suggestive evidence for linkage from D10s1237 (116Mb) to D10s217 (129Mb) in 20 multiplex families. In a genome scan of 75 families from Germany, Israel and Italy, Cichon et al,18 reported suggestive evidence for linkage from D10s1757 (122Mb) to D10s217 (129Mb). Thirdly, Liu et al,20 identified the region flanked by D10s1248 (131Mb) to D10s169 (132Mb) as suggestive for linkage to BD in 57 extended pedigrees from the US and Israel. Finally, the study of Ewald et al,24 also gives supportive evidence for both BD and schizophrenia loci at 10q25.3-q26.3. Using haplotype sharing methods in 15 BD patients and ten with schizophrenia compared to control subjects from a relatively isolated population in the Faroe Islands, Ewald et al,24 showed evidence for association of the region flanked by D10s1230 (123Mb) and D10s2322 (126Mb) to both schizophrenia and BD.
Despite these reports of significant and suggestive linkage, no schizophrenia susceptibility genes mapping within this region have yet been identified. Aiming to do so, we have now examined the data we obtained from a GWAS study of schizophrenia9 for evidence for association to the broad linkage region on 10q. SNPs with evidence for association surpassing nominal thresholds in our sample were genotyped in a series of samples totaling up to nine independent case-control series. Our data provide evidence across ten case-control series for association between a SNP 85kb from the FGFR2 locus.
Materials and Methods
Overview of study design
SNPs from our GWAS sample (479 cases, 2937 controls) mapping within our linkage region (chr10 bases 115,673,689-135,374,737; UCSC March 2006) were inspected for evidence for association. Markers for which there was evidence for association at p≤0.01were genotyped in an additional 163 UK cases and, after confirming genotyping consistency with the GWA data, the data were merged giving a total of 642 cases and 2937 controls. For markers remaining significant at p<0.01 in this extended UK sample, we sought independent replication in a combined analysis (1664 cases, 3541 controls) of 3 additional schizophrenia case control series from Ireland (295 cases, 983 controls), Germany (Munich), (758 cases, 1897 controls) and Bulgaria (611 cases, 661 controls). Markers that showed significant association at p≤0.05 to the same allele that was associated in the UK sample were genotyped in six more case-control series (combined n=6806 cases, n=10102 controls) from Germany (Bonn) (735 cases, 1036 controls), US/Australia (1744 cases, 1938 controls), China (1034 cases, 1034 controls), Japan (748 cases, 831 controls), Israel (741 cases, 1517controls) and Sweden (140 cases, 205 controls). Finally, we also combined in our analysis the data from the The Wellcome Trust Case Control Consortium (WTCCC) study of BD25.
Subjects
Full details of the UK and replication samples from the US/Australia, Germany, Ireland, Bulgaria, Israel, China and Japan are previously described9. All patients from the UK, Ireland, Germany, Bulgaria, China, Japan and the Ashkenazi Jewish samples had a DSMIV diagnosis of schizophrenia. Patients from the US/Australian sample all had a consensus lifetime best estimate diagnosis of DSMIV schizophrenia (90%) or schizoaffective disorder (10%). All patients and controls gave written and informed consent to participate.
Swedish sample
The Swedish sample consisted of 140 unrelated cases (86 male and 54 female) 91 of whom were included in our linkage sample15. Cases were ascertained through the Mental Health inpatient Register and the Swedish Second Generation Register. Patients who had a register diagnosis of “ICD-10 schizophrenia” were included. All had at least 1 affected first degree relative, were white and were born in Sweden. All gave written, informed consent, as approved by the institutional review board of the Karolinska Institute. The control sample consisted of 205 subjects (130 male, 75 female), with a mean age of 40.2, (SD 10.2) years, age range 19 – 86 years. The majority had previously served as healthy controls subjects in biological psychiatric research at the Karolinska Institute and were reassessed for lifetime psychiatric diagnosis using structured interviews, whereas a minority (n=13) was drawn from a representative sample of the population in Stockholm County and assessed as previously described26. None of the controls had a diagnosis of schizophrenia.
Genotyping
The GWAS sample of 479 cases and 2937 controls was genotyped as part of the same pipeline as the Wellcome Trust Case Control Consortium of 7 common phenotypes25. Full details including QC measures are given in our primary GWAS manuscript9. We excluded markers where a) HWE controls p< 0.001 b) HWE cases p< 0.00001, c) MAF in either cases or controls < 0.01. d) the genotyping call rate in either cases or controls was< 0.985. The quality of the genotyping for the markers we report to be associated with schizophrenia was verified by inspection of the genotype clusters. Follow-up genotyping in the additional 163 UK cases, Irish, Bulgarian, German (Munich collection) and Swedish samples was performed using Sequenom iplex Gold chemistry on the Sequenom MassARRAY genotyping platform in Cardiff. Cross-platform studies were performed for 61 SNPs from the chromosome 10 linkage region by re-genotyping on average 179 samples from the 1958 birth cohort sample set (used in the WTCCC study25) on the Sequenom platform. Sequenom assays were also validated by genotyping all follow-up SNPs in the 90 CEPH individuals from the International HapMap project. The Chinese, Japanese, Bonn, Israel and US/Australia samples were genotyped by those groups as previously described and blind to the results from our GWAS9.
Statistical analysis
In the GWAS sample, the primary test of association was the Armitage trend test (1df). For meta-analyses of the replication samples and combined samples, we used the Cochran-Mantel Haenszel (CMH) test as implemented in PLINK27. Hardy Weinberg Equilibrium was calculated by a goodness of fit test.
Results
QC analysis
QC genotyping for 61 SNPs on the Sequenom platform identified six discrepant genotypes from a total of 10,890 giving a discrepancy rate of 0.06% between Sequenom and Affymetrix platforms. The average call rate in the GWAS sample for the SNPs in our target region was 98.9%. The particular SNP emergent as showing strongest evidence for association in this study (rs17101921) was re-genotyped on the Sequenom platform in all GWAS schizophrenia cases and 1426 GWAS controls. Two discrepancies were identified in the case sample. Both were from the only two minor allele homozygous genotypes called in either cases or in controls on the Affymetrix platform. These were assigned heterozygous genotypes on the Sequenom platform. Genotyping of rs17101921 for the two discrepant samples was repeated with a SNaPshot genotyping assay, proving the Sequenom platform to be correct (hence these individuals are called as heterozygotes in all analyses reported for this SNP). 100% concordance was observed between Affymetrix and Sequenom platforms for 1426 GWAS controls. No other minor allele homozygotes were observed for rs17101921 on the Affymetrix platform for any other schizophrenia GWAS cases or any of the 2937 GWAS controls. One WTCCC bipolar case was scored as a minor allele homozygote on the Affymetrix platform and for the purpose of this analysis, this individual was excluded. For the Bulgarian, German, and Irish replication samples typed on the Sequenom platform, average call rates for all chromosome 10 SNPs typed were 98.7, 98.3 and 98.5% respectively. Call rates for rs17101921 in the Bulgarian, Munich, Dublin, Bonn, US/Australian, China, Japan and Swedish samples were 97.8, 95.3, 99.1, 100, 99.9, 98.1, 98.0 and 98.3% respectively. Concordance for all SNPs typed through the same 90 CEPH individuals used in the International HapMap was 99.9%.
Association in the UK sample
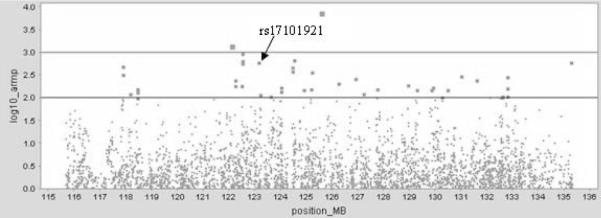
Following QC, we used a total of 3606 SNPs (figure 2) of which 40 showed evidence for association in the GWAS sample p<0.01 (supplementary table 1). Of these 40 markers, 22 remained significant at p<0.01 in the full UK sample (supplementary table 1).
Figure 2.
UK GWAS p-values across chromosome 10 linkage region. position_MB; position in megabases, log10_armp; armitage trend test −log10 p-values. Top horizontal line corresponds to p<0.001, bottom horizontal line corresponds to p<0.01.
Replication studies
Markers that showed nominally significant evidence for association (p<0.01) in the extended UK sample (n=22) were genotyped in samples from Ireland, Bulgaria and Germany. Only 1 SNP showed nominally significant evidence for association to the same allele at p<0.05 in the meta-analysis of the replication samples (supplementary table 2). This was rs17101921 (p=0.002, 1-tailed) located at chr10:123,143,285 (UCSC March 2006) ~85kb 3′ of FGFR2 (table 2).
Table 2.
Meta-analyses of rs17101921
| rs17101921 | No. case | No. con | CMH_P | CMH_OR | 95% CI | 95% CI |
|---|---|---|---|---|---|---|
| rep 1 | 1664 | 3541 | 0.0020 | 1.48 | 1.13 | 1.94 |
| rep all | 6806 | 10102 | 0.0009 | 1.17 | 1.06 | 1.29 |
| meta all+UK | 7448 | 13039 | 0.0002 | 1.19 | 1.09 | 1.31 |
Rep 1 = First set of replication samples from Ireland, Germany (Munich) and Bulgaria.
Rep all = meta-analysis of above, plus additional samples from US/Australia, Germany (Bonn), China, Japan, Israel and Sweden.
Meta all+UK =meta-analysis of all samples including UK.
In the second set of replication samples from Sweden, US/Australia, China, Japan, Germany (Bonn) and Israel (table 1), rs17101921 again showed significant association to the same allele (p=0.011, 1-tailed) and when all replication samples were combined (total 6806 cases, 10102 controls), this marker was associated with schizophrenia (p=0.0009, 1 tailed) with an OR of 1.17 (95% CI 1.06-1.29). When this was subjected to correction for the SNPs taken forward for replication, this finding remained significant, p=0.015, (based on α=0.05 there were 17.1 effective tests from the 22 SNPs calculated as per28. The combined GWAS and replication samples yielded p = 0.0002, OR 1.19, (95% CI 1.09- 1.31) (table 2).
Table 1.
Association analysis of rs17101921.
| rs17101921 | No. Case | No. Cons | CON freq1 |
CASE freq1 |
allelic p | OR | 95% CI | 95% CI |
|---|---|---|---|---|---|---|---|---|
| UK GWAS | 479 | 2937 | 0.020 | 0.035 | 0.006 | 1.73 | 1.17 | 2.55 |
| UK extended | 642 | 2937 | 0.020 | 0.033 | 0.003 | 1.69 | 1.19 | 2.41 |
| Ireland | 295 | 983 | 0.019 | 0.045 | 0.0004 | 2.34 | 1.43 | 3.83 |
| Bulgaria | 611 | 661 | 0.016 | 0.028 | 0.019 | 1.80 | 1.04 | 3.12 |
| German Munich | 758 | 1897 | 0.024 | 0.025 | 0.401 | 1.05 | 0.71 | 1.56 |
| Sweden | 140 | 205 | 0.027 | 0.074 | 0.002 | 2.85 | 1.39 | 5.85 |
| German Bonn | 735 | 1036 | 0.027 | 0.020 | 0.240 | 0.76 | 0.49 | 1.19 |
| US/Australia | 1744 | 1938 | 0.028 | 0.028 | 0.924 | 0.99 | 0.75 | 1.30 |
| China | 1034 | 1034 | 0.138 | 0.165 | 0.008 | 1.24 | 1.04 | 1.47 |
| Japan | 748 | 831 | 0.209 | 0.224 | 0.157 | 1.09 | 0.92 | 1.29 |
| Israel | 741 | 1517 | 0.017 | 0.020 | 0.196 | 1.22 | 0.78 | 1.92 |
CON freq1 = minor allele frequency in controls, CASE freq 1 = minor allele frequency in cases. Allelic p-values are 1-tailed where OR is in the same direction as the GWAS sample. OR = Odds ratio with 95% confidence intervals (CI).
Our GWAS sample contained a total of 108 unrelated individuals with an affected sibling who had been included in our previous linkage study15. In addition we had access to 91 unrelated Swedish cases with an affected sibling who had also been included in our linkage study15, plus a further 49 unrelated Swedish cases with an affected sibling who had been recruited for but did not participate in the linkage study. One of the replication samples (US/Australia) contained a subset of individuals with an affected sibling (n=289) that were also included in a linkage study21 that had shown linkage to chromosome 10q25-q26. We hypothesized that an increased effect size at rs17101921 would be observed in these familial cases. The effect size at rs17101921 was increased in the familial cases from the UK (OR 1.92 [95% CI 1.19-2.41]) and Sweden (OR 2.85 [95% CI 1.39-5.85]) which formed the majority of the sample in which we obtained evidence for linkage. In the US/Australian replication sample, the effect was actually in the opposite direction, but in the subset of familial cases, the trend for association was in the same direction as the GWAS sample, OR 1.12 (95% CI 0.68-1.85). Meta-analyses across all three chromosome 10 linked familial samples showed significant association, p=0.045 (2-tailed) with an OR of 1.52 (95% CI 1.02-2.27) which is greater than that observed in the sample as a whole. Meta-analyses excluding these familial samples was also significant, p=0.001 (2 tailed) and showed a smaller effect size, OR 1.17 (95% CI 1.06-1.29), although the 95% confidence intervals for familial and non-familial samples overlapped.
Sensitivity analysis in which each of one from nine replication samples was dropped from the meta-analysis revealed the evidence for association was not critically dependent on any single sample (supplementary table 3).
Given the observations of linkage of BD to 10q25-q2617,18,20, we also performed a secondary meta-analysis that included the WTCCC BD cases (n=1865) for rs17101921. As the control sample used for the UK schizophrenia GWAS study was the same control sample used in the WTCCC bipolar GWAS study, they were included only once. Inclusion of the BD cases in the meta-analysis (total 9,313 cases, 13,039 controls) increased the evidence for association with rs17101921, p=7.80×10−5, OR 1.20 (95% CI 1.10-1.31).
Discussion
There are seven reports of linkage between chromosome 10q25-q26 and either schizophrenia or BD, but no susceptibility genes within this region have yet been identified. Aiming to map susceptibility variants, we used GWAS data from a UK case-control sample and followed up those variants that provided evidence for association in up to two sample series.
We observed strongest evidence for association between schizophrenia and rs17101921, a marker that maps within 85kb of FGFR2 and is located in the overlapping linked regions of 5/7 studies of schizophrenia and BD (figure 1). Clearly, in our GWAS sample, rs17101921 did not show region-wide significant evidence for association and thus while the meta-analysis of the nine replication samples, excluding the data from the GWAS sample, showed a significant association with schizophrenia (p=0.0009, 1 tailed, OR of 1.17 [95% CI 1.06-1.29]), and this survived correction for all markers taken into the first phase of replication (corrected p=0.015), the finding cannot yet be regarded as definitive, and requires additional evaluation in even larger studies. The results of the meta-analysis suggest a similar conclusion. Based upon an estimated genome wide threshold for significance of 7.2×10−8 29 and that our target region comprises about 2/300ths of the genome the region wide equivalent of genome-wide significance would be 1×10−5. Our overall meta-analysis finding then corresponds to region wide suggestive rather than significant evidence.
The question of whether and to what extent this locus might account for the evidence for linkage to 10q25-q26 is difficult to determine given that the true effect size at this locus remains unknown pending discovery of the underlying functional variant and that the true effect size of the linkage is difficult to measure in an unbiased way. However, we note that the effect size at rs17101921 was greater in the familial cases which formed the majority of the sample in which we obtained evidence for linkage, than that observed in the replication samples, which provides the most un-biased estimate of the true effect size in the remaining samples. Of course, at this stage, we cannot exclude the possibility that the linkage observed at 10q25-26 reflects the combined effects of FGFR2 and another locus or loci within this region. We also note that the evidence for association was increased with the addition of 1865 BD cases from the UK. Given that the increase in evidence was modest, and that while BD was itself significantly associated in the WTCCC sample (p=3.5×10−2), and the controls are shared with the schizophrenia sample, the hypothesis that this SNP is another example of an association across diagnostic boundaries should be taken with additional caution.
In terms of the possible functional implications of the association we report, the nearest gene to rs17101921 is FGFR2 (~85kb 3′, UCSC March 2006 freeze). We note here from two recent studies of gene expression that the majority of significant cis-acting SNP variants occur within 100kb (either 3′ or 5′) of the interrogated expression probe and also that significant cis-acting variants are observed as far as 800kb away30,31. Based on in-silico data, rs17101921 does not appear to reside in a highly conserved or putative functional region. Of additional interest is the variable frequency of the risk allele. In our replication samples, the frequency of the risk allele in Chinese controls is 14% and in Japanese controls, 21% compared to only ~2% in our Caucasian samples. HapMap populations show similar frequencies, with the CEPH sample being non-polymorphic and the Chinese and Japanese samples showing a much higher frequency. The Yoruban population is also non-polymorphic, suggesting perhaps that this variant could be under selection.
FGFR2 is a reasonable functional candidate gene for schizophrenia. Of its multiple functions, perhaps of most plausible relevance to schizophrenia is that it plays roles in presynaptic organization early in development and neuroprotection and repair in adulthood32,33. Recent evidence provides some support for the involvement of the fibroblast growth factor (FGF) system in psychiatric disorders (reviewed in34,35), and, more specifically, some support for the involvement of FGF signaling in schizophrenia. Thus, Hashimoto and colleagues36 reported altered levels of fibroblast growth factor-2 (FGF-2) in blood from drug-free schizophrenia patients, while Gaughran and colleagues37 reported increased measures of FGFR1 mRNA in schizophrenic brain. Interestingly, mice homozygous for a dominant negative mutation at FGFR1 showed increased levels of dopamine in the striatum and impaired pre-pulse inhibition38. While these studies provide some plausibility that altered FGFR2 function might contribute to schizophrenia, our study provides no direct evidence for this. Further studies of genetic association are required, and if confirmed, investigation into the biological significance of the association.
Supplementary Material
Acknowledgments
This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113. The UK research was supported by grants from the MRC and the Wellcome Trust. In Dublin, the research was supported by Science Foundation Ireland, the Health Research Board (Ireland), and the Wellcome Trust. We are grateful to Prof. John Waddington for sample recruitment. Irish controls were supplied by Dr. Joe McPartlin and the Trinity College Biobank. In Bonn and Mannheim, the work was supported by the National Genomic Network of the ‘Bundesministerium für Bildung und Forschung’ (BMBF) and the Alfried Krupp von Bohlen und Halbach-Stiftung. We also thank the Department of Psychiatry, LMU Munich for clinical characterization of the Munich subjects and the processing of the samples. Recruitment in Munich was partially supported by GlaxoSmithKline. The Ashkenazi samples are part of the Hebrew University Genetic Resource, HUGR (www.hugr.org).
References
- 1.Gottesman II. Schizophrenia Genesis, The Origins of madness. WH Freeman and Company; New York: 1991. [Google Scholar]
- 2.Cardno AG, Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–7. [PubMed] [Google Scholar]
- 3.Risch N. Genetic linkage and complex diseases, with special reference to psychiatric disorders. Genet Epidemiol. 1990;7:3–16. doi: 10.1002/gepi.1370070103. [DOI] [PubMed] [Google Scholar]
- 4.Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–23. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 6.Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, et al. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet. 2002;71:337–48. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefansson H, Sarginson J, Kong A, Yates P, Steinthorsdottir V, Gudfinnsson E, et al. Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am J Hum Genet. 2003;72:83–7. doi: 10.1086/345442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, et al. Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Natl Acad Sci U S A. 2002;99:13675–80. doi: 10.1073/pnas.182412499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, et al. Identification of novel schizophrenia loci by genome-wide association and follow-up. Nat Genet. 2008 doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 10.Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–65. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 11.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–43. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 12.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–5. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 13.Stefansson H, Rujescu D, Cichon S, Pietiläinen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008 doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]; The International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008 doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008 doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams NM, Norton N, Williams H, Ekholm B, Hamshere ML, Lindblom Y, et al. A systematic genomewide linkage study in 353 sib pairs with schizophrenia. Am J Hum Genet. 2003;73:1355–67. doi: 10.1086/380206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mowry BJ, Ewen KR, Nancarrow DJ, Lennon DP, Nertney DA, Jones HL, et al. Second stage of a genome scan of schizophrenia: study of five positive regions in an expanded sample. Am J Med Genet. 2000;96:864–9. [PubMed] [Google Scholar]
- 17.Kelsoe JR, Spence MA, Loetscher E, Foguet M, Sadovnick AD, Remick RA, et al. A genome survey indicates a possible susceptibility locus for bipolar disorder on chromosome 22. Proc Natl Acad Sci U S A. 2001;98:585–90. doi: 10.1073/pnas.011358498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cichon S, Schmidt-Wolf G, Schumacher J, Müller DJ, Hürter M, Schulze TG, et al. A possible susceptibility locus for bipolar affective disorder in chromosomal region 10q25-q26. Mol Psychiatry. 2001;6:342–9. doi: 10.1038/sj.mp.4000864. [DOI] [PubMed] [Google Scholar]
- 19.Lerer B, Segman RH, Hamdan A, Kanyas K, Karni O, Kohn Y, et al. Genome scan of Arab Israeli families maps a schizophrenia susceptibility gene to chromosome 6q23 and supports a locus at chromosome 10q24. Mol Psychiatry. 2003;8:488–98. doi: 10.1038/sj.mp.4001322. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Juo SH, Dewan A, Grunn A, Tong X, Brito M, et al. Evidence for a putative bipolar disorder locus on 2p13-16 and other potential loci on 4q31, 7q34, 8q13, 9q31, 10q21-24, 13q32, 14q21 and 17q11- Mol Psychiatry. 2003;8:333–42. doi: 10.1038/sj.mp.4001254. [DOI] [PubMed] [Google Scholar]
- 21.Suarez BK, Duan J, Sanders AR, Hinrichs AL, Jin CH, Hou C, et al. Genomewide linkage scan of 409 European-ancestry and African American families with schizophrenia: suggestive evidence of linkage at 8p23.3-p21.2 and 11p13.1-q14.1 in the combined sample. Am J Hum Genet. 2006;78:315–33. doi: 10.1086/500272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levinson DF, Mahtani MM, Nancarrow DJ, Brown DM, Kruglyak L, Kirby A, et al. Genome scan of schizophrenia. Am J Psychiatry. 1998;155:741–50. doi: 10.1176/ajp.155.6.741. [DOI] [PubMed] [Google Scholar]
- 23.Craddock N, O’Donovan MC, Owen MJ. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull. 2006;32:9–16. doi: 10.1093/schbul/sbj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ewald H, Flint TJ, Jorgensen TH, Wang AG, Jensen P, Vang M, et al. Search for a shared segment on chromosome 10q26 in patients with bipolar affective disorder or schizophrenia from the Faroe Islands. Am J Med Genet. 2002;114:196–204. [PubMed] [Google Scholar]
- 25.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonsson EG, Edman-Ahlbom B, Sillen A, Gunnar A, Kulle B, Frigessi A, et al. Brain-derived neurotrophic factor gene (BDNF) variants and schizophrenia: an association study. Prog Neuropsychopharmacol Biol Psychiatry. 2006;30:924–33. doi: 10.1016/j.pnpbp.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moskvina V, Schmidt KM. On multiple-testing correction in genome-wide association studies. Genet Epidemiol. 2008 doi: 10.1002/gepi.20331. [DOI] [PubMed] [Google Scholar]
- 29.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–34. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–9. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 32.Alzheimer C, Werner S. Fibroblast growth factors and neuroprotection. Adv Exp Med Biol. 2002;513:335–51. doi: 10.1007/978-1-4615-0123-7_12. [DOI] [PubMed] [Google Scholar]
- 33.Umemori H, Linhoff MW, Ornitz DM, Sanes JR, et al. FGF22 and its close relatives are presynaptic organizing molecules in the mammalian brain. Cell. 2004;118:257–70. doi: 10.1016/j.cell.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 34.Turner CA, Akil H, Watson SJ, Evans SJ. The fibroblast growth factor system and mood disorders. Biol Psychiatry. 2006;59:1128–35. doi: 10.1016/j.biopsych.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 35.Riva MA, Molteni R, Bedogni, Racagni G, Fumagalli F. Emerging role of the FGF system in psychiatric disorders. Trends Pharmacol Sci. 2005;26:228–31. doi: 10.1016/j.tips.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Hashimoto K, Shimizu E, Komatsu N, Nakazato M, Okamura N, Watanabe H, et al. Increased levels of serum basic fibroblast growth factor in schizophrenia. Psychiatry Res. 2003;120:211–8. doi: 10.1016/s0165-1781(03)00186-0. [DOI] [PubMed] [Google Scholar]
- 37.Gaughran F, Payne J, Sedgwick PM, Cotter D, Berry MI. Hippocampal FGF-2 and FGFR1 mRNA expression in major depression, schizophrenia and bipolar disorder. Brain Res Bull. 2006;70:221–7. doi: 10.1016/j.brainresbull.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Klejbor I, Myers JM, Hausknecht K, Corso TD, Gambino AS, Morys J, et al. Fibroblast growth factor receptor signaling affects development and function of dopamine neurons - inhibition results in a schizophrenia-like syndrome in transgenic mice. J Neurochem. 2006;97:1243–58. doi: 10.1111/j.1471-4159.2006.03754.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.