Abstract
Common chromosomal fragile sites FRA3B and FRA16D are frequent sites of DNA instability in cancer, but their contribution to cancer cell biology is not yet understood. Genes that span these sites (FHIT and WWOX, respectively) are often perturbed (either increased or decreased) in cancer cells and both are able to suppress tumour growth. While WWOX has some tumour suppressor characteristics, its normal role and functional contribution to cancer has not been fully determined. We find that a significant proportion of Drosophila Wwox interactors identified by proteomics and microarray analyses have roles in aerobic metabolism. Functional relationships between Wwox and either CG6439/isocitrate dehydrogenase (Idh) or Cu–Zn superoxide dismutase (Sod) were confirmed by genetic interactions. In addition, altered levels of Wwox resulted in altered levels of endogenous reactive oxygen species. Wwox (like FHIT) contributes to pathways involving aerobic metabolism and oxidative stress, providing an explanation for the ‘non-classical tumour suppressor’ behaviour of WWOX. Fragile sites, and the genes that span them, are therefore part of a protective response mechanism to oxidative stress and likely contributors to the differences seen in aerobic glycolysis (Warburg effect) in cancer cells.
INTRODUCTION
Common chromosomal fragile sites have been found to coincide with the location of various forms of DNA instability (including homozygous deletions and translocations) in cancer cells (1–3). Common chromosomal fragile sites are found in all individuals and appear to be regions of the genome that are particularly sensitive to environmental damage—they are susceptible to agents in cigarette smoke (4) and their expression levels in cells are modified by dietary factors such as folate level and even caffeine and ethanol (1,5). When induced to appear in cells, >70 different common chromosomal fragile sites exhibit different frequencies of cytogenetic appearance such that FRA3B (located on human chromosome 3) is the most readily observed, followed by FRA16D (on human chromosome 16) and then others distributed over most of the human chromosomes at specific locations that are reproducibly observed (2). The incidence of DNA instability in cancer follows a similar hierarchy (i.e. FRA3B > FRA16D > others) suggesting that cytogenetic ‘fragility’ and DNA instability in cancer are directly related (6,7). Recently, a screen for small homozygous deletions in 746 cancer cell lines detected chromosomal fragile sites FRA3B and FRA16D as the two most frequent sites of this form of mutation (3).
Common chromosomal fragile sites themselves appear to span hundreds of kilobases of DNA and are typically located within lengthy genes (e.g. FHIT 1.5 Mb, WWOX 1.1 Mb, parkin 1.36 Mb; 8). In the case of FRA16D, the fragile site DNA region of ∼260 kb is completely contained within a massive 780 kb intron of the WWOX gene (6,9,10). Just why these genes should be so large (the WWOX spliced mRNA product is only ∼1.1 kb) and contain fragile sites that are susceptible to environmental damage is not known, but is very likely to reflect functional significance since mouse orthologues of FHIT and WWOX also span common chromosomal fragile sites (8). Efforts to find relationships between the different fragile site genes have been reported, but the functional significance of such associations is unclear (8,11,12).
Attention has, therefore, been focussed on the normal functions of common fragile site genes, as it is the perturbation of these functions that is most likely to contribute to cancer. Both FHIT (spanning FRA3B) and WWOX (spanning FRA16D) act to inhibit tumour growth when introduced into cancer cells lacking their expression (13,14); however, they do not exhibit all of the typical characteristics of tumour suppressors. For example, point mutations are extremely rare in WWOX and the protein is quite commonly still present in cancer cells (even sometimes increased in expression; 15–18) indicating absence of the hallmark ‘second-hit’ required to fulfil the Knudsen hypothesis characteristic of tumour suppressors. This ‘non-classical’ tumour suppressor function for WWOX is further evident in analysis of rodent mutants for the WWOX gene. Some (but not all) lines of loss of function WWOX mutant mice have higher incidence of tumours, however tumours from heterozygous mutant mice still express WWOX (19–23). A spontaneous Wwox mutant rat has also been described, but it does not exhibit higher incidence of tumours (24,25). Rodent Wwox mutants typically exhibit early death, which may preclude development of some tumours, in addition to metabolic disorders, but they are otherwise surprisingly different in their phenotypes (19–25). This may reflect the distinct nature of their mutations or their different genetic backgrounds. While these rodent studies have provided some insight into the biological role for WWOX, there is still clearly a need to understand the molecular processes and pathways in which WWOX participates; particularly how WWOX contributes to metabolism and how this altered metabolism can contribute, at least in certain circumstances, to a greater incidence of cancer.
Genetic studies into the function of Wwox have been initiated in Drosophila. Drosophila is ideal for this particular purpose as evident from its unique contributions to understanding human disease pathogenesis pathways where subtle but significant roles of cancer genes and intricate signal transduction pathways have been identified (26,27). We have previously identified the unique Drosophila WWOX orthologue, Wwox (CG7221; 49% identity with human WWOX) and disrupted the Wwox gene by homologous recombination (28). Insertion of stop codons and an altered reading frame at the beginning of the endogenous gene was effective in causing loss of Wwox function by rendering the resultant mRNA from the mutated gene incapable of being translated into the Wwox protein (28). Initially, Drosophila homozygous for this mutation were reported as being sensitive to ionizing radiation. However, further analysis indicated that background mutations, rather than the targeted Wwox mutations, were responsible for this phenotype (29).
The lack of phenotype in Drosophila has precluded direct genetic analysis of the Wwox function. We have, therefore, undertaken a distinct approach to understanding the normal biological functions of Wwox by undertaking genetic and biochemical screens to identify the pathways to which Wwox contributes. Using the combination of proteomic and microarray screens to identify interactive partners and genetic analysis to confirm the functionality of such interactions, we have found that Wwox participates in common pathways with CG6439/isocitrate dehydrogenase (Idh) and Cu–Zn superoxide dismutase (Sod). These interactions indicate a contribution of Wwox to aerobic metabolism and the regulation of reactive oxygen species (ROS) levels within cells.
RESULTS
Biochemical responses to altered Wwox levels
Two-dimensional differential in-gel electrophoresis (2D-DIGE) was used to identify quantitative and qualitative changes in proteins between Drosophila lines with either decreased or increased levels of Wwox. To examine the effect of decreased levels of Wwox, adult flies mutant for Wwox were compared with wild-type (w1118). We used two independent alleles of Wwox for these analyses; Wwox1, a null mutation generated by homologous recombination, and Wwoxf04545, which carries a pBac insertion in exon 2 (28,29). To investigate the effects of increased levels of Wwox, the protein profiles of adult flies carrying a UAS-Wwox transgene together with the da-GAL4 ubiquitous driver (da > Wwox) were compared with adult flies carrying the da-GAL4 driver alone (da>+). A common set of 16 protein spots were identified that showed significant changes in abundance in each of the Wwox mutants, while a further 16 were identified that exhibited significant changes when Wwox was ectopically over-expressed (summarized in Fig. 1 and Supplementary Material, Table S1). Mass spectrometry analysis of peptides was successful in identifying 13 of the 16 candidate proteins identified in Wwox mutants in addition to all of those identified when Wwox was ectopically over-expressed (Supplementary Material, Table S1).
Figure 1.
Proteomic analysis of altered Wwox expression. 2D-DIGE protein spots that exhibited significant changes in abundance either in both of the independent Wwox mutants (Wwox1 and Wwoxf04545) or with ubiquitous ectopic Wwox expression in 0- to 1-day adult Drosophila (see also Supplementary Material, Table S1). Spots 19 and 20 are superoxide dismutase (Sod) isoforms.
In a complementary approach, microarray analysis was also performed on Wwox mutants during early embryonic development of Drosophila. Embryos (4–8 h) from each of the two independent mutant lines were compared with wild-type, revealing a number of transcripts in common that were altered in response to decreased Wwox function. The microarray data have been deposited on the NCBI database (GEO accession number GSE22689). Verification by qPCR showed seven of these were also significantly altered in adults of each of the Wwox mutants as well as flies ectopically over-expressing Wwox (Table 1).
Table 1.
Candidate Wwox interactors identified from proteomic and microarray analyses
| Candidate Wwox Interactor | Spot # | Protein abundance |
Transcript abundance (measured by qPCR) | Molecular function | ||||
|---|---|---|---|---|---|---|---|---|
| w1118 versus Wwox mutants | da>+ versus da>Wwox | w1118 versus Wwox mutants | da>+ versus da>Wwox | |||||
| Wwox1 | Wwoxf04545 | da>Wwox | Wwox1 | Wwoxf04545 | da>Wwox | |||
| Wwox/CG7221 | 1 | n/d | n/d | 1.49 | −2.72 | −17.38 | 172 | Oxidoreductase activity |
| AEROBIC METABOLISM | ||||||||
| TCA cycle | ||||||||
| CG6439 (Idh) | 2 | −1.12 | −1.19 | n/c | – | – | – | Isocitrate dehydrogenase (NAD+) activity |
| CG7998 (Mdh) | 3 | −1.30 | −1.13 | n/c | – | – | – | L-malate dehydrogenase activity |
| Glucose metabolism | ||||||||
| Phosphoglycerate kinase (Pgk)/CG3127 | 4 | n/c | n/c | 1.23 | – | – | – | Phosphoglycerate kinase activity |
| CG7430 | 5 | 1.14 | 1.18 | n/c | – | – | – | Dihydrolipoyl dehydrogenase activity |
| Malic enzyme (Men)/CG10120 | 6 | n/c | n/c | 1.48 | – | – | – | Malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+) activity |
| 7 | n/c | n/c | 1.43 | – | – | – | ||
| Glyceraldehyde-3-phosphate dehydrogenase1 (Gapdh1)/CG12055 | 8 | n/c | n/c | 1.22 | – | – | – | Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) activity |
| CG10638 | – | – | – | – | −2.15 | −1.70 | 2.17 | Aldehyde reductase activity |
| CG10924 (PEPCK) | – | – | – | – | 5.00 | 3.77 | −1.82 | Phosphoenolpyruvate carboxykinase (GTP) activity |
| Ethanol metabolism | ||||||||
| Alcohol dehydrogenase (Adh)/CG3481 | 9 | 1.53 | 1.27 | n/c | – | – | – | Alcohol dehydrogenase (NAD) activity |
| Aldehyde dehydrogenase (Aldh)/CG3752 | 10 | 1.25 | 1.36 | n/c | – | – | – | Aldehyde dehydrogenase (NAD) activity |
| Acetyl Coenzyme A synthase (AcCoAS)/CG9390 | 11 | n/c | n/c | −1.57 | – | – | – | Acetate-CoA ligase activity |
| CG31075 | 12 | 1.14 | 1.23 | n/c | – | – | – | Aldehyde dehydrogenase (NAD) activity |
| 13 | 1.10 | 1.08 | n/c | – | – | – | ||
| Lipid metabolism | ||||||||
| Glycerol-3-phosphate Dehydrogenase (Gpdh)/ CG9042 | 14 | 1.16 | 1.21 | n/c | – | – | – | Glycerol-3-phosphate dehydrogenase (NAD+) activity |
| Fat body protein 1 (Fbp1)/CG17285 | 15 | 2.70 | 2.00 | n/c | – | – | – | Protein transporter activity |
| 16 | n/c | n/c | 3.04 | – | – | – | ||
| 17 | n/c | n/c | 1.60 | – | – | – | ||
| 18 | n/c | n/c | −2.20 | – | – | – | ||
| Oxidation reduction | ||||||||
| Superoxide dismutase (Sod)/CG11793 | 19 | 2.15 | 1.64 | −1.23 | 1.33 | 1.64 | −1.10 | Antioxidant activity; superoxide dismutase activity |
| 20 | −2.23 | −1.53 | n/c | |||||
| CG5590 | 21 | n/c | n/c | −1.37 | – | – | – | Oxidoreductase activity, acting on the CH-OH group of donors |
| OTHER CATEGORIES | ||||||||
| CG7470 | 22 | n/c | n/c | −1.43 | – | – | – | Delta1-pyrroline-5-carboxylate synthetase |
| Suppressor of Profilin 2 (Sop2)/CG8978 | – | – | – | – | −1.08 | 1.13 | −1.23 | Actin binding |
| Hsp60C/CG7235 | 23 | −1.08 | −1.24 | n/c | – | – | – | ATPase activity, coupled |
| CG2852 | 24 | n/c | n/c | 1.31 | – | – | – | Peptidyl-prolyl cis-trans isomerase activity |
| CG11089 | 25 | 1.13 | 1.21 | n/c | – | – | – | IMP cyclohydrolase activity |
| CG14526 | 26 | n/c | n/c | −1.26 | – | – | – | Metalloendopeptidase activity |
| Bancal (bl)/CG13425 | – | – | – | – | 1.10 | 1.01 | −1.29 | mRNA binding; transcription factor binding |
| Prp19/CG5519 | – | – | – | – | 1.47 | 1.78 | −1.52 | Ubiquitin-protein ligase activity |
| Tudor-SN/CG7008 | 27 | n/c | n/c | −1.23 | – | – | – | Transcription coactivator activity |
| CG30152 | – | – | – | – | 1.13 | 1.23 | −1.29 | Unknown |
| CG8193 | 28 | n/c | n/c | 1.22 | – | – | – | Monophenol monooxygenase activity |
Average fold change ratios of protein abundance for 2D-DIGE spots and transcript abundance for candidates that exhibit significant changes in response to altered levels of Wwox in adult flies. n/d, protein was not detected; n/c, no change in protein abundance was detected.
None of the proteins or RNAs detected in these studies, as altered due to increases or decreases in Wwox, is found among those that have previously been reported as having physical interaction with Wwox (reviewed in 30). Indeed none of the proteins detected has PPxY motifs that might indicate a physical interaction with Wwox through either of its WW domains. Therefore, these proteomic and micro-array screens are unlikely to be comprehensive, however they do reveal novel functional characteristics of Wwox that are likely to account for its reported impact on metabolism (19–25).
Candidate Wwox interactors
Proteomic and microarray analyses confirmed an increase in Wwox protein and transcript levels in da>Wwox flies that ectopically over-express Wwox as well as a decrease in Wwox transcript levels in each of the Wwox mutants (Table 1). We have previously shown the absence of Wwox protein in Wwox1 mutant embryos by western analysis and have now confirmed the absence of Wwox protein in both Wwox1 and Wwoxf04545 adult flies (28 and Supplementary Material, Fig. S1).
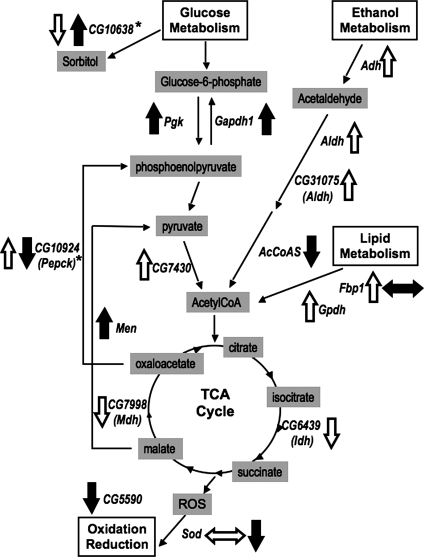
Biochemical analyses of adult flies and embryos with altered levels of Wwox identified a total of 27 different candidate proteins and/or mRNAs that were altered in response to increased and/or decreased levels of Wwox (Table 1). Interestingly, a significant number of these are known or predicted, based on sequence similarities (as listed on FlyBase; 31), to participate in various metabolic pathways. This highlights the impact of altered Wwox levels on metabolic processes; a common feature of all the WWOX knockout rodent animals (19–25). Of significance is the finding that many of these candidates are either directly or indirectly involved in metabolic pathways that converge on the tricarboxylic acid (TCA) cycle (Fig. 2). This is supportive of altered Wwox levels having an impact on aerobic metabolism in this in vivo model. Two of the candidate interactors identified encode proteins with isocitrate dehydrogenase (CG6439) and malate dehydrogenase (CG7998) activities, both of which catalyse enzymatic reactions that are integral to the TCA cycle. Twelve other interactors are predicted to be involved in the regulation of metabolism of various energy sources within cells that ultimately have effects on the TCA cycle. Several candidates are involved in regulation of available glucose levels including phosphoglycerate kinase (Pgk)/CG3127, CG7430 which encodes a predicted dihydrolipoyl dehydrogenase, malic enzyme (Men)/CG10120, glyceraldehyde-3-phosphate dehydrogenase (Gapdh)/CG12055, CG10638 which encodes a predicted aldehyde reductase and CG10924 which encodes a phosphoenolpyruvate carboxykinase (PEPCK). Four other interactors required for metabolism of ethanol —alcohol dehydrogenase (Adh)/CG3481, aldehyde dehydrogenase (Aldh)/CG3752, AcetylCoA synthase (AcCoAS)/CG9390 and CG31075 which also encodes an aldehyde dehydrogenase—were also detected. A further two interactors—fat body protein1 (Fbp1)/CG17285 and glycerol-3-phosphate dehydrogenase (Gpdh)/CG9042—encode enzymes that are involved in the metabolism of lipids.
Figure 2.
Metabolic pathways impacted by altered Wwox levels. Alteration of Wwox levels resulted in changes to many enzymes with roles in the TCA cycle, glucose metabolism, ethanol metabolism, lipid metabolism and oxidation/reduction supportive of a contributing role for Wwox in the maintenance of aerobic metabolism. Arrows indicate up- or down-regulated Wwox interacting candidates: solid black arrows for those altered following ectopic over-expression of Wwox and open arrows for those altered in Wwox loss of function mutants. Side arrows indicate that changes were detected in different isoforms of the protein. Gene abbreviations are listed in Table 1.
Another two of the candidate Wwox interactors, Cu–Zn superoxide dismutase (Sod)/CG11793 and CG5590, encode enzymes that are involved in regulation of oxidative pathways. The formation of ROS is a normal by-product of the electron transport chain, which is tightly linked to the TCA cycle. Wwox also encodes an oxidoreductase suggesting that it has function(s) in similar pathways. CG11089 and CG7470 are involved in the biosynthesis of inosine monophosphate (IMP) and proline, respectively. Other interactors identified are predicted to have various functions as listed in Table 1.
In vivo functional screen for candidate Wwox interactors
The Drosophila genetic model organism enables verification of functional interaction(s) between Wwox and the various candidate Wwox interactors, however a phenotype is required to form the basis of genetic screening. We have previously determined that the loss of Wwox function has no phenotype (29), thus the strategy for our in vivo RNAi screen was first to identify phenotypes produced by altered levels of any of the candidates on their own and then to look for modification of these phenotypes by altered levels of Wwox (Fig. 3A). Candidates were screened for phenotypes resulting from reduced levels of endogenous mRNAs via the ectopic expression of specific homologous RNAi sequences. Given that a significant proportion of the candidates encode genes that are involved in aerobic metabolism, we screened for any effect(s) on survival with ubiquitous expression of RNAi targeted to these candidates.
Figure 3.
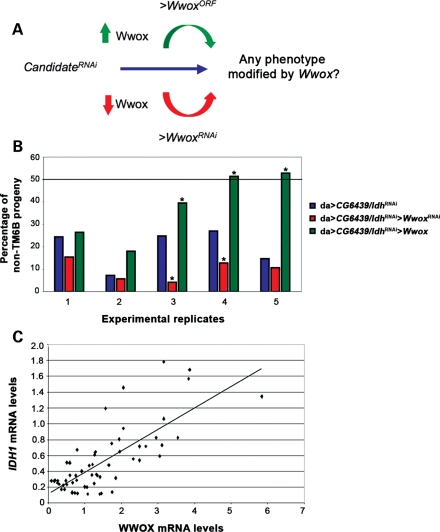
In vivo functional interactions of Wwox with CG6439/Idh. (A) Rationale of the in vivo genetic screens for Wwox functional interactors in Drosophila. Flies with ubiquitous knockdown of the candidate interactors alone were assayed for any resultant phenotypes that could be modified by either decreased or increased levels of Wwox. (B) Wwox interacts genetically with CG6439/Idh in viability assays. A deviation from the expected proportion (50%, indicated by the bold line) of non-TM6B progeny (see Supplementary Material, Fig. S1A) was observed when CG6439/Idh was knocked down ubiquitously (blue), indicative of a decrease in viability. This decrease in viability (blue) was enhanced when Wwox levels were decreased (red) and suppressed when Wwox levels were increased (green). Chi-square test was performed on each of the five separate experimental replicates and ‘*’ denotes statistical significance with P < 0.05. (C) Correlation of WWOX and IDH1 transcript levels in human cancer cell lines. qPCR analyses of WWOX and IDH1 transcript levels were determined for four different time points/confluencies in each of 15 exponentially growing human cancer cell lines. Regression analysis of WWOX and IDH1 mRNA levels revealed a positive correlation with a P-value of 2.5E−12 (see also Supplementary Material, Figs S2, S3 and Table S2).
The binary GAL4-UAS system was used to ectopically express various candidate RNAi constructs (32,33) throughout Drosophila development using the da-GAL4 driver stock. Six of the candidate RNAi lines tested showed a significant decrease in viability when expressed ubiquitously, while a further four were completely lethal (Supplementary Material, Fig. S2B). These reduced viability phenotypes formed the basis of genetic screening to identify functional interaction(s) with Wwox. The use of the GAL4-UAS system to lower candidate gene expression also allowed the simultaneous expression of constructs either to increase (>Wwox) or decrease (>WwoxRNAi) Wwox levels, and thereby assess any modification to the candidate gene phenotype by altered Wwox levels (Fig. 3A). Altered levels of Wwox transcript in each of these genotypes (da > WwoxRNAi and da > Wwox) were confirmed by qPCR analysis (Supplementary Material, Fig. S3) and no effect was observed on viability when levels of Wwox alone were increased or decreased via this GAL4-UAS system (Supplementary Material, Fig. S2C); thus any significant alteration(s) in viability observed for each candidate RNAi would suggest a functional interaction of that candidate with Wwox. The absence of such an interaction, however, does not necessarily rule out a functional interaction between a candidate and Wwox, but rather suggests that within this assay system any interaction between the two proteins is not a rate-limiting requirement for survival of the animal.
Viability assays were thus performed for candidate RNAi lines alone (da > candidateRNAi) and the percentage survival to adulthood was compared with that observed when Wwox levels were either decreased (da > candidateRNAi> WwoxRNAi) or increased (da > candidateRNAi> Wwox) as summarized in Supplementary Material, Table S2. A significant amount of variation was observed in these assays and statistical analyses (as described in the experimental procedures) were used to identify any significant modifications resulting from altered levels of Wwox. Only one of the candidate RNAi lines tested showed a clear and significant interaction with Wwox. This candidate RNAi line is directed against CG6439 which encodes an enzyme with isocitrate dehydrogenase activity. Knockdown of endogenous levels of CG6439 transcript following ectopic expression of this RNAi line was confirmed by qPCR analyses (Supplementary Material, Fig. S3).
Functional interaction of CG6439/isocitrate dehydrogenase (Idh) with Wwox
RNAi knockdown of endogenous levels of CG6439/Idh throughout the development in Drosophila (da > CG6439/IdhRNAi) resulted in a decrease in viability (Fig. 3B). Drosophila with RNAi knockdown of CG6439/Idh as well as altered (increased or decreased) levels of Wwox were tested in five separate experiments to determine whether the observed decrease in viability could be modified by altered levels of Wwox. A trend was observed across all five experimental replicates, where a further decrease in viability was observed when Wwox levels were also decreased in these flies, indicating an enhancement of the CG6439/Idh RNAi phenotype (Fig. 3B). Conversely, ectopic over-expression of Wwox in flies with reduced expression of CG6439/Idh revealed suppression of the decreased viability observed when CG6439/Idh was knocked down alone (Fig. 3B). Together, these results are consistent with an in vivo functional interaction between Wwox and CG6439/Idh.
Independent verification of a genetic interaction between Wwox and Idh/CG6439 was performed using mutant alleles for each of these genes. The Idh/CG6439EY00276 mutation consists of a transposon inserted 174 bp upstream of the initiator ATG codon (and within the reported 5′-UTR) for Idh/CG6439 (34). Drosophila homozygous for this mutation are viable and fertile as previously observed for the Wwox1 mutants. However, we were unable to obtain adult flies that were homozygous for both Idh/CG6439EY00276 and Wwox1, consistent with the enhanced decrease in viability of da > Idh/CG6439RNAi observed when Wwox levels were also decreased by RNAi (Fig. 3B). Although the precise impact of this insertion mutation is not known, the altered activity of both Wwox and Idh/CG6439 has a significant effect on the viability of adult Drosophila, supporting their functional interaction in normal metabolic pathways during development. Thus, decreased levels of Wwox were found to significantly decrease endogenous levels of CG6439/Idh protein in this model and these changes could account for the functional interactions observed between Wwox and CG6439/Idh.
Correlation between WWOX and IDH1 mRNA levels in human cell lines
In order to assess whether WWOX and IDH also exhibit evidence of a functional interaction in human cancer cells, the steady-state levels of their respective mRNA were assessed. Fifteen different human cancer cell lines (AGS, Co-115, HCT116, HeLa, HT29, KATOIII, KM12C, KM12SM, LOVO, LS180, MCF-7, MDA-MB157, MDA-MB436, SK-BR-3, U2-OS) were each grown to different levels of confluence (a factor which affects WWOX mRNA levels, S. Dayan, unpublished data). Each of these samples was then assayed for the mRNA level of WWOX as well as the mRNA levels for isocitrate dehydrogenase family members: IDH1, IDH2 and IDH3. A significant positive correlation was observed between endogenous WWOX and IDH1 mRNA levels (Fig. 3C), consistent with a functional interaction between their respective encoded proteins. This positive correlation between WWOX and IDH1 transcripts in these cells is consistent with the relationship observed between Wwox and CG6439/Idh protein levels in flies. IDH has been found to be altered specifically in human cancers (35–37). Thus, it is clear that there is a relationship between endogenous WWOX and IDH1 levels that is consistent with WWOX contributing to metabolic balance in human cancer cells as well as in the context of the whole organism of Drosophila.
Cu–Zn superoxide dismutase interacts with Wwox
Proteomic analyses identified two different spots of similar molecular weight but different isoelectric point corresponding to Cu–Zn superoxide dismutase (Sod). Spot #19 showed reciprocal changes in abundance in Wwox mutants and ectopic over-expression, while spot #20 showed a decrease in Wwox mutants (Table 1). These results suggest a role for different isoforms and/or differently modified forms of Sod in response to altered levels of Wwox. Since it has previously been shown that Sod mutants are viable but have a decreased lifespan (38), we determined the ability of altered levels of Wwox to modify this phenotype.
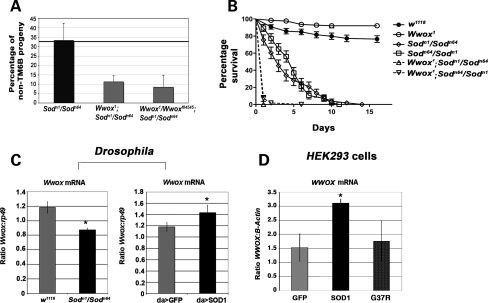
Missense mutations in Sod (Sodn1 and Sodn64) result in the expression of mutant forms of the protein, each with decreased levels of enzyme activity (Sodn1 is also known as Sodn108; 39). The Sodn1 mutation (G50S) disrupts dimer contact, while the Sodn64 mutation (G43E) affects metal ion binding. We generated adult flies that were trans-heterozygous for these two mutations (Sodn1/Sodn64). This mutant combination showed no effect on viability of adult flies as the expected percentage of non-balancer flies was obtained from this cross (Fig. 4A and Supplementary Material, Fig. S4A). However, the introduction of homozygous Wwox1 or trans-heterozygous Wwox1/Wwoxf04545 mutations into this Sod mutant background resulted in a decrease in viability, where the number of adult progeny obtained for each of the Wwox;Sod double mutants was decreased to less than one-third of that expected (Fig. 4A and Supplementary Material, Fig. S4A). Although only low numbers of Wwox;Sod double-mutant flies were able to be collected due to this decreased viability, the effect of the homozygous Wwox1 mutation on the lifespan of these Sod mutants was determined. Sodn1/Sodn64 trans-heterozygous mutants showed a decreased lifespan compared with wild-type or Wwox mutant flies where the Sod mutants were all dead by the tenth day of the lifespan assay (Fig. 4B). This decreased lifespan phenotype was enhanced when the flies were also homozygous mutant for the Wwox1 mutation as Wwox;Sod double-mutant flies all died within the first 24 h of the assay. This was confirmed for Sodn64/Sodn1 trans-heterozygous mutants where crosses were set up in the opposite direction.
Figure 4.
Wwox and Sod interact genetically. (A) A decrease in viability was observed when homozygous Wwox1 or trans-heterozygous Wwox1/Wwoxf04545 mutations were introduced into the Sodn1/Sodn64 mutant background (as indicated by the negative deviation from the expected 33.3% proportion of non-TM6B progeny, see Supplementary Material, Fig. S2). (B) A decreased lifespan was observed for the trans-heterozygous Sod mutations compared with wild-type and Wwox mutants. Each of the Wwox;Sod double mutants showed a further decrease in viability. Survival curves for the two Wwox;Sod double mutants overlap on the graph with all flies dead after 24 h. (C) qPCR showed decreased levels of Wwox transcript in Sodn1/n64 compared with w1118 larvae (P = 0.003) and increased levels of Wwox transcript in SOD1 overexpressing compared with control GFP overexpressing larvae (P = 0.043). (D) qPCR of WWOX levels in a HEK293 cells overexpressing SOD1 showed increased levels of endogenous WWOX transcript compared with the control line overexpressing GFP (P = 0.008, see also Supplementary Material, Fig. S4). No such increase in WWOX transcript was observed when cells were overexpressing a G37R mutant form of SOD1.
Proteomic analysis showed qualitative changes in endogenous Sod in adult flies both with increased or decreased levels of Wwox, while qPCR revealed that Sod transcript levels were increased in Wwox mutants (Table 1). To determine whether alterations in Sod could similarly affect endogenous Wwox transcript levels, qPCR analysis was performed on Sod mutants. Wwox mRNA levels were significantly down-regulated in Sodn1/Sodn64 mutant third instar larvae (Fig. 4C). To check whether endogenous Wwox mRNA levels were directly altered by increased Sod activity, we ubiquitously expressed a full-length human SOD1 construct in Drosophila. Over-expression of SOD1 resulted in an increase in Wwox transcript showing reciprocal changes in endogenous Wwox transcript levels in the presence of Sod mutations compared with ectopically over-expressed human SOD1 (Fig. 4C). We further investigated this relationship in human HEK293 cells and found ectopic expression of human SOD1 also resulted in an increase in endogenous WWOX mRNA expression. This increase in endogenous WWOX mRNA expression was not observed when a G37R mutant form of SOD1 that is associated with ALS (40) was over-expressed as an appropriate negative control (Fig. 4D). Together, these results suggest clear conservation of a functional relationship between these two genes.
Wwox affects endogenous levels of ROS
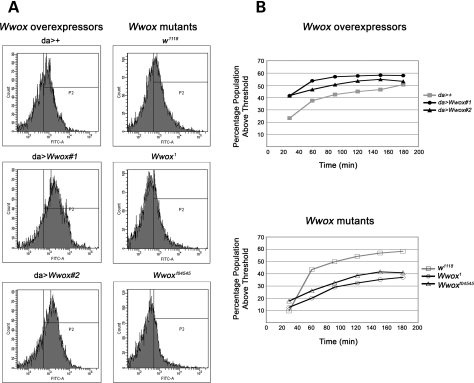
We have shown functional interactions between Wwox and two of the candidate interactors identified: CG6439/Idh and Sod. Given the known role of CG6439/Idh as an integral member of the TCA cycle (a known generator of ROS) and Sod in the direct regulation of ROS levels, we determined the endogenous level of ROS in Drosophila with altered levels of Wwox expression. ROS levels were measured indirectly in dispersed cells from Drosophila by fluorescence-activated cell sorting. In two independent lines of Wwox over-expressing larvae, the percentage of cells with ROS levels above an arbitrary threshold was found to be higher than for control larvae, while Wwox mutants had consistently lower levels of ROS compared with their control larvae (Fig. 5A and B). Thus, these reciprocal results strongly suggest a role for Wwox in the regulation of ROS.
Figure 5.
Altered Wwox expression affects the levels of ROS in Drosophila larvae. (A) Fluorescence-activated cell sorter (FACS) analysis of ROS levels in cells from two independent lines over-expressing Wwox (da > Wwox#1 and da > Wwox#2) compared with control (da>+) and two Wwox mutant lines (Wwox1 and Wwoxf04545) compared with control (w1118) after 1 h incubation with 10 µm CM-H2DCFDA. (B) An arbitrary threshold of fluorescence intensity was set and the percentage of cells from the total population exhibiting fluorescence above that threshold was measured every 30 min for 3 h.
DISCUSSION
Common chromosomal fragile sites are regions that are predisposed to DNA instability in cancer. FRA3B (within the FHIT gene) and FRA16D (within the WWOX gene) are the most susceptible sites (3). This DNA instability varies between different cells, sometimes comprising multiple events (6) that together constitute a scar on the genomic landscape that indicates the passage of the damaged cell through some form of environmental (replicative) stress. The biological consequences of fragile site-mediated DNA instability in cancer are dependent not only upon the frequency of such instability, but also upon the perturbation of the normal function of genes in the vicinity.
Here we report that the highly conserved Drosophila orthologue of WWOX has biologically significant roles in pathways that control aerobic metabolism and ROS. Alterations were observed in a number of genes encoding various members of aerobic metabolic pathways that converge on the TCA cycle as summarized in Figure 2. A common feature of all of the rodent WWOX mutants was decreased size and early death of mutant animals (19–25). These effects could also arise from Wwox mutations leading to altered regulation of aerobic metabolism in these mouse models, perhaps via components of the TCA cycle. Consistent with this is the reported hypoglycaemia of the conditional Wwox knockout (23). In the present study, conserved functional interactions were identified between Wwox and isocitrate dehydrogenase, an integral component of the TCA cycle as well as superoxide dismutase, a known regulator of ROS. Our findings support a protective role for Wwox under conditions of oxidative stress given the observed decrease in survival of the Wwox;Sod double mutants. Significantly, levels of endogenous ROS were found to correlate both with increased and decreased Wwox expression. This role for Wwox in regulating ROS levels could explain previous observations of the biological properties of WWOX, notably its ability when over-expressed to inhibit tumour cell growth and the participation of WWOX in cellular responses to TNF (41), since this pathway is mediated by ROS (42–44).
ROS are effectors of a vast number and range of essential biochemical pathways although they are also inherently dangerous chemicals to cells (45). It is, therefore, not surprising that cells have evolved intricate and integrated strategies to regulate ROS with cellular metabolism. Perturbations in ROS have previously been linked to aerobic glycolysis (Warburg effect) in cancer cells (46,47). Cancer-associated isocitrate dehydrogenase mutations produce 2-hydroxyglutarate that in turn leads to an increase in ROS (48,49). Our finding of a direct correlation between Wwox and ROS levels, together with functional interactions of Wwox with superoxide dismutase and isocitrate dehydrogenase, uncover perturbation in Wwox levels as another contributor to the differences seen in aerobic glycolysis (Warburg effect) between cancer and normal cells.
How could perturbation of WWOX levels (through DNA instability at FRA16D) contribute to cancer cell biology? A plausible explanation for the dramatic inhibition of breast cancer tumour cell growth by WWOX over-expression (14) is that increased WWOX results in higher ROS in these cancer cells. Levels of ROS are known to limit the self-renewal of stem cells (50) which are consistent with mounting evidence that breast cancer cells derive from stem cells (51) and are therefore likely to retain some of their properties. The corollary of this is that decreased WWOX (via FRA16D DNA deletion) will lead to decreased ROS levels that could relieve this limitation on self-renewal and therefore promote tumour cell growth. This latter property of WWOX could explain its behaviour as a non-conventional tumour suppressor in both human cancers and mouse models. Reduction of WWOX levels to 50% may result in a sufficient decrease in endogenous ROS to cause cellular consequences (i.e. further knockout of the second WWOX allele is not necessary).
Our studies on the Wwox function in Drosophila reveal a number of similarities with pathways that involve FHIT identified in human cancer cells (52,53). Both WWOX and FHIT are lost in most cancers and their restoration suppresses tumourigenicity (13,14). FHIT gene transfer into cancer cells increases production of ROS, while Wwox over-expression in Drosophila larval tissue also leads to increases in ROS. FHIT is known to functionally interact with ferredoxin reductase and physically associate with malate dehydrogenase and mitochondrial aldehyde dehydrogenase (as well as other proteins; 52). In addition to Wwox functional interactions with Sod and Idh described herein, we have also detected Wwox associated alterations in malate dehydrogenase and mitochondrial aldehyde dehydrogenase protein levels in Drosophila by proteomics (Table 1). These alterations would affect pathways leading to alterations in aerobic metabolism (Fig. 2), a hallmark of cancer cells, and give further evidence of the role of Wwox as a surveyor of the cell's metabolic status. In addition, WWOX and FHIT also share Hsp60 as a common interactor (52,53) with the Drosophila orthologue, Hsp60c, being detected in the Wwox proteomics analysis (Table 1).
These parallels lead us to propose that there is a functional basis for the association of the tumour suppressor genes WWOX and FHIT with the most unstable common chromosomal fragile sites in humans. The presence of these fragile sites confers the potential for DNA damage within the respective genes. DNA damage at these sites is an early event in tumourigenesis and the variable extent of damage (6) can result in either decreases or increases in gene expression (15–18). Those cells with a resultant increase in expression of WWOX and/or FHIT therefore have increased resilience to further oxidative stress by virtue of increased ROS. We therefore propose that the common pathways in which WWOX and FHIT participate form a network of ‘front-line’ responses to environmental stresses. These responses include altered metabolic status of cells that impact on their likelihood to proceed along the path of tumourigenesis.
MATERIALS AND METHODS
Fly lines and crosses
All Drosophila stocks were maintained in vials containing Fortified (F1) medium and kept at either 18 or 25°C. The F1 medium was composed of 1% agar, 18.75% compressed yeast, 10% polenta, 10% treacle, 1.5% acid mix and 2.5% tegosept. w1118, da-GAL4, CG6439EY00276, UAS-GFP, Sodn1/TM3Sb and Sodn64/TM3Sb were all obtained from the Bloomington stock centre. Wwox1, Wwoxf04545 and UAS-Wwox lines have been previously described, with Wwox1 and Wwoxf04545 having undergone four rounds of backcrossing to w1118 (29). UAS-candidateRNAi lines (listed in Supplementary Material, Table S2) were obtained from the Vienna Drosophila Research Centre. Multiple transgenic lines of UAS-SOD1 were generated by standard P-element transformation techniques. da > Wwox stock was generated by recombination between da-GAL4 and UAS-Wwox #1 both of which are located on the third chromosome. The mutant alleles: Sodn1 and Sodn64 were rebalanced and maintained as stocks over TM6B. The double-mutant stocks: UAS-WwoxRNAi/CyO;da-GAL4/TM6B, Wwox1/CyO:CG6439EY00276/TM6B, Wwox1/CyO:Sodn1/TM6B, Wwox1/CyO:Sodn64/TM6B and Wwoxf04545/CyO:Sodn64/TM6B were all generated by standard genetic techniques and confirmed by PCR and sequencing (for Sod alleles using primers from reference 39) or by western analysis for Wwox alleles.
SOD1 and GFP constructs and expression
SOD1 was amplified from cDNA obtained from HCT116 cells using primers CACCATGGCGACGAAGGCCGTGT and CTACAGCTAGCAGGATAACAG and blunt-end cloned into the pENTR/D-TOPO vector (Invitrogen). The construct was recombined into either pcDNA 3.2-DEST (Invitrogen) containing the CMV promoter for expression into human cells or a pUAST vector containing the Gateway® ccdb cassette (Invitrogen, from H. Dalton) for expression in Drosophila (as described in Gateway cloning methods, Invitrogen). Human SOD1 ALS mutant G37R was made by the QuickChange method as described by Stratagene (QuickChange Site-Directed Mutagenesis Kit). The forward and reverse complement primers were designed with a single-base mutation (underlined), 5′-GTGGGGAAGCATTAAAAGACTGACTGAAGGCC-3′ and used to PCR pENTR/D-TOPO-SOD1 plasmid. Parental methylated and hemimethylated DNA was digested with DpnI and the intact SOD1-mutated plasmids were used for transformation and subsequent cloning into pcDNA 3.2-DEST vector. GFP was amplified from pEGFP-N2 (Clontech) using primers CACCATGGTGAGCAAGGGCG and TGGCTGATTATGATCTAGAGTCG and was similarly cloned into pcDNA 3.2-DEST for expression in human cells. Purified plasmids were used for injecting into Drosophila w1118 embryos or for transfection using standard protocols for Fugene (Roche).
Cell-line culture
The cancer cell lines: AGS, Co-115, HCT116, HeLa, HT29, KATOIII, KM12C, KM12SM, LOVO, LS180, MCF-7, MDA-MB157, MDA-MB436, SK-BR-3 and U2-OS were obtained from the American Type Culture Collection. Cell lines AGS and Lovo were grown in F-12K culture media while Co-115, HCT116, HT29, KatoIII, KM12C, KM12SM, LS180, MCF-7, MDA-MB157 and SK-BR-3 were grown in RMPI. HeLa and U2-OS cell lines were grown in DMEM while MDA-MB436 cells were grown in OPI-MEM. All cell lines were grown in their respective culture media containing 10% foetal calf serum.
Protein sample preparation and DIGE labelling
Ten 0–1 day male flies were homogenized in sample buffer (7 m urea, 2 m thiourea, 30 mm Tris, 4% CHAPS) and protein purification was performed using a Ready Prep™ 2D clean-up kit (BioRad) according to the manufacturer's protocol. Protein concentrations were estimated using an EZQ protein quantitation kit (Molecular Probes). One hundred micrograms of protein were labelled with 200 pmol of Cy Dye. An internal standard sample comprised of a protein pool consisting of 50 μg of each of the adult fly protein samples was labelled with 2.8 nmol Cy2 dye.
2D-electrophoresis and gel imaging
Cy labelled adult fly sample mixtures were applied to the IPG strips (24 cm, pH 3–11, GE Healthcare) and isoelectric focusing was performed on an IPGphor II (GE Healthcare) at 20°C using a stepwise gradient. Second-dimension SDS–PAGE was performed using the Ettan™ DALTtwelve Large Vertical electrophoresis system (GE Healthcare). DIGE gels were scanned using the Ettan DIGE Imager (GE Healthcare) at 100 µm resolution and image analysis was undertaken using DeCyder 2D software (version 6.5, GE Healthcare). Each analytical gel image was processed separately in the differential in-gel analysis module of DeCyder prior to export to the biological variation analysis module.
Mass spectrometry
Spots were excised using an Ettan™ Spot Picker robot (GE Healthcare). Excised gel pieces were dehydrated in 100% acetonitrile then vacuum dried before being rehydrated in 10 ng/μl trypsin in 5 mm ammonium bicarbonate and incubated over night at 37°C. Following digestion, peptides were extracted twice with 50% acetonitrile containing 1% formic acid, concentrated and reconstituted with FA30 (0.1% formic acid, 3% acetonitrile) to a final volume of 5 µl. Samples were then analysed either by LC-ESI-IT-MS where samples were chromatographed using an Agilent Protein ID Chip column assembly (40 nl trap column with 0.075 × 43 mm C-18 analytical column) housed in an Agilent HPLC-Chip Cube Interface connected to an HCT ultra 3D-Ion-Trap mass spectrometer (Bruker Daltonics) or by Matrix Assisted Lazer Desorption Ionisation—Time Of Flight—Mass Spectrometry (MALDI-TOF-MS) using a Bruker ultraflex III MALDI TOF/TOF mass spectometer (Bruker Daltonics).
RNA purification
The equivalent of 50 μl of 4–8 h embryos, five third instar larvae or five 1-day old male flies were used for each biological replicate. Samples were frozen in liquid nitrogen and homogenized in Trizol reagent (Invitrogen). Total RNA was extracted with chloroform and precipitated with isopropanol. RNA was further purified using the RNeasy Mini Kit (Qiagen). For human cell lines, the protocol for total RNA isolation from animal cells as described in Qiagen RNeasy Minikit was used.
Microarray analyses
RNA from an equivalent volume of 50 μl of 4–8 h embryos was processed using the One-Cycle Target Labelling and Control Reagents Kit, as per manufacturer's instructions (Affymetrix, Inc). Briefly 2 μg of total RNA was converted to cDNA (Superscript II, Invitrogen) and an overnight in vitro transcription reaction performed to generate a pool of cRNA carrying a biotin tag (MEGAscript T7 Kit, Ambion, Inc). The Drosophila Genome 2.0 Array was hybridized for 16 h and washed/stained on a FS 450 Fluidics Station using the Midi euk2 v3 script. Data were acquired on a 7G GeneChip Scanner 3000 and data extraction performed in GCOS v1.2. Candidates were identified as those that showed a ‘present’ call in each of three biological replicates. T-tests were performed on raw values to determine samples that showed a significant difference with a P-value < 0.05. The microarray data have been deposited on the NCBI database (GEO accession number GSE22689).
Quantitative real-time PCR (qPCR) assays
The quantitative PCR assays were carried out using protocols provided by the manufacturers. Superscript III (Invitrogen) was used to perform reverse transcription on the RNA and quantitative PCR was carried out using the SYBR Green mix (Applied Biosystems) in an Applied Biosystem ABI Prism 7000 Sequence Detection System (Applied Biosystems). The relative standard curve method for quantification as outlined by Applied Biosystems was used. For Drosophila, mRNA levels were normalized against the house-keeping gene ribosomal protein 49 (rp49). Triplicate reactions for each of three biological replicates were performed for each sample.
Drosophila qPCR primer pairs for Wwox were: ATTGTGCTGTCATCCGAGTCG/ATTCTCCACGGGCAG GTTG, CG6439/Idh: GGTCTACTCCCTCCAGGAGGTCT/TCGAAGTCCACGGGAACG, Sod: GAGACCTTCACGGGCGTA/GGCACGGTTTTCTTCGAACA, rp49: ATCGATATGCTAAGCTGTCGCAC/TGTCGATACCCTTGGGCTTG.
For human cell lines, β-Actin was used as the endogenous control. Human qPCR primers for IDH1 were ACGTGCAGTCGGACTCTGTG/TCATCATGCCGAGAGAGCC, and for WWOX, GAGGCCTTTCACCAAGTCCAT/TCCCAGACCCTCCAGTTCTG.
Viability assays
Drosophila crosses (see Supplementary Material, Fig. S2A) were set up in medium-sized vials containing FI medium and maintained at 25°C. Progeny from the crosses were scored and only the crosses with a minimum of 30 progeny carrying the TM6B balancer were included in the analyses. The ratio of non-TM6B:TM6B progeny was recorded for each cross and chi-square test analysis using GraphPad Prism 5 was carried out to determine whether there was a significant difference between the ratios when Wwox levels were increased or decreased compared with the control in each experiment. The standard value of P < 0.05 was chosen to indicate significance.
Lifespan assays
Male Drosophila 0 to 1 days old were collected and placed in a vial containing fly food. Flies were kept at 25°C with 70% humidity. Flies were turned into fresh vials everyday and deaths recorded at the same time. Statistical analyses and graphing were done using GraphPad Prism 5.
Reactive oxygen species measurements
Single cells from 10 third instar Drosophila larvae were isolated using a modification of Singh et al. (54). Larval cuticle was removed and tissue was digested with 0.5 mg/ml Collagenase (Sigma) in phosphate buffered saline (PBS: 0.8% NaCl, 0.02% KCl, 0.02% KH2PO4, 0.115% Na2HPO4, pH 7.4) for 1 h at 25°C. The digest was filtered through a 100 µm cell strainer and diluted to 1 ml with PBS. Intracellular ROS was determined using 5-(and-6)-chloromethyl-2′,7′-dichlorodihydorfluorescein diacetate (CM-H2DCFDA, Molecular Probes), a non-fluorescent dye, which upon deacetylation by cellular esterases and oxidation by ROS becomes fluorescent. Cells were incubated at 25°C for 30 min with 10 μm CM-H2DCFDA and FACS analysis was performed immediately and every 30 min for 3 h to allow for dye loading differences.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the National Health and Medical Research Council of Australia (519125 to L.V.O. and R.I.R., 207830 to L.V.O.) and the Cancer Council South Australia (to R.I.R.). Funding to pay the Open Access publication charges for this article was provided by the ARC Special Research Centre for the Molecular Genetics of Development.
ACKNOWLEDGEMENTS
We thank Jo Milverton for technical assistance with Drosophila experiments, Tanya Henshall for Western blot analyses, Jack da Silva for advice on statistical analysis and members of the Richards lab for reading drafts of this manuscript and providing valuable suggestions. Rob Richards dedicates his contribution to this work to the memory of David (Happy) Fitzsimons. We also thank Bloomington stock centre and Vienna Drosophila Resource Centre for providing stocks and the Australian Drosophila Biomedical Research Support Facility (OzDros) for their ongoing support of Drosophila research.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Yunis J.J., Soreng A.L. Constitutive fragile sites and cancer. Science. 1984;226:1199–1204. doi: 10.1126/science.6239375. [DOI] [PubMed] [Google Scholar]
- 2.Richards R.I. Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet. 2001;17:339–345. doi: 10.1016/s0168-9525(01)02303-4. [DOI] [PubMed] [Google Scholar]
- 3.Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., et al. Signatures of mutation and selection in cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein C.K., Glover T.W., Palmer J.L., Glisson B.S. Direct correlation between FRA3B expression and cigarette smoking. Genes Chromosomes Cancer. 2002;34:333–340. doi: 10.1002/gcc.10061. [DOI] [PubMed] [Google Scholar]
- 5.Glover T.W., Arlt M.F., Casper A.M., Durkin S.G. Mechanisms of common fragile site instability. Hum. Mol. Genet. 2005;14:R197–R205. doi: 10.1093/hmg/ddi265. [DOI] [PubMed] [Google Scholar]
- 6.Finnis M., Dayan S., Hobson L., Chenevix-Trench G., Friend K., Ried K., Venter D., Woollatt E., Baker E., Richards R.I. Common chromosomal fragile site FRA16D mutation in cancer cells. Hum. Mol. Genet. 2005;14:1341–1349. doi: 10.1093/hmg/ddi144. [DOI] [PubMed] [Google Scholar]
- 7.Durkin S.G., Glover T.W. Chromosome fragile sites. Annu. Rev. Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 8.Smith D.I., McAvoy S., Zhu Y., Perez D.S. Large common fragile site genes and cancer. Sem. Biology. 2007;17:31–41. doi: 10.1016/j.semcancer.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Bednarek A.K., Laflin K.J., Daniel R.L., Liao Q., Hawkins K.A., Aldaz C.M. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3–24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60:2140–2145. [PubMed] [Google Scholar]
- 10.Ried K., Finnis M., Hobson L., Mangelsdorf M., Dayan S., Nancarrow J., Woollatt E., Kremmidiotis G., Gardner A., Venter D., et al. Common chromosomal fragile site FRA16D DNA sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum. Mol. Genet. 2000;9:1651–1663. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- 11.Ishii H., Vecchione A., Furukawa Y., Sutheesophon K., Han S.Y., Druck T., Kuroki T., Trapasso F., Nishimura M., Saito Y., et al. Expression of FRA16D/WWOX and FRA3B/FHIT genes in hematopoietic malignancies. Mol. Cancer Res. 2003;1:940–947. [PubMed] [Google Scholar]
- 12.Guler G., Uner A., Guler N., Han S.Y., Iliopoulos D., Hauck W.W., McCue P., Huebner K. The fragile genes FHIT and WWOX are inactivated coordinately in invasive breast carcinoma. Cancer. 2004;100:1605–1614. doi: 10.1002/cncr.20137. [DOI] [PubMed] [Google Scholar]
- 13.Siprashvili Z., Sozzi G., Barnes L.D., McCue P., Robinson A.K., Eryomin V., Sard L., Tagliabue E., Greco A., Fusetti L., et al. Replacement of Fhit in cancer cells suppresses tumourigenicity. Proc. Natl Acad. Sci. USA. 1997;94:13771–13776. doi: 10.1073/pnas.94.25.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bednarek A.K., Keck-Waggoner C.L., Daniel R.L., Laflin K.J., Bergsagel P.L., Kiguchi K., Brenner A.J., Aldaz C.M. WWOX, the FRA16D gene, behaves as a suppressor of tumour growth. Cancer Res. 2001;61:8068–8073. [PubMed] [Google Scholar]
- 15.Watanabe A., Hippo Y., Taniguchi H., Iwanari H., Yashiro M., Hirakawa K., Kodama T., Aburatani H. An opposing view on WWOX protein function as a tumour suppressor. Cancer Res. 2003;63:8629–8633. [PubMed] [Google Scholar]
- 16.Driouch K., Prydz H., Monese R., Johansen H., Lidereau R., Frengen E. Alternative transcripts of the candidate tumor suppressor gene, WWOX, are expressed at high levels in human breast tumors. Oncogene. 2002;21:1832–1840. doi: 10.1038/sj.onc.1205273. [DOI] [PubMed] [Google Scholar]
- 17.Chang N.-S., Schultz L., Hsu L.-J., Lewis J., Su M., Sze C.-I. 17B-Estradiol upregulates and activates WOX1/WWOXv1 and WOX2/WWOXv2 in vitro: potential role in cancerous progression of breast and prostate to a premetastatic state in vivo. Oncogene. 2005;24:714–723. doi: 10.1038/sj.onc.1208124. [DOI] [PubMed] [Google Scholar]
- 18.Lai F.-J., Cheng C.-L., Chen S.-T., Wu C.-H., Hsu L.-J, Lee J. Y.-Y., Chao S.C., Sheen M.-C., Shen C.-L., Chang N.-S., et al. WOX1 is essential for UVB irradiation-induced apoptosis and down-regulated via translational blockade in UVB-induced cutaneous squamous cell carcinoma in vivo. Clin. Cancer Res. 2005;11:5769–5777. doi: 10.1158/1078-0432.CCR-04-2274. [DOI] [PubMed] [Google Scholar]
- 19.Aqeilan R.I., Trapasso F., Hussain S., Costinean S., Marshall D., Pekarsky Y., Hagan J.P., Zanesi N., Kaou M., Stein G.S., et al. Targeted deletion of Wwox reveals a tumour suppressor function. Proc. Natl Acad. Sci. USA. 2007;104:3949–3954. doi: 10.1073/pnas.0609783104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludes-Meyers J.H., Kil H., Nuñez M.I., Conti C.J., Parker-Thornburg J., Bedford M.T., Aldaz C.M. WWOX hypomorphic mice display a higher incidence of B-cell lymphomas and develop testicular atrophy. Genes Chromosomes Cancer. 2007;46:1129–1136. doi: 10.1002/gcc.20497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aqeilan R.I., Hassan M.Q., de Bruin A., Hagan J.P., Volinia S., Palumbo T., Hussain S., Lee S.-H., Gaur T., Stein G.S., et al. The WWOX tumor suppressor is essential for postnatal survival and normal bone metabolism. J. Biol. Chem. 2008;283:21629–21639. doi: 10.1074/jbc.M800855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aqeilan R.I., Hagan J.P., de Bruin A., Rawahneh M., Salah Z., Gaudio E., Siddiqui H., Volinia S., Alder H., Lian J.B., et al. Targeted ablation of the WW domain-containing oxidoreductase tumor suppressor leads to impaired steroidogenesis. Endocrinology. 2009;150:1530–1535. doi: 10.1210/en.2008-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludes-Meyers J.H., Kil H., Parker-Thornburg J., Kusewitt D.F., Bedford M.T., Aldaz C.M. Generation and characterization of mice carrying a conditional allele of the Wwox tumour suppressor gene. PLoS ONE. 2009;4:e7775. doi: 10.1371/journal.pone.0007775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki H., Takenaka M., Suzuki K. Phenotypic characterization of spontaneously mutated rats showing lethal dwarfism and epilepsy. Comp. Med. 2007;57:360–369. [PubMed] [Google Scholar]
- 25.Suzuki H., Katayama K., Takenaka M., Amakasu K., Saito K., Suzuki K. A spontaneous mutation of the WWOX gene and audiogenic seizures in rats with lethal dwarfism and epilepsy. Genes Brain Behav. 2009;8:650–660. doi: 10.1111/j.1601-183X.2009.00502.x. [DOI] [PubMed] [Google Scholar]
- 26.Brumby A.M., Richardson H.E. Using Drosophila melanogaster to map cancer pathways. Nat. Rev. Cancer. 2005;5:626–639. doi: 10.1038/nrc1671. [DOI] [PubMed] [Google Scholar]
- 27.Vidal M., Cagan R.L. Drosophila models for cancer research. Curr. Opin. Genet. Develop. 2006;16:10–16. doi: 10.1016/j.gde.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 28.O'Keefe L.V., Liu Y.-H., Perkins A., Dayan S., Saint R.B., Richards R.I. FRA16D common chromosomal fragile site oxido-reductase WWOX protects against the effects of ionising radiation in Drosophila. Oncogene. 2005;24:6590–6596. doi: 10.1038/sj.onc.1208806. (2006) 25, 7662. [DOI] [PubMed] [Google Scholar]
- 29.O'Keefe L.V., Smibert P., Colella A., Chataway T.K., Saint R., Richards R.I. Know thy fly. Trends Genet. 2007;23:238–242. doi: 10.1016/j.tig.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Chang J.-Y., He R.-Y., Lin H.-P., Hsu L.-J., Lai F.-J., Hong G., Chen S.-J., Chang N.-S. Signaling from membrane receptors to tumor suppressor WW domain containing oxidoreductase. Exp. Biol. Med. 2010;235:796–804. doi: 10.1258/ebm.2010.009351. [DOI] [PubMed] [Google Scholar]
- 31.Tweedie S., Ashburner M., Falls K., Leyland P., McQuilton P., Marygold S., Millburn G., Osumi-Sutherland D., Schroeder A., Seal R., Zhang H. The FlyBase Consortium. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 33.Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 34.Bellen H.J., Levis R.W., Liao G., He Y., Carlson J.W., Tsang G., Evans-Holm M., Hiesinger P.R., Schulze K.L., Rubin G.M., et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan H., Parsons D., Jin G., McLendon R., Rasheed B.A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G.J., et al. IDH1 and IDH2 mutations in gliomas . N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mardis E.R., Ding L., Dooling D.J., Larson D.E., McLellan M.D., Chen K., Koboldt D.C., Fulton R.S., Delehaunty K.D., McGrath S.D., et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez G.Y., Reitman Z.J., Solomon D., Waldman T., Bigner D.D., McLendon R.E., Samuels Y., Yan H. IDH1(R132) mutations identified in one human melanoma metastasis, but not correlated with metastases to the brain. Biochem. Biophys. Res. Commun. 2010;398:585–587. doi: 10.1016/j.bbrc.2010.06.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkes T.L., Kirby K., Phillips J.P., Hilliker A.J. Transgenic analysis of the cSOD-null phenotypic syndrome in Drosophila. Genome. 1998;41:642–651. [PubMed] [Google Scholar]
- 39.Phillips J.P., Tainer J.A., Getzoff E.D., Boulianne G.L., Kirby K., Hilliker A.J. Subunit-destabilizing mutations in Drosophila copper/zinc superoxide dismutase: neuropathology and a model of dimer dysequilibrium. Proc. Natl Acad. Sci. USA. 1995;92:8574–8578. doi: 10.1073/pnas.92.19.8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cudkowicz M.E., McKenna-Yasek D., Sapp P.E., Chin W., Geller B., Hayden D.L., Schoenfeld D.A., Hosler B.A., Horvitz H.R., Brown R.H. Epidemiology of mutations in superoxide dismutase in amyotrophic lateral sclerosis. Ann. Neurol. 1997;41:210–221. doi: 10.1002/ana.410410212. [DOI] [PubMed] [Google Scholar]
- 41.Chang N.S., Pratt N., Heath J., Schultz L., Sleve D., Carey G.B., Zevotek N. Hyaluronidase induction of a WW domain-containing oxidoreductase that enhances tumour necrosis factor cytotoxicity. J. Biol. Chem. 2001;276:3361–3370. doi: 10.1074/jbc.M007140200. [DOI] [PubMed] [Google Scholar]
- 42.Garg A.K., Aggarwal B.B. Reactive oxygen intermediates in TNF signaling. Mol. Immunol. 2002;39:509–517. doi: 10.1016/s0161-5890(02)00207-9. [DOI] [PubMed] [Google Scholar]
- 43.Brown D.I., Griendling K.K. Nox proteins in signal transduction. Free Rad. Biol. Med. 2009;47:1239–1253. doi: 10.1016/j.freeradbiomed.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandenabeele P., Declercq W., Van Herreweghe F., Vanden Berghe T. The role of the kinases RIP1 and RIP3 in TNF-induced necrosis. Sci. Signal. 2010;3:re4. doi: 10.1126/scisignal.3115re4. [DOI] [PubMed] [Google Scholar]
- 45.Mailloux R.J., Bériault R., Lemire J., Singh R., Chénier D.R., Hamel R.D., Appanna V.D. The tricarboxylic acid cycle, an ancient metabolic network with a novel twist. PLoS ONE. 2007;2:e690. doi: 10.1371/journal.pone.0000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi D.Y., Xie F.Z., Zhai C., Stern J.S., Liu Y., Liu S.L. The role of cellular oxidative stress in regulating glycolysis energy metabolism in hepatoma cells. Mol. Cancer. 2009;8:32–46. doi: 10.1186/1476-4598-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weinberg F., Hamanaka R., Wheaton W.W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G.M., Budinger G.R., Chandel N.S. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumourigenicity. Proc. Natl Acad. Sci. USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kölker S., Pawlak V., Ahlemeyer B., Okun J.G., Hörster F., Mayatepek E., Krieglstein J., Hoffmann G.F., Köhr G. NMDA receptor activation and respiratory chain complex V inhibition contribute to neurodegeneration in d-2-hydroxyglutaric aciduria. Eur. J. Neurosci. 2002;16:21–28. doi: 10.1046/j.1460-9568.2002.02055.x. [DOI] [PubMed] [Google Scholar]
- 49.Latini A., Scussiato K., Rosa R.B., Llesuy S., Belló-Klein A., Dutra-Filho C.S., Wajner M. D-2-hydroxyglutaric acid induces oxidative stress in cerebral cortex of young rats. Eur. J. Neurosci. 2003;17:2017–2022. doi: 10.1046/j.1460-9568.2003.02639.x. [DOI] [PubMed] [Google Scholar]
- 50.Ito K., Hirao A., Arai F., Matsuoka S., Takubo K., Hamaguchi I., Nomiyama K., Hosokawa K., Sakurada K., Nakagata N., et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 51.Visvader J.E., Lindeman G.J. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat. Rev. Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 52.Trapasso F., Pichiorri F., Gaspari M., Palumbo T., Aqeilan R.I., Gaudio E., Okumura H., Iuliano R., Di Leva G., Fabbri M., et al. Fhit interaction with ferredoxin reductase triggers generation of reactive oxygen species and apoptosis of cancer cells. J. Biol. Chem. 2008;283:13736–13744. doi: 10.1074/jbc.M709062200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Okumura H., Ishii H., Pichiorri F., Croce C.M., Mori M., Huebner K. Fragile gene product, Fhit, in oxidative and replicative stress responses. Cancer Sci. 2009;100:1145–1150. doi: 10.1111/j.1349-7006.2009.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh M.P., Reddy M.M., Mathur N., Saxena D.K., Chowdhuri D.K. Induction of hsp70, hsp60, hsp83 and hsp26 and oxidative stress markers in benzene, toluene and xylene exposed Drosophila melanogaster: Role of ROS generation. Toxicol. Appl. Pharmacol. 2009;235:226–243. doi: 10.1016/j.taap.2008.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.