Abstract
Macromolecular transport systems in bacteria currently are classified by function and sequence comparisons into five basic types. In this classification system, type II and type IV secretion systems both possess members of a superfamily of genes for putative NTP hydrolase (NTPase) proteins that are strikingly similar in structure, function, and sequence. These include VirB11, TrbB, TraG, GspE, PilB, PilT, and ComG1. The predicted protein product of tadA, a recently discovered gene required for tenacious adherence of Actinobacillus actinomycetemcomitans, also has significant sequence similarity to members of this superfamily and to several unclassified and uncharacterized gene products of both Archaea and Bacteria. To understand the relationship of tadA and tadA-like genes to those encoding the putative NTPases of type II/IV secretion, we used a phylogenetic approach to obtain a genealogy of 148 NTPase genes and reconstruct a scenario of gene superfamily evolution. In this phylogeny, clear distinctions can be made between type II and type IV families and their constituent subfamilies. In addition, the subgroup containing tadA constitutes a novel and extremely widespread subfamily of the family encompassing all putative NTPases of type IV secretion systems. We report diagnostic amino acid residue positions for each major monophyletic family and subfamily in the phylogenetic tree, and we propose an easy method for precisely classifying and naming putative NTPase genes based on phylogeny. This molecular key-based method can be applied to other gene superfamilies and represents a valuable tool for genome analysis.
Keywords: conjugation, ATPase, Archaea, molecular key, Walker box
Bacteria use a variety of systems to transport proteins and other macromolecules out of the cytoplasm. These secretion systems are classified into five types based on function and sequence (1–6). The type I, II, III, and IV systems are unified by the presence of at least one potential NTP hydrolase (NTPase) protein. The putative NTPases from type II and type IV secretion systems are remarkably similar to one another in function and sequence. Although they are required in as diverse processes as pilus biogenesis, toxin secretion, and DNA transport in conjugation and natural transformation (7–10), they are thought to play similar functional roles as the energizers of macromolecular transport necessary in all of these activities.
The putative NTPases of type II/IV secretion systems are soluble, found in the cytoplasm, and usually associated with the inner membrane (10–18). Recent electron microscopic images and cross-linking experiments show that several superfamily members homodimerize and form similar toroidal structures (15, 19). Protein sequence alignments have disclosed the presence of four conserved domains in all superfamily members—two canonical nucleotide-binding motifs designated as Walker boxes A and B and two conserved regions designated as the Asp and His boxes (10, 16, 20). Several representative members of the type IV family of NTPases bind and hydrolyze ATP, and mutations in the Walker A motif abolish both this activity and macromolecular secretion (11, 13, 16, 21). Similar mutations in putative NTPase genes of type II secretion systems also disrupt macromolecular secretion, and ATPase activity has been demonstrated in at least one member of this family (10, 17). Thus, NTP binding and/or hydrolysis very likely is essential to the function of all of these proteins.
It has been suggested from strong identity and similarity scores in sequence comparisons that putative NTPases from the type II/IV superfamily are evolutionarily related (8, 16, 22, 23). However, currently there exist no precise descriptions of the evolutionary relationships of type II/IV NTPase gene superfamily members. Although pairwise sequence similarities and alignments provide important clues to understanding the relationship between genes, they do not give a clear representation of gene grouping, history, or homology (24, 25). Because they have evolved largely by a process of lineage splitting and divergence, genes can be organized into nested hierarchies. Phylogenetic analysis is designed to reconstruct such hierarchies, and it offers a powerful and precise tool for describing the groupings and histories of genes. Recent attempts to classify type II/IV secretion systems on the basis of the functional history of component genes (26) also would be improved by more precise understanding of phylogenetic relationships.
We recently described a cluster of genes (tadABCDEFG) in Actinobacillus actinomycetemcomitans required for nonspecific, tenacious adherence of the bacterium to surfaces (27). The predicted product of the tadA gene showed significant sequence similarity to the putative NTPases of type II and type IV secretion systems, and recent work has confirmed that TadA hydrolyzes ATP (M. K. Battacharjee, S.C.K., and D.H.F., unpublished data). ORFs bearing significant sequence similarities to tadA are present in many other Bacteria and Archaea (27–29).
To better understand the relationship of tadA to genes for putative NTPases of type II and IV secretion systems, we searched public databases for genes and ORFs of Bacteria and Archaea with predicted protein products similar to this superfamily of putative NTPases. We used phylogenetic methods to infer the relationships of the resulting 148 genes and reconstruct a gene history for these putative NTPases. The phylogeny shows that putative NTPase genes from type II and type IV secretion systems form two distinct groups and that bacterial tadA and tadA-like genes group with a cluster of Archaeal NTPase genes to form a novel and widespread gene subfamily.
We also developed a method for gene classification based on a diagnostic algorithm or “key” that defines each major phylogenetic grouping in the type II/IV NTPase gene tree. Although keys for the classification of organisms often are used in evolutionary biology, we are unaware of any sequence-based keys available for gene classification. This method will allow other researchers to more accurately identify and classify newly discovered NTPase genes, and it can be applied generally to other gene superfamilies.
Methods
Sequence Similarity Searching.
Nucleotide sequences similar to type II and IV NTPase genes were identified by using the blast alignment program (30) to search both GenBank and the Microbial Finished and Unfinished Genomes Database (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html). Representative protein products of each major named gene group were used to search both databases. The following protein sequences were used as representatives: Aeromonas hydrophila ExeE and TapB; Agrobacterium tumefaciens pTiC58 VirB11; RK2 TrbB; Escherichia coli HofB, PilB, and GspE; A. actinomycetemcomitans TadA; Pseudomonas aeruginosa PilT; Klebsiella pneumoniae PulE; Erwinia carotovora OutE; and pKM101 TraG. Default settings of the blast program were used except for the low-complexity filter, as this option often excluded the glycine-rich Walker box A from similarity and identity calculations. Because of the continual increase in sequence data, only sequences present in public databases before March 15, 2000 were included in this study. We excluded sequences from unfinished genomes in which the ORF was within 500 bp of the end of a contig as well as hits with sequence similarity only to Walker box A. The Bordetella pertussis tadA ORF was found to be fused with a downstream tadB-like ORF. We made a −1 frame shift at nucleotide 846 in the sequence where similarity of the predicted product with other TadA proteins broke down to restore a separate 450-aa protein product whose C terminus was similar to other TadA proteins. Haemophilus aphrophilus tadA was sequenced after PCR amplification by using primers designed for A. actinomycetemcomitans (27). We attempted to include the functionally analogous cytoplasmic NTPases of type III secretion, but we were unable to convincingly align these with the putative NTPases of type II/IV secretion. Therefore, type III secretion NTPases were not included in the analysis. All 148 sequences included in the analysis (Fig. 1) significantly exceeded blast default criteria. We retained the names of previously identified sequences obtained from GenBank or finished annotated genomes. We named ORFs in unfinished genomes from the phylogenetic analysis by using the name of the smallest subfamily or clear subgrouping to which that sequence belonged. All inteins or intein-like sequences were virtually excised and not included in the analysis.
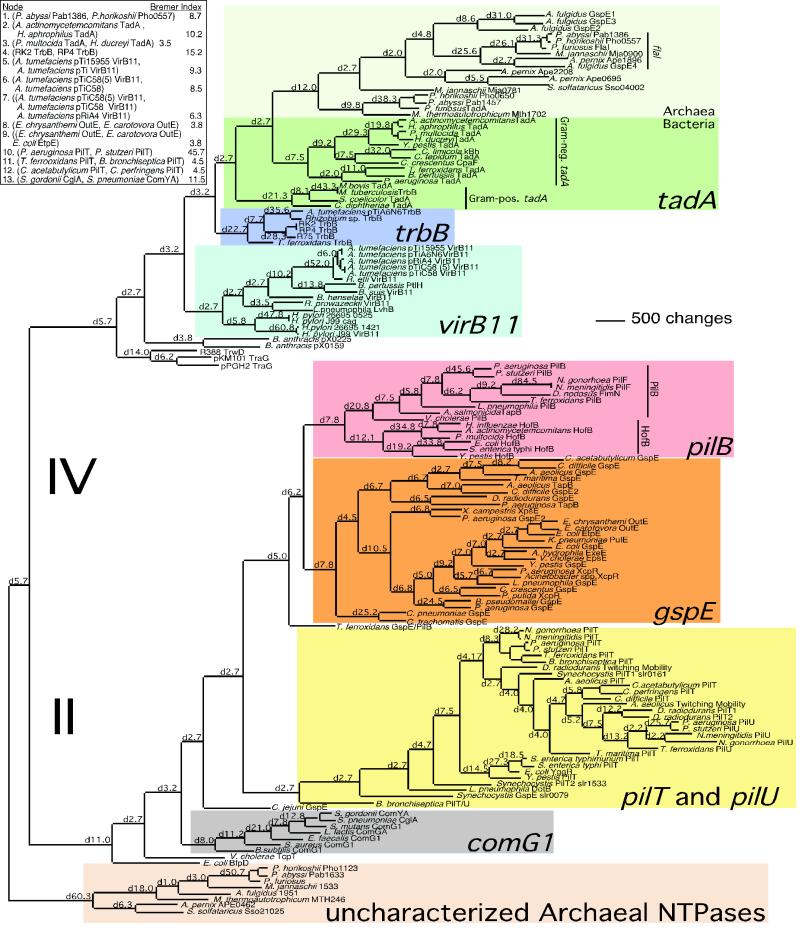
Figure 1.
A phylogenetic tree of putative NTPase genes of type II/IV secretion systems constructed with maximum parsimony algorithms. Gene subfamilies are designated by colored boxes, and subgroupings, by vertical lines. Archaeal and Bacterial divisions in the tadA gene subfamily are shown in two shades of green. The numbers on branches are Bremer indices of the nodes at the end of the designated branch (see Methods). Bremer indices for some organisms did not fit onto the tree and are supplied in the box in the upper left corner. Accession numbers for all genes are listed on the web site http://cpmcnet.columbia.edu/dept/figurski.
Preliminary sequence data for Neisseria meningitidis and Enterococcus faecalis were obtained from The Institute for Genomic Research (http://www.tigr.org.); Clostridium difficile, Mycobacterium bovis, M. tuberculosis, and Salmonella enterica subsp. typhi, from the respective Sanger Centre sequencing groups (http://www.sanger.ac.uk/); Neisseria gonorrhoeae, Staphylococcus aureus, and Streptococcus mutans, from the University of Oklahoma's Advanced Center for Genome Technology (http://www.genome.ou.edu); S. enterica subsps. typhimurium, from Washington University Genome Sequencing Center (http://genome.wustl.edu/gsc/bacterial/salmonella.shtml); Clostridium acetabutylicum, from Genome Therapeutics Center (http://www.cric.com/genesequences/clostridium/clospage.html); and Legionella pneumophila, from Columbia Genome Center (http://genome3.cpmc.columbia.edu/∼legion). Preliminary sequence data from all remaining unfinished genomes were obtained as in Kachlany et al. (27). A compilation of intein sequences is found in the New England BioLabs Intein Database (http://www.neb.com/inteins).
Alignment.
CLUSTAL X 1.63 (32) was used to align all 148 putative NTPase sequences. We varied gap parameters in six independent alignments of the 148 sequences and combined the six resulting matrices for analysis—a method referred to as “elision” (33). Elision gives greater weight to amino acid positions that consistently align under a wide range of gap/change costs. Gap/change ratios were varied in increments of 2 to yield the following values: 2, 4, 6, 8, 10, and 12.
Phylogenetic Analysis.
The combined matrix produced by elision then was analyzed by using parsimony algorithms of PAUP*4.0β (34). Because of the large size of the data set, a maximum-likelihood approach was prohibitive. We performed an aggressive heuristic search with 5,000 replicates of random addition of taxa followed by the subtree pruning and regrafting function of paup, saving only one tree at each replicate. All characters and state transformations were given equal weight, and columns with gaps were retained as phylogenetically informative characters (35). The resulting 5,000 trees were tested with the more rigorous tree–branch reconnection (TBR) technique. This yielded a single-most parsimonious tree of 145,028 steps. We used the program autodecay in conjunction with paup to calculate Bremer supports of confidence for nodes in the tree (36). Bremer support indices show how many additional evolutionary steps are required to lose the groupings represented by the phylogeny (37). Ten TBR replicates were done at each node in the phylogeny to obtain the Bremer index. Because Bremer indices were calculated by using the elided data set of six independent alignments, each value then was divided by 6 and rounded to the nearest 10th to yield the numbers presented in Fig. 1.
Rooting the Tree and Identifying Ancestral Sequences.
Rooting the phylogenetic tree presented the common methodological difficulty of identifying an outgroup in widespread gene family phylogenies. Others have addressed this problem by using paralog rooting strategies (38, 39). However, it is difficult to recognize the true ancestral root when the phylogeny includes more than two paralagous groups and possible horizontal transfer events. In such cases it may be necessary to use external criteria to root the phylogeny.
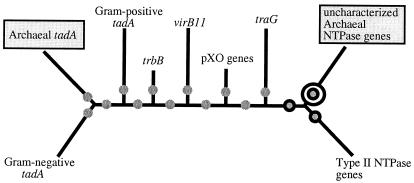
We assigned a tentative root in the branch composed of uncharacterized Archaeal NTPase genes for the following reasons. (i) This root maintains a phylogenetic distinction between type II and type IV NTPase genes as monophyletic groups. (ii) This root separates at least one Archaeal subfamily from Bacterial sequences. (iii) Finally, although the uncharacterized Archaeal NTPases align with other members of the type II/IV ATPase superfamily and contain identifiable Walker A boxes, they do not share identifiable Walker B, His, or Asp boxes (Fig. 2).
Figure 2.
Scheme for rooting the tree. The network shows the two divisions in the superfamily tree between Bacteria and Archaea. Archaeal groups are shown in shaded boxes. Shaded circles show roots that would conserve likely splits between the Archaea and Bacteria. Single solid circles represent roots that maintain a phylogenetic difference between type II and IV secretion NTPase genes. The double solid circle creates a root that unites all proteins with all four conserved domains (Walker A and B, and the His and Asp boxes).
Identifying this root allowed us to reconstruct an evolutionary scenario for type II/IV secretion NTPase genes. We also used the rooted phylogeny to reconstruct the most parsimonious ancestral sequence (MPAS) at each major node in the phylogeny. We chose the portion of the elided alignment with a gap/change ratio of 8 in combination with the most parsimonious tree to reconstruct ancestral states by using accelerated transformation criteria. Ambiguous positions present in all ancestral sequences were deleted.
Identifying Diagnostic Sequences.
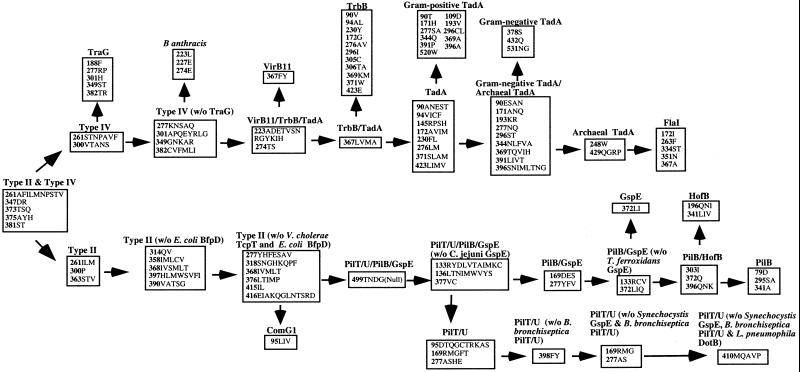
Diagnostic amino acid characters were identified by using macclade (40) to trace character changes on the most parsimonious phylogenetic tree by using the portion of the elided alignment with a gap/change ratio of 8. At each alignment position, only those amino acids that were specific (i.e., absent in sister taxa but present in all members of the group) for a particular group were recorded. In several cases, multiple states of a specific amino acid position were identified as diagnostic for a group. In these cases each member of the group must have one of the multiple states. Diagnostic amino acids were mapped to specific positions on the MPASs. A list of diagnostic sequences along with the number indicating their position on the MPAS alignment is represented in Fig. 3 as a nested hierarchy.
Figure 3.
Diagnostic key for classification of putative NTPase genes of type II/IV secretion. Starting with the diagnostic sequences for type II/IV putative NTPases, one can place the newly discovered gene into a family, subfamily, and, in some cases, subgroup. Numbers represent standardized positions on the MPASs. Any one of the amino acids listed after the position number is diagnostic for that position. The boxes represent major monophyletic groups in the tree. To advance to the next box, a sequence must contain a diagnostic amino acid at each position listed in that box. Our web page (http://cpmcnet.columbia.edu/dept/figurski) performs the diagnosis and classifies new genes.
Results
A Gene Tree for Putative NTPases of Type II and IV Secretion Systems.
Searches of public sequence databases yielded 148 genes that met our criteria for inclusion in this analysis (see Methods). The deduced protein sequences of these genes were aligned by using a technique that deemphasizes alignment ambiguities and preferentially weights stable portions of the alignment (33). Using a maximum parsimony-based algorithm, we constructed a phylogenetic tree depicting the relationships of putative NTPase genes of type II and IV secretion systems. Because of the very large size of the data set, we used an aggressive heuristic search to find the most parsimonious tree topology. Other search techniques were unable to find a tree with 145,028 or fewer steps. The resulting tree was fully resolved and strongly supported by Bremer indices (see Methods) (Fig. 1).
To reconstruct an evolutionary scenario and organize genes into hierarchical families, one must first root the phylogeny with an outgroup. We placed a tentative root in the branch leading to the uncharacterized Archaeal NTPase genes (see Methods).
The tadA Subfamily.
The phylogeny shows that tadA from A. actinomycetemcomitans is situated in a monophyletic clade of genes that includes the putative NTPase genes from the Archaea along with those of several Gram-negative and Gram-positive Bacteria. This group is located at the apex of the type IV secretion NTPase gene lineage, and it represents a subfamily distinct from other type IV NTPase genes such as virB11, trbB, and traG. Therefore, we have designated this cluster as the tadA subfamily of the type IV family of putative NTPase genes. Many of the genes in the Gram-negative and Archaeal subgroups of the tadA subfamily are followed immediately downstream by an ORF encoding a product with significant similarity to TadB (27). In this collection, none of the type IV NTPase genes outside of the tadA clade has an ORF downstream or in the immediate vicinity that encodes a TadB-like product.
Diagnostic Sequences.
Using our phylogenetic tree as a foundation, we reconstructed the MPAS at each node in the phylogeny. Each ancestral sequence represents the best reconstruction of the predicted protein product of the founding member of a gene cluster. We then determined a set of amino acid residues that specifically defines each gene clade to the exclusion of all others in the phylogeny. Each diagnostic amino acid position maps to a specific position on every MPAS. Therefore, alignments with any MPAS can be used to locate diagnostic characters and place genes in families and subfamilies.
Discussion
We examined the evolution of a superfamily of genes for putative cytoplasmic NTPases involved in type II and IV secretion systems by using phylogenetic analysis. The resulting genealogy describes the history and relationships of members of this superfamily and demarcates families and subfamilies. This phylogeny serves as a high-resolution template for developing an accessible and precise method of gene classification.
The tadA Subfamily and the tad Locus.
The tadA group had not been identified previously as a distinct gene subfamily. This subfamily is present in organisms representing the Gram-negative proteobacteria and green sulfur bacteria, the high G + C Gram-positive bacteria, and both major divisions of the Archaea—the Crenarcheota and Euryarcheota. The data available suggest that this subfamily spans many more groups of organisms than do any of the other type IV NTPase gene subfamilies. Consistent with the monophyly of this group is that genes from the tadA group often are associated with similar gene clusters (27, 28). In the Gram-negative Bacteria that are known to have tadA, ORFs encoding products with significant similarity to TadB and TadC are always found in sequential order immediately downstream, and Archaeal tadA genes usually are situated immediately upstream of ORFs whose products resemble TadB (27). Furthermore, ORFs with predicted products that are significantly similar to those of A. actinomycetemcomitans genes flp, orfA, orfB, and rcpA consistently are found upstream of tadA in Gram-negative bacteria. These genes are homologous to components of type II secretion systems that are involved in type IV pilus biogenesis (unpublished data). Flp is the major component of fibrils in A. actinomycetemcomitans (41), and the gene encoding Flp recently has been shown to be required for fibril production (S.C.K., P.J.P., R.D., D. H. Fine, D.H.F., and J. B. Kaplan, unpublished data). Skerker and Shapiro (28) recently reported that the locus including the tadA homolog cpaF in C. crescentus is required for pilus assembly. These findings suggest that the tad locus constitutes a novel pilus secretion system that combines a type IV NTPase gene with genes related to those of type II secretion systems along with other uncharacterized genes.
A Method for Gene Classification.
As more sequences of putative NTPase genes of type II/IV secretion become available, it will be important to conserve a logical nomenclature and classification scheme to clearly understand the relationships between different systems and genes. The recent flood of genomic data produced by microbial genome projects presents an unparalleled opportunity to understand the relationships among genes. Many genes currently are classified on the basis of pairwise similarity. Although similarity-based techniques are essential tools for searching enormous databases and making first approximations of gene homology, phylogeny-based methods offer a more precise and accurate way to classify and name genes (24, 25).
We therefore present an algorithm, based on diagnostic sequences and phylogeny, that will allow researchers to easily classify newly discovered genes into one of the designated type II/IV subgroups (Fig. 3). This technique combines the ease of pairwise similarity methods with the accuracy of phylogenetic analysis. It is composed of two steps. First, a newly discovered sequence is aligned with the most similar MPAS. Then, using the MPAS as a standard to locate diagnostic amino acid positions, the newly discovered gene can be placed in a family and subfamily by using a sequence-based key for gene classification (Fig. 3). We have designed a program available on our web site that carries out both of these operations and classifies putative type II/IV NTPase genes (http://cpmcnet.columbia.edu/ dept/figurski). Because this method is based on the distribution of characters mapped on a phylogenetic tree, any gene superfamily for which a phylogenetic tree can be constructed will be amenable to this approach.
This technique is not a replacement for phylogenetic analyses, as it cannot describe the relationships between individual genes; it can only place genes in a group. Also, with new sequence data, some diagnostic residues may need to be altered or excluded. Diagnostic residues with few possible amino acid states from groups with many taxa are less likely to change in the future.
Evolutionary Scenarios: Complex Histories of Gene Duplication, Horizontal Transfer, and Loss.
The phylogeny presents the following evolutionary scenario. A common gene ancestor of all genes for putative NTPases of type II/IV secretion duplicated and diverged into the two major families—type II and type IV. The type IV NTPase family then gave rise to the traG, virB11, trbB, and tadA subfamilies in that order. The type II lineage gave rise to four major subfamilies: comG1, gspE, pilB, and pilT/U.
Overall, the type II/IV secretion NTPase gene superfamily phylogeny represents several duplication events as evidenced by multiple sets of paralagous genes. For instance, the genome of P. aeruginosa contains seven representative genes distributed among the tadA (1), pilB (1), pilT/U (2), and gspE (3) subfamilies. The genomes of 16 other organisms included in our analysis contain three or more superfamily member genes.
Of interest is the total absence of certain gene subfamilies in several organisms whose genomes are completely sequenced. For instance, E. coli and H. influenzae have no genes in the tadA family in contrast to their relatives Y. pestis, H. ducreyi, A. actinomycetemcomitans, P. multocida, and H. aphrophilus. These absences either represent losses of tadA subfamily member genes in E. coli and H. influenzae or acquisition of tadA by the others or their ancestors.
Further, each subfamily does not always display relationships that are congruent with other subfamilies or other traditional Bacterial or Archaeal phylogenies. This may be the result of horizontal transfer—a very possible explanation because many of these genes are located on genetic elements that may be transferred between distantly related organisms. However, claims for transfer events will require statistical tests of incongruence and the elimination of other possible explanations such as gene duplication with subsequent lineage sorting, convergence, incorrect rooting, and long-branch attraction. In general, considering the recent reappraisal of the role of horizontal gene transfer in organismal evolution, the relationship between the type II/IV NTPase gene phylogeny and organismal evolution awaits specific assessments of the extent of gene exchange between distantly related organisms (42, 43).
Nevertheless, we note one transfer event that may have occurred at the division between the Bacteria and Archaea in the tadA subfamily. If the division between the uncharacterized Archaeal NTPase genes and those of type II/IV secretion represents the initial division between the Archaeal and Bacterial organismal lineages, tadA would have been introduced from the Bacterial lineage into the Archaeal lineage. Because of the complexity of other possible explanations, but pending a more rigorous statistical analysis of incongruence, we favor the hypothesis of an interdomain horizontal transfer within the tadA family.
Evolution of Conjugative Transfer and Protein Export.
Precise descriptions of the evolutionary history of putative NTPase genes of type II/IV secretion systems test the hypothesis that type IV secretion of macromolecules is an adaptation of a conjugative DNA transfer system (44). It has been suggested recently that type IV protein secretion systems should be defined by their derivation from conjugative transfer systems (26). Such a definition requires explicit statements about the polarity or direction of evolutionary change: Did conjugative transfer or protein secretion arise first?
We can use the phylogeny of putative NTPase genes as a guide to reconstruct functional evolution if we assume that it represents the phylogeny of type IV secretion. The traG subfamily is the first major group to diverge from the main phylogenetic trunk. This, and the presence of other early branching superfamily members on transmissible elements (e.g., tcpT, bfpD, and the putative NTPase genes from B. anthracis virulence plasmids), may indicate that the earliest type IV NTPase was indeed part of a DNA transfer system (45). However, the NTPase genes of known type IV conjugative transfer systems [trbB (46), traG (45), and virB11 from A. tumefaciens (47)] are scattered throughout the tree and separated by branches representing protein transport systems. Indeed, the A. tumefaciens virB11 genes arise from a phylogenetic branch that is dominated, before the emergence of virB11, by NTPases that are known or predicted to be involved in protein transport (48–51). In this case, the most parsimonious reconstruction suggests that the A. tumefaciens virB system is derived from a protein export system, not vice versa. Likewise, a role in conjugative DNA transfer could have arisen independently in the traG and trbB lineages, allowing for the possibility that protein secretion was the earliest type IV function.
Because of the ability of some type IV NTPases to functionally substitute for one another (23), it is possible that recombination events, in which NTPases were exchanged between systems, have resulted in a phylogeny that does not represent the evolution of secretion systems. However, phylogenies of genes linked to type IV NTPase family genes, such as the homologs of virB4 and virB9, are similar to the NTPase gene phylogeny (51). This should be confirmed by more extensive comparative phylogenetic analysis with statistical tests of congruence (25) between other type IV secretion genes and the NTPase gene phylogeny.
At present, no simple statement can be made about the derivation of type IV protein secretion systems from conjugative transfer systems. More appropriate would be to note that type IV secretion systems are known for their ability to take part in both processes. Further, because DNA transfer requires proteins coupled to the DNA (9), the distinction between protein transport and conjugative DNA transfer may be artificial.
In addition to understanding gene relationships, precise classification of genes will benefit other aspects of biological study. Functional or structural predictions from primary structure are strengthened when they can take into account the groupings of genes and the history of portions of their sequences (31). In the case of the putative NTPase genes studied here, many are known to be important in pathogenic processes between bacteria and their hosts. A clear understanding of relationships among these genes may help in the design of both broad-range and more directed chemotherapeutics as well as other interventions.
Acknowledgments
We thank Indra Neil Sarkar for writing the gene classification program and designing our web page. P.J.P. and S.C.K. were supported by Columbia University and National Institutes of Health training grants, respectively.
Abbreviation
- MPAS
most parsimonious ancestral sequence
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF317389). Further information concerning these data may be found at http://cpmcnet.columbia.edu/dept/figurski.
References
- 1.Binet R, Letoffe S, Ghigo J M, Delepelaire P, Wandersman C. Gene. 1997;192:7–11. doi: 10.1016/s0378-1119(96)00829-3. [DOI] [PubMed] [Google Scholar]
- 2.Burns D L. Curr Opin Microbiol. 1999;2:25–29. doi: 10.1016/s1369-5274(99)80004-6. [DOI] [PubMed] [Google Scholar]
- 3.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salmond G P C. Annu Rev Phytopathol. 1994;32:181–200. [Google Scholar]
- 5.Henderson I R, Nataro J P, Kaper J B, Meyer T F, Farrand S K, Burns D L, Finlay B B, St. Geme J W., III Trends Microbiol. 2000;8:352. [PubMed] [Google Scholar]
- 6.Jose J, Jahnig F, Meyer T F. Mol Microbiol. 1995;18:378–380. doi: 10.1111/j.1365-2958.1995.mmi_18020378.x. [DOI] [PubMed] [Google Scholar]
- 7.Chung Y S, Dubnau D. J Bacteriol. 1998;180:41–45. doi: 10.1128/jb.180.1.41-45.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hobbs M, Mattick J S. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 9.Pansegrau W, Lanka E. Prog Nucleic Acids Res Mol Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 10.Possot O, Pugsley A P. Mol Microbiol. 1994;12:287–299. doi: 10.1111/j.1365-2958.1994.tb01017.x. [DOI] [PubMed] [Google Scholar]
- 11.Christie P J, Ward J E, Jr, Gordon M P, Nester E W. Proc Natl Acad Sci USA. 1989;86:9677–9681. doi: 10.1073/pnas.86.24.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grahn A M, Haase J, Bamford D H, Lanka E. J Bacteriol. 2000;182:1564–1574. doi: 10.1128/jb.182.6.1564-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krause S, Pansegrau W, Lurz R, de la Cruz F, Lanka E. J Bacteriol. 2000;182:2761–2770. doi: 10.1128/jb.182.10.2761-2770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lory S. Curr Opin Microbiol. 1998;1:27–35. doi: 10.1016/s1369-5274(98)80139-2. [DOI] [PubMed] [Google Scholar]
- 15.Rashkova S, Zhou X R, Chen J, Christie P J. J Bacteriol. 2000;182:4137–4145. doi: 10.1128/jb.182.15.4137-4145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivas S, Bolland S, Cabezon E, Goni F M, de la Cruz F. J Biol Chem. 1997;272:25583–25590. doi: 10.1074/jbc.272.41.25583. [DOI] [PubMed] [Google Scholar]
- 17.Sandkvist M, Bagdasarian M, Howard S P, DiRita V J. EMBO J. 1995;14:1664–1673. doi: 10.1002/j.1460-2075.1995.tb07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner L R, Lara J C, Nunn D N, Lory S. J Bacteriol. 1993;175:4962–4969. doi: 10.1128/jb.175.16.4962-4969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krause S, Barcena M, Pansegrau W, Lurz R, Carazo J M, Lanka E. Proc Natl Acad Sci USA. 2000;97:3067–3072. doi: 10.1073/pnas.050578697. . (First Published March 14, 2000; 10.1073/pnas.050578697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitchurch C B, Hobbs M, Livingston S P, Krishnapillai V, Mattick J S. Gene. 1991;101:33–44. doi: 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]
- 21.Stephens K M, Roush C, Nester E. J Bacteriol. 1995;177:27–36. doi: 10.1128/jb.177.1.27-36.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmond G P, Reeves P J. Trends Biochem Sci. 1993;18:7–12. doi: 10.1016/0968-0004(93)90080-7. [DOI] [PubMed] [Google Scholar]
- 23.Segal G, Russo J J, Shuman H A. Mol Microbiol. 1999;34:799–809. doi: 10.1046/j.1365-2958.1999.01642.x. [DOI] [PubMed] [Google Scholar]
- 24.Reeck G R, de Haen C, Teller D C, Doolittle R F, Fitch W M, Dickerson R E, Chambon P, McLachlan A D, Margoliash E, Jukes T H, et al. Cell. 1987;50:667. doi: 10.1016/0092-8674(87)90322-9. [DOI] [PubMed] [Google Scholar]
- 25.Thornton J W, DeSalle R. Annu Rev Genomics Hum Genet. 2000;1:41–73. doi: 10.1146/annurev.genom.1.1.41. [DOI] [PubMed] [Google Scholar]
- 26.Christie P J, Vogel J P. Trends Microbiol. 2000;8:354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kachlany S C, Planet P J, Bhattacharjee M, Kollia E, DeSalle R, Fine D H, Figurski D H. J Bacteriol. 2000;182:6169–6176. doi: 10.1128/jb.182.21.6169-6176.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skerker J M, Shapiro L. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bayley D P, Jarrell K F. J Mol Evol. 1998;46:370–373. [PubMed] [Google Scholar]
- 30.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisen J A. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- 32.Thompson J D, Higgins D G, Gibson T J. clustal x 1.63 Multiple Sequence Alignment Program. Hamburg, Germany: European Molecular Biology Organization; 1997. [Google Scholar]
- 33.Wheeler W C, Gatesy J, DeSalle R. Mol Phylogenet Evol. 1995;4:1–9. doi: 10.1006/mpev.1995.1001. [DOI] [PubMed] [Google Scholar]
- 34.Swofford D L. paup 4.02b. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 35.Phillips A, Janies D, Wheeler W. Mol Phylogenet Evol. 2000;16:317–330. doi: 10.1006/mpev.2000.0785. [DOI] [PubMed] [Google Scholar]
- 36.Eriksson T. Hypercard stack distributed by the author, Botaniska instiutionen. Stockholm: Stockholm University; 1997. [Google Scholar]
- 37.Bremer K. Cladistics. 1995;10:295–304. [Google Scholar]
- 38.Gogarten J P, Kibak H, Dittrich P, Taiz L, Bowman E J, Bowman B J, Manolson M F, Poole R J, Date T, Oshima T, et al. Proc Natl Acad Sci USA. 1989;86:6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T. Proc Natl Acad Sci USA. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maddison W P, Maddison D R. macclade, version 3. Sunderland, MA: Sinauer; 1992. [Google Scholar]
- 41.Inoue T, Tanimoto I, Ohta H, Kato K, Murayama Y, Fukui K. Microbiol Immunol. 1998;42:253–258. doi: 10.1111/j.1348-0421.1998.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 42.Doolittle W F. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 43.Woese C R. Proc Natl Acad Sci USA. 2000;97:8392–8396. doi: 10.1073/pnas.97.15.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winans S C, Burns D L, Christie P J. Trends Microbiol. 1996;4:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winans S C, Walker G C. J Bacteriol. 1985;161:402–410. doi: 10.1128/jb.161.1.402-410.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lessl M, Balzer D, Lurz R, Waters V L, Guiney D G, Lanka E. J Bacteriol. 1992;174:2493–2500. doi: 10.1128/jb.174.8.2493-2500.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christie P J. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segal E D, Cha J, Lo J, Falkow S, Tompkins L S. Proc Natl Acad Sci USA. 1999;96:14559–14564. doi: 10.1073/pnas.96.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stein M, Rappuoli R, Covacci A. Proc Natl Acad Sci USA. 2000;97:1263–1268. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiss A A, Johnson F D, Burns D L. Proc Natl Acad Sci USA. 1993;90:2970–2974. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli M L, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]