Abstract
Approximately 90% of humans are right-handed. Handedness is a heritable trait, yet the genetic basis is not well understood. Here we report a genome-wide association study for a quantitative measure of relative hand skill in individuals with dyslexia [reading disability (RD)]. The most highly associated marker, rs11855415 (P = 4.7 × 10−7), is located within PCSK6. Two independent cohorts with RD show the same trend, with the minor allele conferring greater relative right-hand skill. Meta-analysis of all three RD samples is genome-wide significant (n = 744, P = 2.0 × 10−8). Conversely, in the general population (n = 2666), we observe a trend towards reduced laterality of hand skill for the minor allele (P = 0.0020). These results provide molecular evidence that cerebral asymmetry and dyslexia are linked. Furthermore, PCSK6 is a protease that cleaves the left–right axis determining protein NODAL. Functional studies of PCSK6 promise insights into mechanisms underlying cerebral lateralization and dyslexia.
INTRODUCTION
The population bias in right-handedness is a characteristic feature of humans (1). There is some variance across cultures (2,3); however, there are no known populations in the world in which left-handers are the majority.
Handedness is correlated with cerebral asymmetries and right-handedness implies a dominance of the left hemisphere for motor function. Since Paul Broca reported in 1861 the case of a patient who had aphasia caused by a lesion in the left hemisphere (4), there has been significant interest in the idea that language laterality and handedness are linked. It has been proposed that handedness emerged as a consequence of the evolution of language (5). Moreover, theories dating back almost a century posit a connection between handedness and neurodevelopmental disorders such as specific language impairment (SLI) and dyslexia (6), two disorders affecting language and reading skills, respectively, with a prevalence between 5 and 10% (7). However, no convincing association has been found between either hand preference or hand skill and neurodevelopmental disorders (8,9). Functional brain imaging studies have shown a weak correlation between handedness and cerebral dominance for language; with 96% of strong right-handers, compared with 73% of strong left-handers, showing left-hemisphere dominance for language (10).
There is suggestive, but mixed, evidence indicating that there may be atypical cerebral asymmetry in patients with dyslexia or SLI (reviewed in 11,12). With respect to reading disability (RD), Galaburda et al. (13,14) reported eight consecutive post-mortem specimens with RD that had reduced planum temporale asymmetry. Subsequent structural MRI studies have been inconsistent in replicating this finding (reviewed in 11). However, a meta-analysis of 17 functional neuroimaging studies comparing RD individuals to controls during reading tasks suggests that underactivation of the left hemisphere is found in the inferior parietal, superior temporal, middle and inferior temporal and fusiform regions (15). Conversely, there were no abnormalities of activation observed in the right hemisphere or cerebellum (15). One study included in this meta-analysis demonstrated that adults with RD have reduced activation of the left middle and inferior temporal regions during reading tasks (16). The same authors subsequently showed using voxel-based morphometry that these same regions show cortical structural disorganization (17), providing a link between functional and structural differences of the left hemisphere in individuals with RD.
Understanding the genetic basis of handedness can help define the relationships between handedness, language, cerebral asymmetry and neurodevelopmental disorders. Handedness is a heritable trait, with additive genetic effects accounting for about a quarter of the variance (9,18). Molecular studies are consistent with a polygenic model of handedness. Linkage analyses have identified several loci including 2p12-q11 (19,20), 10q26 (21) and 12q21-23 (22). To date, only two specific genes have been suggested as candidates for handedness. The imprinted gene LRRTM1 in 2p12-q11 has been associated with handedness and schizophrenia (23), and a candidate gene approach identified the X-linked androgen receptor (24). However, a recent genome-wide association study (GWAS) (25) found no single nucleotide polymorphisms (SNPs) were associated with handedness at P-values below 5 × 10−8, the standard genome-wide threshold for significance.
Here we report a GWAS for a quantitative measure of relative hand skill (peg-board task). The most highly associated SNP is in an intron of PCSK6, a gene that encodes a protein involved in left-right axis determination (26). We then replicate this association in two other independent samples with RD reaching overall genome-wide significance.
RESULTS
We genotyped 197 unrelated individuals with RD (stage 1) on the Illumina 550k SNP array and used imputation to test over 2 million SNPs for association with a quantitative measure of relative hand skill (peg-board task) (27). The samples were selected as part of an ongoing study of dyslexia from a large sample of families, each with at least two siblings who show symptoms of dyslexia (28,29). The peg-board task measures the time taken by the subjects to move a row of 10 pegs from one location to another with the left hand (L) and right hand (R) separately (30). From these data, we derived the measure PegQ [2(L − R)/(L + R)], which adjusts for overall differences in hand skill between subjects (19). This task produces an approximately normally distributed variable with a positive mean (Supplementary Material, Fig. S1); a positive PegQ indicates superior relative right-hand skill, and a negative PegQ indicates superior relative left-hand skill.
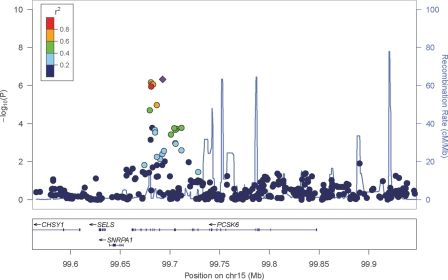
After the initial filtering and quality control stages, we tested 192 individuals for association with PegQ. There was no evidence of population structure and the genomic control statistic (λ) was 1.0031. No SNPs gave P-values below 5 × 10−8 in stage 1 (Supplementary Material, Table S1, Figs S2 and S3). The strongest association signal in the genome for directly genotyped SNPs was with rs9806256 (P = 1.1 × 10−6), while for imputed SNPs peak association was observed with rs11855415 (P = 4.7 × 10−7). Both SNPs are located within a cluster of five highly correlated SNPs, spanning 12 kb from introns 14–18 of PCSK6 (proprotein convertase subtilisin/kexin type 6, also known as PACE4, Fig. 1) on chromosome 15q26. Individuals with the minor (derived) allele have significantly greater relative right-hand skill compared with those carrying the major (ancestral) allele. The mean effect size of each copy of the minor allele is 0.60 standard deviations (SD) to the positive (right-handed) end of the PegQ distribution (see ‘β’ in Table 1).
Figure 1.
Visualization of P-values at the PCSK6 locus. Negative log10 of the P-values for all genotyped and imputed SNPs around PCSK6 plotted in LocusZoom (53). Linkage disequilibrium (r2) with the most highly associated SNP, rs11855415 (purple diamond), is calculated based on the HapMap phase II CEU population and is shown by the colour of the SNPs. The recombination rate is shown by the blue line and the locations of genes in this locus are shown in the panel below the plot.
Table 1.
Association analysis results for the two markers in PCSK6 in individuals with RD
| Marker | Stage 1 |
Stage 2 |
Stage 3 |
Meta-analysis | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | P-value | βa | SEb | n | P-value | βa | SE | n | P-value | βa | SE | n | P-value | βa | β 95Lc | β 95Uc | |
| rs11855415 | 191 | 4.7 × 10−7 | 0.60 | 0.12 | 368 | 0.033 | 0.19 | 0.09 | 185 | 0.0025 | 0.36 | 0.12 | 744 | 1.99 × 10−8 | 0.35 | 0.23 | 0.47 |
| rs9806256 | 192 | 1.1 × 10−6 | 0.56 | 0.12 | 364 | 0.18 | 0.12 | 0.09 | 182 | 0.00067 | 0.40 | 0.12 | 738 | 2.34 × 10−7 | 0.31 | 0.19 | 0.43 |
aThe mean effect size of each copy of the minor allele measured in standard deviations.
bStandard error.
c95% confidence intervals for β (L, lower, U, upper).
PCSK6 is known to play a key role in regulating left-right axis specification (26), making it a highly attractive candidate gene for involvement in handedness. We therefore followed up the initial association by genotyping rs11855415 and rs9806256 in an independent sample of individuals with RD who had also performed the same peg-board task (stage 2, n = 376). The results showed the same trend for increased right-hand skill in carriers of the minor allele; the association was nominally significant with rs11855415 (P = 0.033, β = 0.19, Table 1), but did not meet significance for rs9806256. Next, we genotyped these same two SNPs in individuals from the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort who have performed a similar peg-board task, and have been characterized by similar reading measures to the two previous stages. We selected individuals (stage 3, n = 197) to closely match the ascertainment criteria for RD used in stages 1 and 2. The analyses showed a significant increase in relative right-hand skill associated with the minor allele of both rs11855415 (P = 0.0025, β = 0.36, Table 1) and rs9806256 (P = 0.00067, β = 0.40, Table 1). Meta-analysis of all three stages yielded a P-value of 2.0 × 10−8 (Table 1, Supplementary Material, Fig. S4) for rs11855415, exceeding accepted genome-wide thresholds for significance. The data indicated that in the total sample of RD individuals, the average effect size of the minor allele is 0.35 SD towards the positive end of the PegQ distribution.
Then we investigated whether the effect of rs11855415 that we had observed in people with RD could be detected in the general population. We analyzed all the available ALSPAC children with a PegQ score (n = 2666), excluding those with neurodevelopmental disorders and performance IQ < 85 (see Supplementary Methods). The PegQ distribution for this sample compared with the ALSPAC children with RD in stage 3 shows neither a significant difference in the mean (P = 0.54), consistent with a previous study (9), nor a difference in the likelihood of being left- or right-handed (P = 0.69). In this general population sample, we did not detect significant association between rs11855415 and relative hand skill (P = 0.25, β = −0.038). However, the distribution of PegQ for each genotype in this general population sample shows that the PegQ scores for carriers of the minor allele appear clustered tightly around the mean (Supplementary Material, Fig. S5). This suggests that carriers of the minor allele in the general population cohort show reduced variability in relative hand skill, centered on the mean, in contrast to the RD sample which shows an increase in relative right-hand skill. To test this, we ran the association analysis for rs11855415 using the absolute value of the standardized PegQ score as a quantitative trait (Supplementary Material, Fig. S6). This compared scores closer to the mean against scores at both tails of the PegQ distribution. This analysis showed a nominally significant trend towards the mean of the PegQ distribution for carriers of the minor allele (P = 0.0020). Future studies will establish whether a similar pattern of findings is seen in other population cohorts.
DISCUSSION
We have performed the first GWAS for a quantitative measure of human handedness, as assessed by the peg-board task (30). The most highly associated SNP, rs11855415, is within an intron of PCSK6. The minor allele of this SNP confers increased relative right-hand skill. This association replicated in two independent groups with RD and is the first SNP for handedness to be identified at a genome-wide significant level. This effect was specific for people with RD, while the minor allele of rs11855415 showed a significant trend towards reduced laterality of hand skill in the general population.
PCSK6 encodes a proprotein convertase that processes proteins such as NODAL from their latent precursors into biologically active products (31). NODAL is a TGFβ-related protein that specifies both the anteroposterior and left–right axes (32). In a proportion of mouse embryos null for Pcsk6, the normally asymmetrically expressed Nodal, Lefty and Pitx2 mRNAs are bilaterally expressed (26). Some embryos subsequently display laterality defects such as situs ambiguous (26). PCSK6 is widely expressed in humans, with particularly high expression in the liver, corpus callosum (the band of axon fibers that connect the left and right hemispheres) and spinal cord (33). Mutations in other NODAL pathway members have been linked to lateralization defects such as holoprosencephaly, which is an incomplete separation of the forebrain into discrete hemispheres (34,35).
The observation that the increase in relative right-hand skill associated with rs11855415 in PSCK6 is specific to RD suggests there may be epistatic interaction between PCSK6 and RD susceptibility genes. There have been a number of candidate genes proposed for RD, including DCDC2 (36,37), DYX1C1 (38), KIAA0319 (39,40) and ROBO1 (41). These genes are all involved in neuronal migration or axon guidance (42). It may be that the functional change in PCSK6 subtly alters the initial left–right patterning of the early embryo, and this has a downstream effect during neuronal migration on the development of cerebral asymmetry. Future studies will seek to explain the difference between RD and general population subjects.
The genome-wide significance of SNP associations in PCSK6, coupled with the role of this gene in a known biological pathway for left–right asymmetry, make it a strong candidate for involvement in cerebral asymmetry and thus handedness. Further work will seek to identify the functional genetic change in PCSK6, along with its mechanism of action. Our results are consistent with the emerging view that handedness is determined by multiple interacting genetic and environmental factors. Interestingly, PCSK6 has been found to be a direct target of FOXP2 (43), while PCSK6 may interact with SRPX2 (44). Mutation in either FOXP2 or SRPX2 may lead to severe forms of language or speech impairment. It would therefore be interesting to investigate the effect of PCSK6 on handedness in individuals with other neurodevelopmental disorders such as SLI. Understanding the genes that modify the effects of PCSK6 in RD individuals will offer us a significant insight into the etiology of both cerebral asymmetry and dyslexia.
MATERIALS AND METHODS
Study participants
Individuals were initially selected from our collection of families with RD (45) for severity of phenotype to undergo a GWAS for RD. Genotype data were generated for 197 individuals (stage 1). Families were recruited from the Dyslexia Clinic of the Royal Berkshire Hospital and range in age from 6 to 25 years. A second sample of 376 unrelated individuals with RD (stage 2) was recruited from either the Dyslexia Research Centre clinics in Oxford and Reading or the Aston Dyslexia and Development Clinic in Birmingham. The majority of these individuals are between 8 and 18 years old. The third sample of unrelated individuals came from the Avon Longitudinal Study of Parents and Children (ALSPAC). For ALSPAC, all pregnant women in the Bristol, UK, area with a then expected delivery date between 1 April 1991 and 31 December 1992 were approached for participation in the study that has today resulted in a general population cohort of about 14 000 children (46). For the present study, we assigned individuals from the ALSPAC cohort into a sub-group with RD (n = 197, stage 3), and a sub-group representing the general population without a neurodevelopmental disorder (n = 2667). Complete details of the ascertainment criteria for the RD and general population sub-groups are given as Supplementary Material.
Relative hand skill phenotype
The Pegboard test (30) was administered to the individuals who were analyzed in stages 1 and 2. The test involved the measurement of the time taken by the subjects to move, with each hand, a row of 10 pegs on a board from one location to another. This test was repeated five times for each hand in all the subjects. From these data, we derived the measure of relative hand skill (PegQ) for each subject, which was calculated as the difference between the average times for the left hand (L) and the right hand (R), L − R, divided by the average time for both hands combined, (L + R)/2, to correct for the overall difference in hand skill between the subjects (19). It has previously been shown that although overall hand skill improves with age (i.e. the peg test is performed in a shorter time with both hands), relative hand proficiency measured in this way does not change significantly between the ages of 3 and 15 (47). Therefore, although there is a large range of ages in our study (6–25 years), we do not expect this to significantly affect our results. The distribution of PegQ in stage 1 had a mean of 0.0796 (SD = 0.099), and in stage 2 it was 0.061 (SD = 0.091). The mean PegQ score is similar to Francks’ (19) sample, which had a mean of 0.072. The PegQ distribution is unimodal, continuous and approximately normal, making it suitable as a quantitative phenotype for a GWAS (Supplementary Material, Fig. S1).
A peg test similar to that used in stages 1 and 2 was performed in the ALSPAC sample as part of a battery of manual dexterity tests known as Movement ABC (48). In this case, there were 12 pegs on the table that the child picked up one-at-a-time and placed in a peg board. After initial practice, children performed the test once (due to time constraints) with each hand. The mean PegQ was slightly different in stage 3 at 0.113 (SD = 0.199), while the distribution remains unimodal, continuous and approximately normal. One outlier was removed from further analysis (PegQ = 1.50).
Genotyping and quality control
The 197 individuals (stage 1) were genotyped on the Illumina® 550k Platform according to the manufacturer's instructions. One individual was excluded based on having a call rate <95%. We filtered out all SNPs with either minor allele frequency (MAF) < 0.05 (due to the small sample size), or successful genotype call rate < 0.95, or violation of Hardy–Weinberg equilibrium (P < 5.7 × 10−7; in autosomes). In total, 477 347 SNPs passed quality control. Principal component analysis (PCA) to test for individuals with divergent ancestry compared with CEU HapMap3 samples was performed as detailed previously (49), and as a result four further individuals were excluded (Supplementary Material, Fig. S7). In order to investigate population structure within our sample, we repeated our PCA without the HapMap3 samples. We tested for association between PegQ and the first two principal components (PC, adjusting for age and sex) in a linear regression model. There was no evidence of association with either component (P = 0.244 for PC1 and P = 0.184 for PC2). We thus did not adjust for these components in our subsequent GWAS association analysis.
The 376 unrelated cases (stage 2) were genotyped for the two SNPs, rs11855415 and rs9806256, as part of a larger study using iPLEX assays from Sequenom® (San Diego, CA, USA), according to the manufacturer's instructions. We also re-genotyped the original 197 individuals (stage 1) with these assays, and found 99% concordance for the directly genotyped rs9806256 and 98% concordance for the imputed SNP rs11855415. The Hardy–Weinberg equilibrium P-values for the SNPs were rs11855415 P = 0.243 and rs9806256 P = 0.820. The integrated genotyping system (50) was used to store genotypes.
The same two SNPs were genotyped in the ALSPAC children cohort by KBiosciences, Hertfordshire, UK (www.kbioscience.co.uk date last accessed 12/11/10), using a fluorescence-based competitive allele-specific PCR (KASPar) assay. The Hardy–Weinberg P-values for the two SNPs were rs11855415 P = 0.018 and rs9806256 P = 0.248.
Imputation
We used IMPUTE [v2.0.5 for autosomes, (27) and v1.0.0 for the X chromosome, (51)] to estimate the genotypes of SNPs not directly genotyped on the Illumina 550k platform. This involved combining information from (i) the directly genotyped SNPs, (ii) a densely genotyped reference panel (60 CEPH founders from HapMap Phase II, build 35) and (iii) a fine-scale recombination map, to infer the missing genotypes. Imputed SNPs were filtered out if they had an MAF < 0.05 or proper info <0.5, allowing for us to test a further 1 709 209 SNPs for association.
Statistical analysis
Directly genotyped and imputed SNPs were tested for association with the quantitative measure PegQ under an additive model using SNPTEST (v1.1.4, www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html date last accessed 12/11/10) simultaneously specifying sex and age as covariates. For the X chromosome, analysis in stage 1 was conducted by running the association test separately for the two sexes in SNPTEST and combined the results using GWAMA [v1.4, (52)], which performs fixed effects meta-analysis using inverse-variance weighting of the parameter estimates. The results for the three stages were then meta-analyzed using GWAMA (52). The values of PegQ were standardized in all stages separately to give a mean of 0, and a SD of 1. For the two SNPs genotyped in ALSPAC in the general population, the absolute value of the standardized PegQ score was also run as a quantitative phenotype.
To rule out any underlying confounding effects, we tested rs11855415 and rs9806256 for association with measures of hand motor skill, IQ and reading ability (CCI, CCN, OLSON, READ, SPELL, SPOON, defined in Supplementary Material), in each of the three stages as well as in the general population sample whenever data were available, and found no significant association (Supplementary Material, Table S2).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This research was specifically funded by the Wellcome Trust (076566/Z/05/Z); (075491/Z/04), the Medical Research Council (G0800523/86473) and the EU (Neurodys, 018696). Core support for ALSPAC is provided by the UK Medical Research Council (Grant ref: 74882), the Wellcome Trust (Grant ref: 076467) and the University of Bristol. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
ACKNOWLEDGEMENTS
We are extremely grateful to all the families who took part in this study. We thank Janet Walter for collecting many of the samples and carrying out many of the psychometric tests in stages 1 and 2. For the ALSPAC study, we wish to thank the midwives for their help in recruiting the families, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. We thank Laura Miller and Bryan Howie for technical assistance, and Dorothy Bishop, Simon Fisher and Jonathan Flint for their constructive comments regarding the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Gilbert A.N., Wysocki C.J. Hand preference and age in the United States. Neuropsychologia. 1992;30:601–608. doi: 10.1016/0028-3932(92)90065-t. doi:10.1016/0028-3932(92)90065-T. [DOI] [PubMed] [Google Scholar]
- 2.Marchant L.F., McGrew W.C. Human handedness: an ethological perspective. Hum. Evol. 1998;13:221–228. doi:10.1007/BF02436506. [Google Scholar]
- 3.Raymond M., Pontier D. Is there geographical variation in human handedness? Laterality. 2004;9:35–51. doi: 10.1080/13576500244000274. [DOI] [PubMed] [Google Scholar]
- 4.Broca P. Remarques sur le siége de la faculté du language articulé, suivies d'une observation d'aphémie (perte de la parole) Bull. Soc. Anatom. 1861;6:330–357. [Google Scholar]
- 5.Corballis M.C. The Lopsided Ape: Evolution of the Generative Mind. New York: Oxford University Press; 1991. [Google Scholar]
- 6.Orton S.T. Word-blindness in School Children and Other Papers on Strephosymbolia (Specific Language Disability-Dyslexia) 1925–1946. Pomfret, Conn: Orton Society; 1966. [Google Scholar]
- 7.Pennington B.F., Bishop D.V. Relations among speech, language, and reading disorders. Annu. Rev. Psychol. 2009;60:283–306. doi: 10.1146/annurev.psych.60.110707.163548. doi:10.1146/annurev.psych.60.110707.163548. [DOI] [PubMed] [Google Scholar]
- 8.Bishop D.V.M. Handedness and Developmental Disorder. Lavenham: Mac Keith Press; 1990. [Google Scholar]
- 9.Francks C., Fisher S.E., Marlow A.J., MacPhie I.L., Taylor K.E., Richardson A.J., Stein J.F., Monaco A.P. Familial and genetic effects on motor coordination, laterality, and reading-related cognition. Am. J. Psychiatry. 2003;160:1970–1977. doi: 10.1176/appi.ajp.160.11.1970. doi:10.1176/appi.ajp.160.11.1970. [DOI] [PubMed] [Google Scholar]
- 10.Knecht S., Drager B., Deppe M., Bobe L., Lohmann H., Floel A., Ringelstein E.B., Henningsen H. Handedness and hemispheric language dominance in healthy humans. Brain. 2000;123:2512–2518. doi: 10.1093/brain/123.12.2512. doi:10.1093/brain/123.12.2512. [DOI] [PubMed] [Google Scholar]
- 11.Leonard C.M., Eckert M.A. Asymmetry and dyslexia. Dev. Neuropsychol. 2008;33:663–681. doi: 10.1080/87565640802418597. doi:10.1080/87565640802418597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friederici A.D. The neural basis of language development and its impairment. Neuron. 2006;52:941–952. doi: 10.1016/j.neuron.2006.12.002. doi:10.1016/j.neuron.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Galaburda A.M., Sherman G.F., Rosen G.D., Aboitiz F., Geschwind N. Developmental dyslexia: four consecutive patients with cortical anomalies. Ann. Neurol. 1985;18:222–233. doi: 10.1002/ana.410180210. doi:10.1002/ana.410180210. [DOI] [PubMed] [Google Scholar]
- 14.Galaburda A.M. Ordinary and extraordinary brain development: anatomical variation in developmental dyslexia. Ann. Dyslexia. 1989;39:65–80. doi: 10.1007/BF02656901. doi:10.1007/BF02656901. [DOI] [PubMed] [Google Scholar]
- 15.Richlan F., Kronbichler M., Wimmer H. Functional abnormalities in the dyslexic brain: a quantitative meta-analysis of neuroimaging studies. Hum. Brain Mapp. 2009;30:3299–3308. doi: 10.1002/hbm.20752. doi:10.1002/hbm.20752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paulesu E., Demonet J.F., Fazio F., McCrory E., Chanoine V., Brunswick N., Cappa S.F., Cossu G., Habib M., Frith C.D., et al. Dyslexia: cultural diversity and biological unity. Science. 2001;291:2165–2167. doi: 10.1126/science.1057179. doi:10.1126/science.1057179. [DOI] [PubMed] [Google Scholar]
- 17.Silani G., Frith U., Demonet J.F., Fazio F., Perani D., Price C., Frith C.D., Paulesu E. Brain abnormalities underlying altered activation in dyslexia: a voxel based morphometry study. Brain. 2005;128:2453–2461. doi: 10.1093/brain/awh579. doi:10.1093/brain/awh579. [DOI] [PubMed] [Google Scholar]
- 18.Medland S.E., Duffy D.L., Wright M.J., Geffen G.M., Hay D.A., Levy F., Van-Beijsterveldt C.E.M., Willemsen G., Townsend G.C., White V., et al. Genetic influences on handedness: data from 25,732 Australian and Dutch twin families. Neuropsychologia. 2009;47:330–337. doi: 10.1016/j.neuropsychologia.2008.09.005. doi:10.1016/j.neuropsychologia.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francks C., Fisher S.E., MacPhie I.L., Richardson A.J., Marlow A.J., Stein J.F., Monaco A.P. A genomewide linkage screen for relative hand skill in sibling pairs. Am. J. Hum. Genet. 2002;70:800–805. doi: 10.1086/339249. doi:10.1086/339249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francks C., DeLisi L.E., Fisher S.E., Laval S.H., Rue J.E., Stein J.F., Monaco A.P. Confirmatory evidence for linkage of relative hand skill to 2p12-q11. Am. J. Hum. Genet. 2003;72:499–502. doi: 10.1086/367548. doi:10.1086/367548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Agtmael T., Forrest S.M., Williamson R. Parametric and non-parametric linkage analysis of several candidate regions for genes for human handedness. Eur. J. Hum. Genet. 2002;10:623–630. doi: 10.1038/sj.ejhg.5200851. doi:10.1038/sj.ejhg.5200851. [DOI] [PubMed] [Google Scholar]
- 22.Warren D.M., Stern M., Duggirala R., Dyer T.D., Almasy L. Heritability and linkage analysis of hand, foot, and eye preference in Mexican Americans. Laterality. 2006;11:508–524. doi: 10.1080/13576500600761056. [DOI] [PubMed] [Google Scholar]
- 23.Francks C., Maegawa S., Lauren J., Abrahams B.S., Velayos-Baeza A., Medland S.E., Colella S., Groszer M., McAuley E.Z., Caffrey T.M., et al. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol. Psychiatry. 2007;12:1129–1139. doi: 10.1038/sj.mp.4002053. doi:10.1038/sj.mp.4002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medland S.E., Duffy D.L., Spurdle A.B., Wright M.J., Geffen G.M., Montgomery G.W., Martin N.G. Opposite effects of androgen receptor CAG repeat length on increased risk of left-handedness in males and females. Behav. Genet. 2005;35:735–744. doi: 10.1007/s10519-005-6187-3. doi:10.1007/s10519-005-6187-3. [DOI] [PubMed] [Google Scholar]
- 25.Eriksson N., Macpherson J.M., Tung J.Y., Hon L.S., Naughton B., Saxonov S., Avey L., Wojcicki A., Pe'er I., Mountain J. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6:e1000993. doi: 10.1371/journal.pgen.1000993. doi:10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Constam D.B., Robertson E.J. SPC4/PACE4 regulates a TGFbeta signaling network during axis formation. Genes Dev. 2000;14:1146–1155. [PMC free article] [PubMed] [Google Scholar]
- 27.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. doi:10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher S.E., Marlow A.J., Lamb J., Maestrini E., Williams D.F., Richardson A.J., Weeks D.E., Stein J.F., Monaco A.P. A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am. J. Hum. Genet. 1999;64:146–156. doi: 10.1086/302190. doi:10.1086/302190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marlow A.J., Fisher S.E., Richardson A.J., Francks C., Talcott J.B., Monaco A.P., Stein J.F., Cardon L.R. Investigation of quantitative measures related to reading disability in a large sample of sib-pairs from the UK. Behav. Genet. 2001;31:219–230. doi: 10.1023/a:1010209629021. doi:10.1023/A:1010209629021. [DOI] [PubMed] [Google Scholar]
- 30.Annett M. Left, Right, Hand and Brain: The Right Shift Theory. London: Psychology Press; 1985. [Google Scholar]
- 31.Kiefer M.C., Tucker J.E., Joh R., Landsberg K.E., Saltman D., Barr P.J. Identification of a second human subtilisin-like protease gene in the fes/fps region of chromosome 15. DNA Cell Biol. 1991;10:757–769. doi: 10.1089/dna.1991.10.757. doi:10.1089/dna.1991.10.757. [DOI] [PubMed] [Google Scholar]
- 32.Beck S., Le Good J.A., Guzman M., Ben Haim N., Roy K., Beermann F., Constam D.B. Extraembryonic proteases regulate Nodal signalling during gastrulation. Nat. Cell Biol. 2002;4:981–985. doi: 10.1038/ncb890. doi:10.1038/ncb890. [DOI] [PubMed] [Google Scholar]
- 33.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. doi:10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 34.Roessler E., Ouspenskaia M.V., Karkera J.D., Velez J.I., Kantipong A., Lacbawan F., Bowers P., Belmont J.W., Towbin J.A., Goldmuntz E., et al. Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am. J. Hum. Genet. 2008;83:18–29. doi: 10.1016/j.ajhg.2008.05.012. doi:10.1016/j.ajhg.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohapatra B., Casey B., Li H., Ho-Dawson T., Smith L., Fernbach S.D., Molinari L., Niesh S.R., Jefferies J.L., Craigen W.J., et al. Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum. Mol. Genet. 2009;18:861–871. doi: 10.1093/hmg/ddn411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng H., Smith S.D., Hager K., Held M., Liu J., Olson R.K., Pennington B.F., DeFries J.C., Gelernter J., O'Reilly-Pol T., et al. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc. Natl Acad. Sci. USA. 2005;102:17053–17058. doi: 10.1073/pnas.0508591102. doi:10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumacher J., Anthoni H., Dahdouh F., Konig I.R., Hillmer A.M., Kluck N., Manthey M., Plume E., Warnke A., Remschmidt H., et al. Strong genetic evidence of DCDC2 as a susceptibility gene for dyslexia. Am. J. Hum. Genet. 2006;78:52–62. doi: 10.1086/498992. doi:10.1086/498992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taipale M., Kaminen N., Nopola-Hemmi J., Haltia T., Myllyluoma B., Lyytinen H., Muller K., Kaaranen M., Lindsberg P.J., Hannula-Jouppi K., et al. A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc. Natl Acad. Sci. USA. 2003;100:11553–11558. doi: 10.1073/pnas.1833911100. doi:10.1073/pnas.1833911100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Francks C., Paracchini S., Smith S.D., Richardson A.J., Scerri T.S., Cardon L.R., Marlow A.J., MacPhie I.L., Walter J., Pennington B.F., et al. A 77-kilobase region of chromosome 6p22.2 is associated with dyslexia in families from the United Kingdom and from the United States. Am. J. Hum. Genet. 2004;75:1046–1058. doi: 10.1086/426404. doi:10.1086/426404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cope N., Harold D., Hill G., Moskvina V., Stevenson J., Holmans P., Owen M.J., O'Donovan M.C., Williams J. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am. J. Hum. Genet. 2005;76:581–591. doi: 10.1086/429131. doi:10.1086/429131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hannula-Jouppi K., Kaminen-Ahola N., Taipale M., Eklund R., Nopola-Hemmi J., Kaariainen H., Kere J. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet. 2005;1:e50. doi: 10.1371/journal.pgen.0010050. doi:10.1371/journal.pgen.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galaburda A.M., LoTurco J., Ramus F., Fitch R.H., Rosen G.D. From genes to behavior in developmental dyslexia. Nat. Neurosci. 2006;9:1213–1217. doi: 10.1038/nn1772. doi:10.1038/nn1772. [DOI] [PubMed] [Google Scholar]
- 43.Vernes S.C., Spiteri E., Nicod J., Groszer M., Taylor J.M., Davies K.E., Geschwind D.H., Fisher S.E. High-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am. J. Hum. Genet. 2007;81:1232–1250. doi: 10.1086/522238. doi:10.1086/522238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Royer-Zemmour B., Ponsole-Lenfant M., Gara H., Roll P., Leveque C., Massacrier A., Ferracci G., Cillario J., Robaglia-Schlupp A., Vincentelli R., et al. Epileptic and developmental disorders of the speech cortex: ligand/receptor interaction of wild-type and mutant SRPX2 with the plasminogen activator receptor uPAR. Hum. Mol. Genet. 2008;17:3617–3630. doi: 10.1093/hmg/ddn256. doi:10.1093/hmg/ddn256. [DOI] [PubMed] [Google Scholar]
- 45.Fisher S.E., Francks C., Marlow A.J., MacPhie I.L., Newbury D.F., Cardon L.R., Ishikawa-Brush Y., Richardson A.J., Talcott J.B., Gayan J., et al. Independent genome-wide scans identify a chromosome 18 quantitative-trait locus influencing dyslexia. Nat. Genet. 2002;30:86–91. doi: 10.1038/ng792. doi:10.1038/ng792. [DOI] [PubMed] [Google Scholar]
- 46.Golding J., Pembrey M., Jones R. ALSPAC—the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatr. Perinat. Epidemiol. 2001;15:74–87. doi: 10.1046/j.1365-3016.2001.00325.x. doi:10.1046/j.1365-3016.2001.00325.x. [DOI] [PubMed] [Google Scholar]
- 47.Annett M. The growth of manual preference and speed. Br. J. Psychol. 1970;61:545–558. doi: 10.1111/j.2044-8295.1970.tb01274.x. [DOI] [PubMed] [Google Scholar]
- 48.Henderson S.E., Sugden D.A. Movement Assessment Battery for Children Manual. Sidcup: The Psychological Corporation; 1992. [Google Scholar]
- 49.Anderson C.A., Pettersson F.H., Clarke G.M., Cardon L.R., Morris A.P., Zondervan K.T. Data quality control in genetic case-control association studies. Nat. Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. doi:10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fiddy S., Cattermole D., Xie D., Duan X.Y., Mott R. An integrated system for genetic analysis. BMC Bioinformatics. 2006;7:210. doi: 10.1186/1471-2105-7-210. doi:10.1186/1471-2105-7-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. doi:10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 52.Magi R., Morris A.P. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. doi:10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. doi:10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.