Abstract
Rapid evolution driven by positive Darwinian selection is a recurrent theme in male reproductive protein evolution. In contrast, positive selection has never been demonstrated for female reproductive proteins. Here, we perform phylogeny-based tests on three female mammalian fertilization proteins and demonstrate positive selection promoting their divergence. Two of these female fertilization proteins, the zona pellucida glycoproteins ZP2 and ZP3, are part of the mammalian egg coat. Several sites identified in ZP3 as likely to be under positive selection are located in a region previously demonstrated to be involved in species-specific sperm-egg interaction, suggesting the selective pressure is related to male-female interaction. The results provide long-sought evidence for two evolutionary hypotheses: sperm competition and sexual conflict.
Keywords: zona pellucida, ZP3, fertilization, sperm competition, sexual conflict
One of the striking features that has emerged from the study of reproduction is diversity (1–17). Diversity has been observed in several morphological traits that are involved in reproduction, including sperm (14) and reproductive organ (15) morphology. Recently, male-derived molecules involved in reproduction have been shown to be extraordinarily divergent between closely related species (1–13). These molecules include proteins involved in signaling between males and females (3–5), fertilization (6–11), sperm chromosome condensation (1, 12), and sex-specific transcription (16, 17). However, only the divergence of molecules from males has been shown to be driven by positive selection. The divergence of female reproductive proteins has never been demonstrated to be promoted by selection. This finding is surprising because many of the models that account for rapid evolution of reproductive proteins are based on male-female interaction and therefore would predict that female proteins also would be subjected to positive selection. These models include sperm competition, a process in which it has been demonstrated that females play a significant role (19, 20), and sexual conflict, a model that explicitly assumes adaptive evolution of female proteins (2, 20). To examine whether female reproductive molecules undergo adaptive evolution, we performed phylogeny-based statistical tests of positive selection on three mammalian female reproductive proteins. Our goal was to determine whether any female reproductive proteins were subject to positive selection. Because such selection is most likely to act on extracellular or surface proteins involved in male-female (sperm-egg or oviduct) recognition (rather than on all reproductive proteins) we concentrated on those reproductive proteins that are known to play important roles in sperm-egg interaction [ZP2 and ZP3, vertebrate egg coat (zona pellucida) proteins; ref. 21] or that have been suggested to be involved in fertilization [oviductal glycoprotein (OGP); ref. 22].
A stringent and unequivocal signal of positive Darwinian selection in molecular evolution is a significantly higher nonsynonymous (dN; amino acid replacing) than synonymous (dS; silent) substitution rate (23). The ratio of the two rates, dN/dS, denoted ω herein, measures the magnitude and direction of selective pressure on a protein, with ω = 1, <1, and >1 indicating neutral evolution, purifying selection, and positive diversifying selection, respectively. This criterion has been used to demonstrate rapid evolution of male reproductive proteins in a variety of invertebrate and vertebrate species (1–17). Recently, Wyckoff et al. (1) used the ω ratio and other criteria to demonstrate the rapid evolution of male reproductive proteins in primates. In their study, female reproductive proteins, including OGP and ZP3, were placed within a control group of nonrapidly evolving genes expressed in a variety of tissues (1). To elucidate the selective forces underlying the rapid divergence of reproductive proteins, it is important to analyze female, as well as male, reproductive proteins.
The diversity of female reproductive proteins was previously analyzed by two-dimensional denaturing PAGE (24). Both female and male reproductive proteins were more diverse than proteins from other tissues. However, because DNA sequences were unavailable for these proteins, it was not possible to test for positive selection and therefore impossible to rule out that the divergence was simply caused by lack of constraint and neutral drift. In the one case of a female reproductive protein for which ω was estimated, an egg coat gene in abalone, ω was <1, indicating no signs of positive selection (25). Additional tests of neutrality for that gene, including methods presented herein and analyses of a polymorphism survey, are consistent with that conclusion (52). However, the repetitive nature of the abalone female protein suggested a hypothesis for the rapid evolution of its male ligand that did not require positive selection acting on the female receptor (25) and was consistent with a previous model for the evolution of species-specific interaction of gametes (6, 26).
Here we present an analysis suggesting that positive Darwinian selection promotes the divergence of three mammalian female reproductive proteins (ZP3, ZP2, and OGP) from a variety of mammalian species. ZP3 is the primary species-specific binding site for sperm, and species-specifically induces sperm to undergo the acrosome reaction (21). ZP2 binds acrosome-reacted sperm and has been termed the secondary sperm receptor (22). The role of OGP in fertilization remains unclear, but estrous oviductal fluid, which would contain OGP, plays a role in species-specific gamete interaction (28). For comparison, we reanalyzed the three male reproductive proteins (protamine 1, protamine 2, and transition protein 2) identified as being under positive selection by Wyckoff et al. (ref. 1; see also refs. 12 and 29). Furthermore, to demonstrate the reliability of these new analyses, we perform calculations on two control proteins: (i) the class I MHC glycoprotein, which is known to be subjected to positive selection (thus serving as a positive control) and whose structure and variable regions are known (30) and (ii) a housekeeping gene, carbonic anhydrase I, for which there is no evidence of positive selection (thus serves as a negative control). With the control proteins confirming the robustness of the methodology used, we demonstrate that ZP3, ZP2, and OGP are subjected to positive Darwinian selection. Furthermore, we identify specific residues of these proteins that may be the targets of selection and are perhaps involved in species-specific gamete interaction.
Materials and Methods
Sequences Analyzed.
GenBank accession numbers (organisms) for sequences used are as follows:
ZP2: M90366 (Homo sapiens), Y10690 (Macaca radiata), Y10767 (Callithrix jacchus), U05776 (Felis catus), D45064 (Sus scrofa), D45069 (Canis familiaris), M34148 (Mus musculus), AB000929 (Rattus norvegicus).
ZP3: M20026 (M. musculus), Y10823 (Rattus rattus), X56777 (H. sapiens), S71825 (marmosets), X82639 (M. radiata), D45070 (C. familiaris), D45068 (F. catus), D45065 (S. scrofa).
OGP: U09550 (H. sapiens), M59903 (Papio hamadryas anubis), U87259 (Macaca mulatta), D16639 (Bos taurus), U16719 (Ovis aries), U15048 (Mesocricetus auratus), U43490 (S. scrofa), D32137 (M. musculus).
Protamine P1: Y00443 (H. sapiens), L14587 (Gorilla gorilla), L14588 (Hylobates lar), L14591 (Pan troglodytes), L14589 (Pongo pygmaeus), X61678 (Saguinus imperator), L10654 (Equus caballus), X13381 (S. scrofa), Z11544 (Cavia porcellus), M18395 (B. taurus).
Protamine P2: X72968 (P. troglodytes), AF215723 (P. paniscus), X71336 (G. gorilla), AF215713 (H. sapiens), X71339 (H. lar), X71337 (P. pygmaeus), X71340 (Macaca nemestrina), X85371 (C. jacchus), X71335 (Alouatta seniculus).
Transition protein 2: L03378 (H. sapiens), AF071208 (C. familiaris), X14776 (R. norvegicus), J03494 (M. musculus) X56401 (B. taurus), AF215720 (M. mulatta), AF215719 (P. pygmaeus), AF215718 (G. gorilla), AF215717 (P. paniscus), AF215716 (P. troglodytes).
Class I MHC: M94053 (H. sapiens), M24039 (H. sapiens), M24034 (H. sapiens), Z27120 (H. sapiens), X13111 (H. sapiens), X61701 (H. sapiens).
Carbonic anhydrase I: L11621 (P. troglodytes), L11622 (G. gorilla), M33987 (H. sapiens), L25082 (M. nemestrina), L42178 (O. aries), M32452 (M. musculus).
Statistical Analyses.
Amino acid sequences were aligned by using clustalw (http://www2.ebi.ac.uk/clustalw/), and the aligned proteins were used to generate nucleotide alignments (http://bioweb.pasteur.fr/seqanal/interfaces/protal2dna-simple.html). We applied the maximum likelihood method of Nielsen, Yang, and coworkers (31–33) to test for positive selection and to infer amino acid sites under positive selection. The codeml program in paml (31) was used. The major advantage of these likelihood models over previous analysis is that they account for variable selective pressures among sites by assuming that there are different classes of sites in the gene with different ω ratios. The analysis consists of two major steps. The first step uses the likelihood ratio test to test for positive selection, that is, for presence of sites with ω > 1. This is achieved by comparing a null model that does not allow for sites with ω > 1 and a more-general model that does. We used two likelihood ratio tests. The first compares model M0, which assumes one ω for all sites, with M3 (discrete), which assumes three site classes with independent ωs estimated from the data. Because M0 involves one parameter ω whereas M3 involves five (two proportions and three ω ratios), twice the log-likelihood difference (2Δl) is compared with the χ2 distribution with d.f. = 5 − 1 = 4. This is a test for variation of ω among sites, but when estimates of ω under M3 are >1, positive selection is implicated. The second test is a refined test for presence of sites under positive selection. The null model (M7 beta) assumes a beta distribution B(p, q), with ω limited in the interval (0, 1). This is a flexible distribution, but ω is limited to the interval (0, 1), where 0 indicates complete constraint and 1 is the expectation under no selective constraint. The alternative model M8 (beta and ω) adds an extra class of sites with ω estimated, so that a proportion p0 of sites come from the beta distribution B(p, q) and the remaining sites (p1 = 1 − p0) have a ω ratio estimated from the data that can be greater than 1. M8 has two more parameters than M7, so d.f. = 2. Likelihood analyses using other models of variable ω ratios among sites also were performed and produced consistent results with those presented here (data not shown).
The second major step of the analysis is to identify residues under positive selection when the likelihood ratio test suggests their presence. This is achieved by using the Bayes theorem to calculate the (posterior) probabilities that each site, given the data at that site, are from the different ω classes (32, 33). Sites with a high probability of coming from the class with ω > 1 are likely to be under positive selection. Positions predicted to be under positive selection for the MHC were mapped onto the crystal structure (PDB file 1QLFA) using molscript (34).
These methods analyze variation in the ω ratio within a phylogenetic context. Previous analyses have demonstrated the methods are fairly robust to different tree topologies (33). The phylogenetic trees used in our analyses are seen in Fig. 2, which is published as supplemental material on the PNAS website, www.pnas.org. We also performed the analyses by using alternative tree topologies and found results consistent with those presented here (data not shown).
We note that the statistical analysis used here is able to demonstrate adaptive molecular evolution by revealing unequivocal evidence for positive diversifying selection, but cannot provide a mechanism for the positive selection. However, by pinpointing amino acid sites likely to be involved in positive selection, such analysis might provide important clues for further laboratory investigation.
Results
The average dN/dS ratios (ω) averaged across lineages and sites are smaller than one in all reproductive and control proteins used here (Table 1). However, these proteins could contain constrained amino acid sites subjected to purifying selection with ω close to zero as well as sites that could be subjected to positive selection. A large number of constrained sites would mask a signal of positive selection when the ω ratio was averaged over all sites, and therefore the average ω is not a sensitive measure of selection (32, 33). Thus, we tested for positive selection at individual amino acid sites using maximum likelihood models that account for variable selective pressures among sites indicated by the ω ratio (32, 33). Similar methods also have been developed to measure variation in the ω between sites (35), but they require more data than is currently available for the female reproductive proteins ZP2, ZP3, and OGP.
Table 1.
Likelihood ratio test of positive selection in female and male reproductive proteins
| Gene | n | Lc | S | dN/dS | 2Δℓ M3 vs. M0 | 2Δℓ M8 vs. M7 | Parameter estimates under M8 (beta & ω) | Positively selected sites |
|---|---|---|---|---|---|---|---|---|
| Female reproductive proteins | ||||||||
| ZP2 | 8 | 676 | 2.6 | 0.53 | 138.8** | 14.8** | p1 = 0.047, ω = 2.5p0 = 0.953, B(0.66, 0.66) | 38, 57, 117, 161, 170, 198, 241, 287, 339, 342, 665, 674, 710, 711 |
| ZP3 | 8 | 365 | 2.9 | 0.27 | 219.6** | 8.6* | p1 = 0.076, ω = 1.7p0 = 0.924, B(0.40, 1.24) | 25, 28, 31, 32, 33, 34, 47, 50, 84, 185, 194, 331, 333, 340, 341, 345, 347, 348, 372, 373 |
| OGP | 8 | 447 | 1.7 | 0.35 | 108.4** | 8.1* | p1 = 0.017, ω = 4.2p0 = 0.983, B(0.31, 0.56) | 122, 150, 280, 446, 448, 449 |
| Male reproductive proteins | ||||||||
| Protamine P1 | 10 | 48 | 3.7 | 0.93 | 69.5** | 27.5** | p1 = 0.456, ω = 3.6p0 = 0.544, B(131.7, 686.3) | 10, 12, 16, 18, 19, 20, 22, 23, 29, 30, 36, 37, 38, 42, 43, 44, 45, 46, 48, 49, 51 |
| Protamine P2 | 9 | 99 | 1.3 | 0.58 | 12.0* | 0.2 | p1 = 0.560, ω = 1.2p0 = 0.440, B(1.43, 17.77) | 4, 5, 6, 9, 12, 13, 15, 16, 17, 18, 20, 21, 22, 23, 25, 28, 29, 30, 36, 37, 38, 40, 41, 43, 45, 48, 49, 50, 51, 52, 57, 59, 63, 64, 66, 69, 70, 71, 75, 81, 84, 87, 88, 90, 94, 96, 98, 101, 102 |
| Transition protein 2 | 10 | 101 | 3.5 | 0.29 | 39.0** | 2.0 | p1 = 0.106, ω = 1.6p0 = 0.893, B(0.91, 2.32) | 36, 64, 66, 71, 73, 88, 98 |
| Controls | ||||||||
| Class I MHC | 6 | 362 | 0.6 | 0.55 | 78.6** | 14.7** | p1 = 0.073, ω = 4.0 p0 = 0.927, B(0.10, 0.27) | 45, 48, 51, 52, 55, 56, 82, 99, 101, 121, 126 |
| Carbonic anhydrase I | 6 | 261 | 1.2 | 0.34 | 45.8** | 0.6 | B(0.14, 0.28) | None detected |
The data have n sequences, each of Lc codons after alignment gaps are removed. S is the tree length, measured as the number of nucleotide substitutions per codon, and dN/dS is the average ratio over sites and branches, both calculated under a codon model with one ω for all sites. The proportion of sites under positive selection (p1), or under selective constraint (p0), and parameters p and q for the beta distribution B(p, q) are given under M8. Parameter estimates for carbonic anhydrase I come from M7, because M8 is not a significantly better fit of the data. Parameters indicating positive selection are in bold.
: significant at 5% level;
: significant at 1% level. Sites potentially under positive selection identified under model M8 are listed according to the mouse sequence numbering except for protamine P1 and P2, which are numbered according to the human sequence, and class I MHC, which is numbered according to the protein data bank file 1QLFA. Positively selected sites with posterior probability >0.9 are underlined, 0.8–0.9 in bold, 0.7–0.8 in italics, and 0.5–0.7 in plain text.
Control Proteins: Class I MHC and Carbonic Anhydrase.
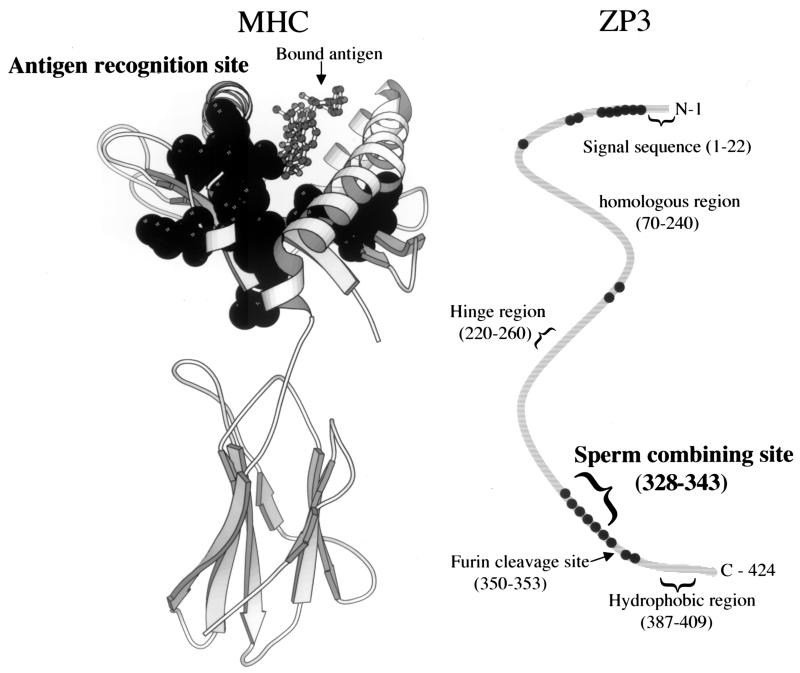
To demonstrate the reliability of the method in detecting positive selection and inferring sites potentially subjected to selection, we first performed likelihood ratio tests on control proteins. The ω ratio calculated across all sites for the class I MHC protein is <1 (Table 1). However, it has previously been shown that the ω ratio calculated for the residues located in the antigen recognition site of this protein is >1, indicating positive selection (30). Therefore, the class I MHC protein is a good test case for the likelihood models used herein. The MHC data includes sequences from the HLA-A and HLA-B loci. Similar results are obtained with 12 sequences (tree length of 0.4) from only the HLA-A locus. Both likelihood ratio tests (comparing M0 vs. M3 and M7 vs. M8, respectively) for the class I MHC protein indicate variation in ω between sites, with a class of sites having ω > 1, indicating positive selection (Table 1). Furthermore, all of the residues identified as likely to be under positive selection are located in the antigen recognition site when mapped onto the crystal structure of MHC (Fig. 1 Left). This result is as predicted, because it is the antigen recognition site that binds foreign peptides and is subjected to diversifying selection (30). Note that the likelihood analysis does not use the structural information. To demonstrate that the likelihood methods do not spuriously detect selection, we performed likelihood ratio tests for a housekeeping gene that presumably evolves neutrally (a negative control protein). We use carbonic anhydrase I because it has a comparable overall level of divergence as the other genes used herein, as measured by the tree length (nucleotide substitutions per codon: Table 1). Comparison of model M0 with M3 suggests variation in the ω ratio between sites; however no sign of positive selection (as indicated by ω > 1) was detected (Table 1). Comparison of model M7 with M8 additionally does not detect selection (Table 1). These control analyses support the view that the likelihood ratio tests are reliable for studying the selective pressures affecting protein evolution. This reliability also is supported by results obtained from extensive computer simulations, which show that the likelihood ratio test used here tends to be conservative but is nevertheless very powerful in detecting positive selection (M. Anisimova and Z.Y., unpublished data).
Figure 1.
Location of the residues identified as likely to be under positive selection in class I MHC and ZP3. (Left) MHC (protein data bank file 1QLFA) with a bound antigen shown in stick-and-ball format. MHC residues identified as under selection are in black spacefill, and all fall in the domain containing the antigen recognition site. Chain C, present in structure but not used in sequence analysis, is not shown. (Right) A schematic representation of the ZP3 molecule indicating functional regions (adapted from ref. 21; no ZP3 three-dimensional structure has been determined). Black dots represent relative positions of sites under positive selection, a cluster of which fall in the region identified as the sperm-combining site and another cluster in a region immediately following the signal sequence.
Female and Male Reproductive Proteins.
Next, models M0 and M3 were compared for female and male reproductive genes. This test is significant for all female and male genes examined (Table 1), indicating that the selective pressure indeed varies among amino acid sites in each protein. Furthermore, parameter estimates under M3 suggest the presence of sites under positive selection in each protein. For the three female proteins, the proportions of sites potentially under positive selection are 4.0% with ω = 2.7 for ZP2, 8.9% with ω = 1.7 for ZP3, 1.7% with ω = 4.2 for OGP. For the three male proteins, the proportions are 47% with ω = 3.6 for protamine 1, 56% with ω = 1.2 for protamine 2, and 13% with ω = 1.5 for transition protein 2.
As a refined test for selection, model M8 was compared with M7. M8 fits the data significantly better than M7 for all of the three female proteins and the estimated ω ratios for the extra class are all >1, suggesting that allowing for sites with ω > 1 significantly improves the fit of the model to data (Table 1). The proportions of sites under selection estimated under M8 are listed in Table 1. These proportions are similar to estimates from model M3 presented above. Thus, there is clear statistical evidence that divergence of these female mammalian reproductive proteins is promoted by positive Darwinian selection.
For the male proteins, comparison of M8 against M7 is significant for protamine P1, but not for protamine P2 and transition protein 2. The lack of significance appears to be due to the sparse sampling of species and low sequence divergence, resulting in low information content in the data. Parameter estimates under both M3 and M8 suggest high proportions of sites under diversifying selection in all three male proteins (Table 1). Similar high proportions have been seen in other small sperm molecules under selection (36), which might be because these small proteins consist of only one domain upon which selection acts. Larger molecules often contain multiple domains, and selection often acts on a subset of the domains. It is likely that including more mammalian species in such an analysis would provide even stronger evidence of adaptive evolution in the male proteins. These results are consistent with the analysis of Wyckoff et al. (1), demonstrating positive selection on these male reproductive proteins; however, some controversy exists regarding the exact nature of the selection (12, 29).
Sperm competition (1, 18, 19) and sexual conflict (2, 20) are among possible selective forces (15) driving the divergence of these female and male reproductive proteins. The sexual conflict hypothesis assumes rapid adaptive evolution of female reproductive proteins to avoid mating-induced deleterious effects (2, 20, 37), such as polyspermy, sensory exploitation, seminal fluid toxicity, and mating-induced reduction in female lifespan (16, 38–40). Our demonstration that positive selection promotes the divergence of female reproductive proteins lends support to this hypothesis.
Identification of Amino Acid Positions under Positive Selection and Their Role in Species Specificity.
As with the class I MHC protein, sites subjected to positive selection may be involved in ligand recognition. Thus, identification of selected sites may shed light on the selective agents and identify regions that are functionally important for female-male (egg-sperm) interaction. To identify such regions, we used the Bayes theorem to calculate the posterior probabilities of ω classes for each site. Sites with high probabilities of coming from the class with ω > 1 are likely to be under positive selection (32, 33). These sites are indicated in Table 1 for the female proteins (ZP2, ZP3, and OGP), male proteins (protamine P1, protamine P2, and transition protein 2) and the class I MHC control protein. Their locations are represented schematically in Fig. 1 for ZP3 and class I MHC protein.
The molecular basis for species-specific interaction between mouse ZP3 and sperm has been extensively characterized (41–43). Therefore, we can compare our predictions of sites potentially under positive selection to sites identified as important from detailed biochemical and genetic characterizations of this gene (Fig. 1 Right). ZP2 and OGP have not been as well characterized as ZP3, so similar comparisons cannot be performed. For the ZP3 gene, many of the sites identified here as likely to be under positive selection fall in the region 331–373 (Fig. 1 Right; Table 1). This region has previously been identified as potentially governing species specificity in sperm-egg interaction by exon swapping and site-directed mutagenesis (41–43). Although in vitro tests of expressed site-directed mutants suggest that the glycosylation sites Ser-332 and Ser-334 in mouse ZP3 are important for function, it has been suggested amino acids surrounding the glycosylation sites could influence their glycosylation pattern (41–43). Studies of glycosylation patterns of residues in different amino acid backbone contexts support this hypothesis (44–47). However, transgenic experiments replacing mouse with human ZP3 in mouse eggs did not result in the expected switch of species specificity, suggesting factors in addition to ZP3's primary sequence also may affect the final conformation or glycosylation of the protein, leading to species specificity (48). We also note that mouse site Ser-334 has been substituted to Ala in the marmoset (49). Substitution from Ser to Ala would obliterate glycosylation of this residue and would provide means for substitutions at the primary sequence level to alter the glycosylation pattern. Region 331–373 also was found to be divergent in analysis of vertebrate (including nonmammalian) ZP3 molecules (50). Our analysis suggests that the high divergence in this region results not from lack of functional constraint, but rather from positive selection promoting rapid evolution.
Discussion
We have demonstrated that the female reproductive proteins ZP2, ZP3, and OGP are subjected to positive Darwinian selection. These results lend support to the models of sperm competition (1, 18, 19), sexual conflict (2, 20, 37), and cryptic female choice (15) driving the evolution of reproductive proteins, because these models involve male-female interactions. It is important for functional as well as evolutionary studies to examine the rapid evolution of both female and male reproductive proteins. Functional studies can glean important information not only from conserved regions of the molecules but also from the divergent regions under positive selection, because the latter may be functionally important for specificity. Our analysis identified several sites in ZP3 under positive selection. These include a region previously implicated as functionally important in sperm-egg interaction (41–43). Additionally, a region in ZP3 immediately following the signal sequence was identified (Fig. 1 Right) for which tests of functional importance have not been reported and which our data predict might also play a role in species specificity. The sites we identified in ZP2 as likely to be under positive selection are candidates to test for functional importance in ZP2's role as receptor for acrosome-reacted sperm (21, 27).
It is likely that the evolution of additional female and male reproductive proteins also are promoted by positive Darwinian selection. For example, many reproductive proteins (including ZP2, ZP3, and the sperm protamines analyzed here, but not OGP) are found in the 10% most divergent sequences from an aligned set of 2,820 human-rodent orthologs (ref. 51 and our unpublished analyses). These reproductive molecules are as divergent as many genes involved in immune response. Another ZP glycoprotein (ZP1) is also among these rapidly evolving proteins, but insufficient phylogenetic sampling to date precluded its analysis by using likelihood ratio tests. Future sequencing and phylogenetic analyses of these reproductive proteins are necessary to determine whether their rapid divergence is promoted by positive selection or caused by lack of constraint. It also will be important to determine in general what proportion of reproductive proteins show signs of selectively driven rapid evolution seen herein.
Our demonstration of positive Darwinian selection in female as well as male reproductive proteins lends support for models of sexual conflict and sperm competition driving the divergence of reproductive proteins (2, 20, 37). Although the nature of the selective pressure remains unclear, our observation that selection acts to diversify a region in ZP3 previously identified as functionally important for species specificity suggests that the selective pressure may be related to male-female interaction, in this case sperm-egg interaction.
Supplementary Material
Acknowledgments
We thank V. D. Vacquier, R. Durrett, S. D. Tanksley, J. D. Calkins, and V. L. Bauer for comments on the manuscript. This work was supported by a National Science Foundation/Alfred P. Sloan postdoctoral fellowship in molecular evolution to W.J.S. Support also was provided by Biotechnology and Biological Sciences Research Council Grant 31/G10434 (to Z.Y.), National Science Foundation Grant IBN 97–23356 (to M.F.W.), and National Institutes of Health Grants HD38921 (to M.F.W.) and GM36431 (to C.F.A.).
Abbreviations
- ZP
zona pellucida
- OGP
oviductal glycoprotein
References
- 1.Wyckoff G J, Wang W, Wu C-I. Nature (London) 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- 2.Gavrilets S. Nature (London) 2000;403:886–889. doi: 10.1038/35002564. [DOI] [PubMed] [Google Scholar]
- 3.Aguadé M. Genetics. 1999;152:543–551. doi: 10.1093/genetics/152.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karn R C, Nachman M W. Mol Biol Evol. 1999;16:1192–1197. doi: 10.1093/oxfordjournals.molbev.a026209. [DOI] [PubMed] [Google Scholar]
- 5.Tsaur S C, Wu C-I. Mol Biol Evol. 1997;14:544–549. doi: 10.1093/oxfordjournals.molbev.a025791. [DOI] [PubMed] [Google Scholar]
- 6.Metz E C, Palumbi S R. Mol Biol Evol. 1996;13:397–406. doi: 10.1093/oxfordjournals.molbev.a025598. [DOI] [PubMed] [Google Scholar]
- 7.Swanson W J, Vacquier V D. Proc Natl Acad Sci USA. 1995;92:4957–4961. doi: 10.1073/pnas.92.11.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hellberg M E, Moy G W, Vacquier V D. Mol Biol Evol. 2000;17:458–466. doi: 10.1093/oxfordjournals.molbev.a026325. [DOI] [PubMed] [Google Scholar]
- 9.Metz E C, Robles-Sikisaka R, Vacquier V D. Proc Natl Acad Sci USA. 1998;95:10676–10681. doi: 10.1073/pnas.95.18.10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellberg M E, Vacquier V D. Mol Biol Evol. 1999;16:839–848. doi: 10.1093/oxfordjournals.molbev.a026168. [DOI] [PubMed] [Google Scholar]
- 11.Ferris P J, Pavlovic C, Fabry S, Goodenough U W. Proc Natl Acad Sci USA. 1997;94:8634–8639. doi: 10.1073/pnas.94.16.8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rooney A P, Zhang J, Nei M. Mol Biol Evol. 2000;17:278–283. doi: 10.1093/oxfordjournals.molbev.a026307. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y-H, Ota T, Vacquier V D. Mol Biol Evol. 1995;12:231–238. doi: 10.1093/oxfordjournals.molbev.a040200. [DOI] [PubMed] [Google Scholar]
- 14.Pitnick S. Am Nat. 1996;148:57–80. [Google Scholar]
- 15.Eberhard W G. Female Control: Sexual Selection by Cryptic Female Choice. Princeton: Princeton Univ. Press; 1996. [Google Scholar]
- 16.Sutton K A, Wilkinson M F. J Mol Evol. 1997;45:579–588. doi: 10.1007/pl00006262. [DOI] [PubMed] [Google Scholar]
- 17.Ting C T, Tsaur S C, Wu M L, Wu C-I. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- 18.Price C S C. Nature (London) 1997;388:663–666. doi: 10.1038/41753. [DOI] [PubMed] [Google Scholar]
- 19.Clark A G, Begun D J, Prout T. Science. 1999;283:217–220. doi: 10.1126/science.283.5399.217. [DOI] [PubMed] [Google Scholar]
- 20.Rice W R. Nature (London) 1996;381:232–234. doi: 10.1038/381232a0. [DOI] [PubMed] [Google Scholar]
- 21.Wassarman P M. Cell. 1999;96:175–183. doi: 10.1016/s0092-8674(00)80558-9. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt A, Mavrogianis P A, O'Day-Bowman M B, Jaffe R C, Verhage H G. Am J Reprod Immun. 1997;38:377–383. doi: 10.1111/j.1600-0897.1997.tb00316.x. [DOI] [PubMed] [Google Scholar]
- 23.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 24.Civetta A, Singh R S. J Mol Evol. 1995;41:1085–1095. doi: 10.1007/BF00173190. [DOI] [PubMed] [Google Scholar]
- 25.Swanson W J, Vacquier V D. Science. 1998;281:710–712. doi: 10.1126/science.281.5377.710. [DOI] [PubMed] [Google Scholar]
- 26.Wu C-I. Evolution. 1985;39:66–82. doi: 10.1111/j.1558-5646.1985.tb04080.x. [DOI] [PubMed] [Google Scholar]
- 27.Greenhouse S, Rankin T, Dean J. Am J Hum Genet. 1998;62:1282–1287. doi: 10.1086/301893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slavik T, Fulka J. Folia Biol (Prague) 1999;45:53–58. [PubMed] [Google Scholar]
- 29.Clark A G, Civetta A. Nature (London) 2000;403:261–263. doi: 10.1038/35002236. [DOI] [PubMed] [Google Scholar]
- 30.Hughes A L, Nei M. Nature (London) 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 31.Yang Z. Phylogenetic Analysis by Maximum Likelihood (PAML) (University College, London), version 2.0j. 1999. [Google Scholar]
- 32.Nielsen R, Yang Z. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Z, Nielsen R, Goldman N, Pedersen A-M K. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraulis P R. J App Crystallogr. 1991;24:946–950. [Google Scholar]
- 35.Suzuki Y, Gojobori T. Mol Biol Evol. 1999;16:1315–1328. doi: 10.1093/oxfordjournals.molbev.a026042. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z, Swanson W J, Vacquier V D. Mol Biol Evol. 2000;17:1446–1455. doi: 10.1093/oxfordjournals.molbev.a026245. [DOI] [PubMed] [Google Scholar]
- 37.Rice W R, Holland B. Behav Ecol Soc. 1997;41:1–10. [Google Scholar]
- 38.Chapman T, Liddle L F, Kalb J M, Wolfner M F, Partridge L. Nature (London) 1995;373:241–244. doi: 10.1038/373241a0. [DOI] [PubMed] [Google Scholar]
- 39.Gems D, Riddle D L. Nature (London) 1996;379:723–725. doi: 10.1038/379723a0. [DOI] [PubMed] [Google Scholar]
- 40.Hsin H, Kenyon C. Nature (London) 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- 41.Chen J, Litscher E S, Wassarman P M. Proc Natl Acad Sci USA. 1995;95:6193–6197. doi: 10.1073/pnas.95.11.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wassarman P M, Litscher E V. Curr Top Dev Biol. 1995;30:1–19. doi: 10.1016/s0070-2153(08)60562-1. [DOI] [PubMed] [Google Scholar]
- 43.Kinloch R A, Sakai Y, Wassarman P M. Proc Natl Acad Sci USA. 1995;92:263–267. doi: 10.1073/pnas.92.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elhammer A P, Poorman R A, Brown E, Maggiora L L, Hoogerheide J G, Kezdy F J. J Biol Chem. 1993;268:10029–10038. [PubMed] [Google Scholar]
- 45.Gooley A A, Williams K L. Glycobiology. 1994;4:413–417. doi: 10.1093/glycob/4.4.413. [DOI] [PubMed] [Google Scholar]
- 46.Nehrke K, Hagen F K, Tabak L A. J Biol Chem. 1996;271:7061–7065. doi: 10.1074/jbc.271.12.7061. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Agrwal N, Eckhardt A E, Stevens R D, Hill R L. J Biol Chem. 1993;268:22979–22983. [PubMed] [Google Scholar]
- 48.Rankin T L, Tong Z B, Castle P E, Lee E, Gore L R, Nelson L M, Dean J. Development (Cambridge, UK) 1998;25:2415–2424. doi: 10.1242/dev.125.13.2415. [DOI] [PubMed] [Google Scholar]
- 49.Thillai-Koothan P, van Duin M, Aitken R J. Zygote. 1993;1:93–101. doi: 10.1017/s0967199400001350. [DOI] [PubMed] [Google Scholar]
- 50.Yang J C, Hedrick J L. Dev Growth Differ. 1997;39:457–467. doi: 10.1046/j.1440-169x.1997.t01-3-00007.x. [DOI] [PubMed] [Google Scholar]
- 51.Makalowski W, Boguski M S. Proc Natl Acad Sci USA. 1998;95:9407–9412. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swanson, W. J., Aquadro, C. F. & Vacquier, V. D. (2001) Mol. Biol. Evol., in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.