Abstract
More than 12 neurogenetic disorders are caused by unstable expansions of (CTG)•(CAG) repeats. The expanded repeats are unstable in germline and somatic cells, with potential consequences for disease severity. Previous studies have shown that contractions of (CAG)95 are more frequent when the repeat tract is transcribed. Here we determined whether transcription can promote repeat expansion, using (CTG)•(CAG) repeat tracts in the size range that is typical for myotonic dystrophy type 1. We derived normal human fibroblasts having single-copy genomic integrations of 800 CTG repeats. The repeat tract showed modest instability when it was not transcribed, yielding an estimated mutation rate of 0.28% per generation. Instability was enhanced several-fold by transcription in the forward or reverse transcription, and 30-fold by bidirectional transcription, yielding many expansions and contractions of more than 200 repeats. These results suggest that convergent bidirectional transcription, which has been reported at several disease loci, could contribute to somatic instability of highly expanded (CTG)•(CAG) repeats.
INTRODUCTION
Many neurogenetic disorders are caused by unstable expansions of simple tandem repeats (1). The tendency of unstable repeats to expand or contract in the germline can lead to marked phenotypic variability within families. Instability of expanded repeats also occurs in somatic cells, which may affect the age of symptom onset or rate of disease progression (2).
In the case of myotonic dystrophy type 1 (DM1), the expanded repeat is particularly large and unstable. Individuals with DM1 usually inherit alleles with hundreds of CTG repeats in the 3′-untranslated region (UTR) of DMPK, whereas unaffected people have fewer than 37 repeats (3,4).
Evidence suggests that instability of expanded (CTG)•(CAG) repeats is associated with cellular processes, such as DNA replication, repair or transcription, that entail strand separation, thus promoting the formation of extrahelical looped-out structures that are substrates for error-prone repair (reviewed in 2,5,6). The effects of transcription on repeat stability are particularly pertinent for DM1 because (i) expansion of the repeat does not silence transcription of the mutant allele (7); (ii) the organs most affected—skeletal muscle, heart and brain—are those with low rates of cell proliferation yet highest levels of DMPK expression (3,8); (iii) the somatic instability ultimately generates enormous alleles in cardiac and skeletal muscle, ranging from 2000 to 6000 repeats (9–15); and (iv) the DM1 (CTG)•(CAG) tract is transcribed in both directions (16).
Previous studies in bacteria using plasmids that carried an interrupted (CAG)175 repeat showed that deletions were more frequent when the repeat tract was transcribed, an effect suggested to result from collision of the transcription complex with the replication fork (17). In mammalian cells, the evidence for transcriptional enhancement of repeat instability comes from studies of human fibrosarcoma cells, using a selection system to detect large contractions that reduce (CAG)95 down to fewer than 39 repeats (18). The basal frequency of such events was low, around 4 × 10−7 per generation, but was stimulated 15-fold by transcription across the CAG repeat. Subsequent studies using the contraction selection assay revealed the involvement of various nucleotide excision repair factors including CSB, linking the process to transcription-coupled repair (19). Recent studies have also shown that CAG transcription induced the formation of RNA:DNA hybrids (R-loops) that stimulated repeat contractions (20). However, it has not been determined whether transcription can stimulate (CTG)•(CAG) expansions, the genetic events that are most relevant clinically.
Here we examined repeat stability by introducing highly expanded (CTG)•(CAG) repeats in normal human fibroblasts. We tested the effects of unidirectional and bidirectional transcription on the frequency and size of repeat contractions and expansions.
RESULTS
Instability of CTG repeats that are not transcribed
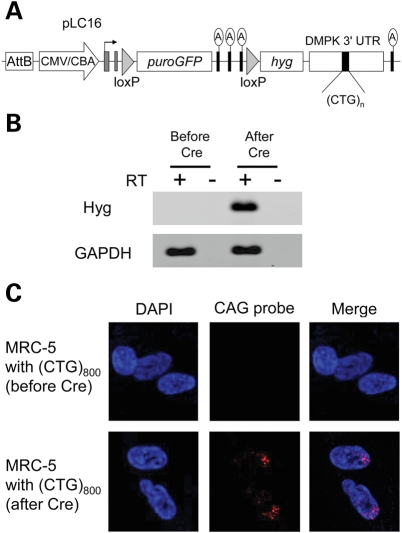
To introduce highly expanded CTG repeats into normal human cells, we inserted 800 CTG repeats into plasmid pLC16, using rolling circle amplification and cell-free cloning as previously described (21). MRC-5 fibroblasts were co-transfected with plasmid pLC16 (Fig. 1A), which expresses a puromycin resistance-green fluorescent protein (GFP) selectable marker (puroGFP), and plasmid pPhiC31o, for transient expression of phiC31 integrase. The integrase catalyzes single-copy genomic integrations of circular DNA at any of >70 pseudo-attP attachment sites in the human genome (22). Following puromycin selection, we obtained three independent clones showing robust transgene expression, as reflected by GFP activity. As initially derived, these cells did not express the (CTG)800 tract because it lies downstream from a strong transcription terminator. The absence of CTG expression was confirmed by reverse transcription polymerase chain reaction (RT–PCR) and fluorescent in situ hybridization (FISH) (Fig. 1B and C).
Figure 1.
(A) Diagram of pLC16, a construct for conditional expression of expanded CTG repeats. The attB site supports genomic integration by PhiC31 integrase. Expression is driven by the CMV/chicken beta-actin enhancer/promoter (block arrow). Initially this construct expresses the puromycin resistance-GFP (puroGFP) selectable marker. The triple ‘A' denotes the transcription-terminator element, consisting of three consecutive SV40 polyadenylation signals. Upon Cre-mediated excision of the selection cassette and transcription terminator, the construct expresses the hygromycin selectable marker (hyg), fused to the 3′-UTR from DMPK. (B) RT–PCR analysis of transgene expression. After Cre-mediated excision of the transcription terminator, hyg was expressed. (C) FISH analysis for RNA containing an expanded CUG repeat (CUGexp). Nuclear CUGexp foci were present in clones after transient expression of Cre recombinase.
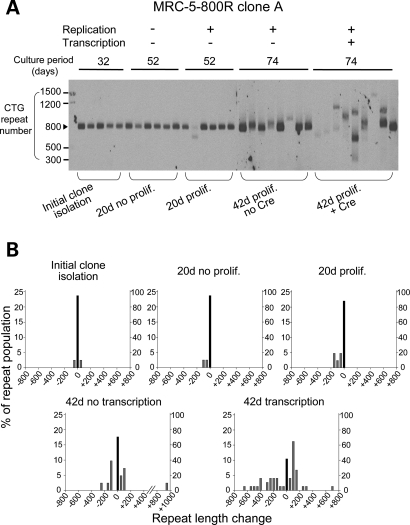
After 32 days of clonal growth, the stability of (CTG)800 was examined by small-pool PCR followed by Southern blot. At least 40 alleles were analyzed for each clone, using a gel system that resolves changes of 25 or more repeats. The (CTG)800 tract exhibited modest instability during the initial 32 days of clone expansion (Fig. 2A and B and Supplementary Material, Fig. S1). The frequency of unstable alleles ranged from 2.5 to 7.5% in different clones, with no significant difference between clones (Table 1 and Supplementary Material, Table S1). The overall mutation rate was estimated at 0.28% per generation across all clones.
Figure 2.
(A) Small-pool PCR followed by Southern blot for analysis of CTG repeat length. DNA from fibroblast clones was diluted to produce, on average, one amplifiable expanded repeat allele per PCR tube. The repeat tract was amplified by 24 cycles of PCR. The amplified product was detected by Southern blot probed with a CAG repeat oligonucleotide. The scale on the left shows the molecular weight markers converted into numbers of CTG repeats. (B) Histograms showing repeat length distributions in MRC-5-800R clone A (conditional expression of expanded repeat). The distribution of unstable alleles is shown by gray bars (left vertical axis). The frequency of stable alleles is shown by black bars (right vertical axis). Allele lengths are grouped in bins spanning 50 repeats.
Table 1.
Repeat instability in MRC-5 fibroblasts with conditional expression of 800 or 250 CTG repeats
| MRC-5-800R | % unstable allelesa (average change of repeat sizeb) |
||||
|---|---|---|---|---|---|
| Initial clonally derived cells | Proliferation (−), transcription (−) | Proliferation (+), transcription (−) | Proliferation (+), transcription (−) | Proliferation (+), transcription (+) | |
| Clone A | 5.0% (2.6) | 5.0% (3.8) | 11.9% (11) | 29.3% (27c) | 58.1%* (124) |
| Clone B | 7.5% (11) | 8.7% (14) | 22.2% (27) | 34.1% (32) | 59.6%** (99c) |
| Clone C | 2.5% (1.6) | 5.0% (5.0) | 9.1% (20) | 14.3% (20) | 38.3%* (79) |
| Culture period | 32 days | 52 days | 52 days | 74 days | 74 days |
| MRC-5-250R | % unstable allelesd (average change of repeat sizeb) | ||||
| Initial clonally derived cells | Proliferation (+), transcription (−) | Proliferation (+), transcription (+) | |||
| Clone D | 0% (0) | 4.9% (9.5) | 16.7% (11) | ||
| Clone E | 0% (0) | 7.0% (11) | 16.4% (24) | ||
| Clone F | 0% (0) | 4.4% (6.9) | 10.0% (3.6) | ||
| Culture period | 40 days | 75 days | 75 days | ||
aA cut-off point of ±25 repeats was used for counting as unstable.
bAmong all alleles (stable + unstable), the average change of repeat size is expressed as number of repeats.
cAn outlier allele that changed by over 800 repeats was excluded.
dA cut-off point of ±10 repeats was used for counting as unstable.
*P < 0.01, **P < 0.05, Chi-square test.
Clones were then split and continuously passaged for 10 additional doublings (20 days), or maintained in contact inhibition for the same interval. Bromodeoxyuridine (BrdU) incorporation confirmed an absence of cell proliferation in the contact-inhibited cells. Confluent cells showed no incorporation after 3 h, whereas proliferating cells showed 35% nuclear staining, consistent with previous studies of MRC-5 fibroblasts (23). In confluent cells, there was no significant increase in the frequency of unstable alleles during the 20-day interval (0–2.5% increase, Table 1), whereas proliferating cells continued to accrue unstable alleles (6.6–14.8% increase, P < 0.05 after the 20-day interval, Table 1 and Fig. 2B, and Supplementary Material, Fig. S1). These results suggest that repeat instability is enhanced by proliferation of these cells.
There was ongoing instability during a final 22 days of cell growth (32 + 20 + 22 = 74 days or 37 doublings), producing many expansions or contractions of more than 200 repeats (Fig. 2B and Supplementary Material, Fig. S1). The cumulative frequency of unstable alleles in proliferating cells was 25.2%, comprising 9.2% expansions and 16.0% contractions (Table 1 and Supplementary Material, Table S1).
Transcription enhances instability of expanded CTG repeats
Next we activated transcription of the expanded repeat by Cre-mediated excision of the puroGFP-transcription-terminator cassette (Fig. 1A). The recombined allele expresses hygromycin resistance (hyg) fused to the 3′-UTR of DMPK, in which the (CTG)800 is inserted. The fibroblast clones were transfected with plasmid encoding Cre, followed by transient selection in hygromycin. Although transcripts with expanded CUG repeats (CUGexp) largely are retained in the nucleus (24,25), enough hygromycin resistance protein was translated to support the selection of recombinant cells. Excision of the puroGFP-transcription-terminator cassette was confirmed by PCR of genomic DNA, loss of GFP activity (data not shown), transcription of hyg (Fig. 1B) and accumulation of CUGexp RNA in nuclear foci (Fig. 1C).
We cultured the cells for 42 days after transfection of Cre, and then compared the repeat instability with non-Cre-transfected cells that were cultured for the same interval. In each of the three clones, the frequency of unstable alleles was significantly increased in recombined cells (P < 0.05 for each clone, Table 1 and Supplementary Material, Table S1). The estimated mutation rate was 3.7% per generation across all clones. Many alleles exhibited changes of more than 200 repeats (Fig. 2B and Supplementary Material, Fig. S1 and Table S1), comprising 23% expansions and 30% contractions across all alleles. The increase in instability did not result from increased proliferation, as the rate of cell doubling was not higher in recombined than non-recombined cells (data not shown). Moreover, the level of transgene expression in recombined cells, as assessed by quantitative RT–PCR (qRT–PCR), was similar to native DMPK in human myoblasts (Supplementary Material, Fig. S2A). Also, the nuclear CUGexp foci in fibroblast clones were less conspicuous than in DM1 cardiac tissue (Supplementary Material, Fig. S2B). These results indicate that transcription-induced instability in the fibroblast clones was not driven by extreme levels of CTG expression.
Previous work has shown that repeat instability depends on the length of a (CTG)•(CAG) tract (5). To determine whether the repeat length may affect transcription-induced destabilization of expanded CTG repeats, we transfected fibroblasts with a (CTG)250 version of plasmid pLC16. We obtained three stably transfected clones for conditional expression of the (CTG)250 repeat. Again we observed repeat instability during cell proliferation that was further enhanced by transcription (Table 1). However, the frequency of unstable alleles and size of the length change were less than in (CTG)800 clones, indicating that transcription stimulates repeat instability in a size-dependent manner.
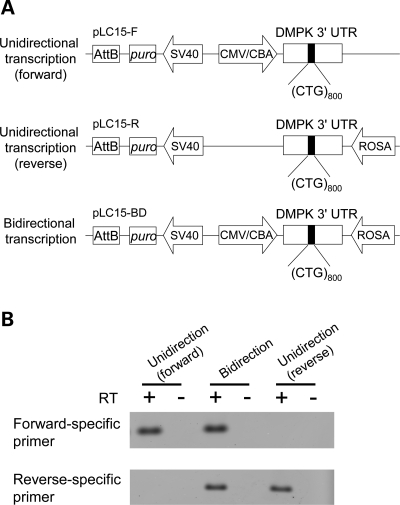
Taken together, these results show that transcription aggravates the instability of (CTG)800 or (CTG)250 at a particular integration site, yielding both expansion and contraction events. It is possible, however, that Cre recombination or hygromycin selection has contributed to repeat instability in these experiments. Since we did not observe growth inhibition in fibroblasts that express CTG repeats, we removed the transcription-terminator and hyg gene from our construct (plasmid pLC15-F, Fig. 3A) and then derived fibroblast clones with constitutive expression of (CTG)800. The transcription of the expanded repeat was confirmed by strand-specific RT–PCR (Fig. 3B). Within the initial 32 days of clonal expansion (∼18 doublings), the instability was much greater in four clones with constitutive expression than in clones without expression of CTG repeats (30–50% unstable alleles for constitutive clones versus 2.5–7.5% for conditional clones prior to Cre activation; P < 0.0001, Fig. 4B compared with Fig. 2B, Supplementary Material, Table S1). As the proliferation rates for conditional and constitutive clones were similar, these results support the conclusion that instability of (CTG)800 is markedly enhanced by transcription.
Figure 3.
(A) Diagram of constructs for constitutive expression of expanded repeats. Forward transcription is driven by the CMV/chicken beta-actin enhancer/promoter. Reverse transcription is driven by the ROSA26 promoter. (B) Strand-specific RT–PCR analysis of transgene expression.
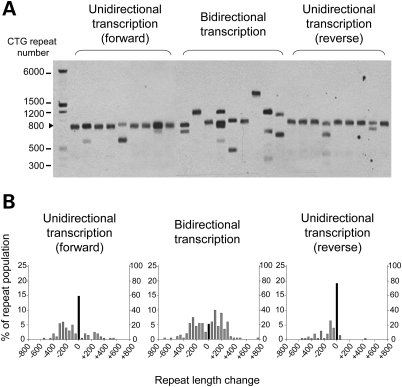
Figure 4.
(A) Small-pool PCR analysis of repeat length in fibroblast clones that have constitutive expression of (CTG)•(CAG) repeats. Representative lanes of small-pool PCR are shown. The scale on the left shows the molecular weight markers converted into numbers of CTG repeats. Cells were harvested after 32 days of clonal expansion. (B) Histograms of repeat length distributions in clones with constitutive expression of (CTG)•(CAG) repeats. The distribution of unstable alleles is shown by gray bars (left vertical axis). The frequency of the stable allele is shown by black bars (right vertical axis). Fifty alleles from four different clones in a same group were analyzed for each construct. Allele lengths are grouped in bins spanning 50 repeats.
These studies have addressed the instability of DM1-like expansions in which the CTG repeat is located in a non-protein coding region. The pathogenic (CTG)•(CAG) expansions that are located in protein coding regions are oriented for expression of CAG repeats and polyglutamine proteins. While these expansions usually have fewer than 100 repeats, evidence suggests that somatic instability can generate alleles with hundreds of CAG repeats in selected neuronal populations (26), although cells with highly expanded alleles may be continuously depleted by neuronal dropout. We therefore examined instability of highly expanded (CTG)•(CAG) repeats with constitutive transcription in the CAG-repeat direction, in the absence of a coding region for polyglutamine translation, so that polyglutamine toxicity would not be limiting. Expression of CAGexp RNA in plasmid pLC15-R is driven by the ROSA26 promoter (Fig. 3A). Transcription in the CAG-repeat direction was confirmed by strand-specific RT–PCR (Fig. 3B). Analysis of clones 32 days after transfection showed 23% unstable alleles. In contrast to cells with conditional expression, where expansion and contraction events occurred with similar frequency, the clones with constitutive expression showed a preponderance of contraction events, especially with transcription in CAG-repeat direction (2.0% expansion versus 21.0% contraction for all alleles, Supplementary Material, Table S1).
Bidirectional transcription further aggravates repeat instability
The (CTG)•(CAG) repeat at the DM1 locus is transcribed in the sense and antisense directions (16). To model this situation, we prepared a construct for convergent bidirectional transcription of (CTG)800 (pLC15-BD, Fig. 3A). We obtained four independent stably transfected fibroblast clones, and confirmed transcription in both directions by strand-specific RT–PCR (Fig. 3B). After 32 days of clonal expansion, 79% of alleles showed instability (P < 0.001 for comparison with forward or reverse transcription, Table 2, Fig. 4B). The estimated mutation rate was 8.3% per generation across all alleles, 30-fold greater than in cells not expressing the (CTG)800 repeat (Supplementary Material, Table S1). Forty-two percent of alleles showed expansion and 37% showed contraction (Fig. 4B and Supplementary Material, Table S1). The average size of the length change was greater with bidirectional transcription than in cells not expressing (CTG)800 (Table 2). These results indicated that bidirectional transcription caused a marked aggravation of repeat instability, including expansions and contractions, producing unstable alleles at a higher frequency than the sum of the frequencies with unidirectional transcription in either direction (Table 2).
Table 2.
Repeat instability in cells with constitutive expression
| % unstable allelesa | Average change of repeat sizeb | |
|---|---|---|
| Unidirectional transcription (forward) | 40.5% | 95 |
| Unidirectional transcription (reverse) | 23.0% | 44 |
| Bidirectional transcription | 79.0%* | 165 |
aA cut-off point of ±25 repeats was used for counting as unstable.
bAmong all alleles (stable + unstable), the average change of repeat size is expressed as the number of repeats.
*P < 0.001, Chi-square test.
DISCUSSION
Progenitor alleles in classical DM1 are usually between 100 and 800 CTG repeats (27,28). In contrast, adult biopsy and autopsy samples typically show 2000 to 6000 CTG repeats in the muscle or heart, with marked variability of the repeat length in different tissues of an individual (9,10,12,14,15). Since the heterogeneity in fetal or neonatal tissues is much less extensive (9,29), it seems likely that somatic instability producing these hyperexpanded alleles has mainly occurred during postnatal life.
The functional significance of somatic instability remains unclear (30), but several lines of evidence suggest that it may have a role in determining the onset or progression of symptoms. In most of the repeat expansions disorders, there is a correlation between disease severity and size of the expanded repeat in peripheral blood cells (31). While the repeat size in blood is not representative of other tissues, the existence of this correlation does suggest that pathogenicity is modulated at some level by the length of the expanded repeat. Microdissection studies in Huntington disease (HD) have shown that somatic expansions are most conspicuous in neuronal populations that are selectively vulnerable to degeneration (26). In HD transgenic mice, the somatic instability was suppressed by ablation of mismatch repair protein MSH2, which caused a delay in the appearance of neuropathologic changes (32,33). In DM1, the mechanisms that underlie RNA dominance would suggest that RNA toxicity is modulated by size of the expanded RNA repeat. Since the toxicity of RNA is determined partly by sequestration of poly(CUG) binding proteins (34), it seems likely that the capacity of a transcript to titrate binding proteins is proportional to the size of its expanded repeat.
If the theory that severity of DM1 is modulated by somatic expansion proves to be correct, it is the transition from hundreds to thousands of repeats, causing a many-fold rise in the cellular burden of CUGexp RNA, which may serve as a critical driver of disease progression. It is this aspect of repeat dynamics that has been difficult to study in experimental systems, owing to the strong tendency of expanded repeats to undergo contraction in conventional cloning vectors and many model systems (30,35). To our knowledge, this is the first experimental system to integrate highly expanded uninterrupted (CAG)•(CTG) repeats in mammalian cells. We focused our initial studies on transcription, because previous work has suggested that transcription-induced instability is particularly important in non-dividing cells (18), the cells that are most affected in DM1. Also, considering that interventions to stabilize the repeat may have the potential to delay or ameliorate symptoms (2), the effect of transcription on repeat instability has important therapeutic implications. For example, it seems possible that suppression of either sense or antisense transcription may provide a safer approach to stabilize repeats than the direct targeting of factors that control DNA replication or repair.
Previous studies of transcription-induced repeat instability used a selectable marker to detect rare contractions of (CAG)95 down to fewer than 39 repeats (18). The basal frequency of contraction events was low, around 4 × 10−7 per generation, rising 15-fold when transcription of the repeat was induced. As contractions of this nature are rarely observed in human (CAG)•(CTG) expansion disorders, and cannot account for somatic growth of repeat expansions in tissue, the generalization of these observations to expansion events, to larger repeat sizes, to non-transformed cells and to other flanking sequence contexts was uncertain. The rate of instability in the current study was higher (up to 8 × 10−2 per generation), which may result from several factors, including the larger size of the expanded repeat, the detection of expansion as well as contraction events, the lower size threshold for detection of instability and the inclusion of flanking sequences from the human DM1 locus. Despite these experimental differences, it is noteworthy that results from the two systems substantially agree. An advantage of the current study is that it quantifies the frequency and size of variant alleles, including expansions as well as contractions, under circumstances (non-transformed cells, highly expanded repeats) that may resemble conditions in DM1 patients. However, the throughput is limited—the current study required the analysis of more than 1800 lanes of Southern blots. Also, the use of primary cells, and the requirement for extended propagation, limits the ability to study the contribution of specific factors. By comparison, the contraction selection assay is more efficient and has clear advantages for studying a broader range of genetic or pharmacologic conditions.
Cells from DM1 individuals and DM1 model systems exhibit different patterns of CTG instability. We observed many discreet changes of more than 200 repeats, suggesting a saltatory process that creates large expansions or contractions in a single step. DM1 patient-derived lymphoblastoid cell lines also showed ‘step-wise' patterns of repeat expansion (36). In contrast, DM1 fetal fibroblasts that carried 216 CTG repeats showed a ‘synchronous' pattern instability and a consistent bias for further expansion (37). The synchronous expansions depended on cell proliferation, and probably resulted from steady accrual of small expansions (one or several repeats) during DNA replication. Similar patterns of ‘synchronous' expansion were previously observed by other investigators (38–40). The underlying mechanisms responsible for synchronous versus stepwise patterns of instability, and the relative frequency of expansion or contraction events, are presently unknown. It is noteworthy, however, that DM1 fibroblasts were converted from synchronous to saltatory patterns when exposed to agents causing arrest of replication fork progression and/or DNA polymerases, triggering step-wise increases of as many as 170 CTG repeats (37). Since the level of DMPK expression in fibroblasts is quite low (41), we speculate that in the present study the transcription of a longer repeat, driven by a different promoter, may have led to collapse of replication forks, perhaps by a mechanism similar to bacteria (17,42), resulting in saltatory patterns of repeat instability. Recent studies of DM1 patient-derived cells mapped two potential replication origins, one upstream and one downstream of the CTG/CAG repeat (43).
A limitation of our study is that every clone represents a different integration event. The phiC31 integrase mediates integration at pseudo-attP sites, with minimal bias for intra-genic versus inter-genic insertions (22). The number of these sites in the human genome is fairly large (>70), with no strong preference for utilization of a single site. For example, none of the pseudo-attP sites accounted for more than 8% of integration events in a previous study (22). Therefore, while we have not mapped integration sites, it is likely that our fibroblast clones have CTG repeat insertions at different chromosomal locations. Despite this variability, it is interesting that instability was a consistent feature among many different clonal isolates, suggesting that marked instability of highly expanded CTG repeats does not have a stringent requirement for specific local chromatin environments, provided that the repeat is transcribed. Alternatively, it is possible that the immediate flanking sequence contained within the construct, and common to all of our clones, has dominated the instability behavior. Of note, our constructs contain the CTCF sites that flank the CTG repeat in DMPK. Recent studies have established CTCF as a key modifier of (CAG)•(CTG) repeat instability at the SCA7 locus (44). Finally, an additional limitation of the study is that the activity of promoters that drive expression of the expanded repeat may differ from activity of native promoters at the DM1 locus.
A key finding of the current study is that bidirectional convergent transcription of an expanded (CTG)•(CAG), a phenomenon observed at several expanded repeat loci (44–46), has markedly increased the frequency of repeat expansions. This finding is consistent with a recent study showing that bidirectional transcription increases the frequency of contractions for a (CAG)95 repeat (47). The mechanism for this effect has not been determined. In S. cerevisiae, convergent bidirectional transcription, resulting in ‘transcriptional collision', can inhibit or stall elongation by RNA polymerase II (48). Such an effect may potentiate the formation of extrahelical loops of CAG or CTG repeats, or trigger the formation of persistent RNA:DNA hybrids (20,49), and thereby promote instability. In the case of immunoglobulin gene variable regions, bidirectional transcription is proposed to make ssDNA more accessible to proteins that carry out somatic recombination (50). This raises the possibility that enhanced access of repair factors to expanded repeat DNA could similarly promote instability (51). Bidirectional transcription of tandem repeats may also recruit factors that mediate epigenetic modifications, such as CpG methylation. In turn, these factors or modifications may modulate the process of repeat instability. Finally, convergent bidirectional transcription would increase positive supercoiling ahead of the respective transcription complexes (52), which may promote the instability of expanded (CAG)•(CTG) repeats (53). Further studies will be needed to determine which of these mechanisms, if any, are operating in DM1.
MATERIALS AND METHODS
Plasmids
Plasmid pLC16 for conditional transcription of expanded CTG repeats (Fig. 1A) was created from plasmid LLC9 (54). pLC16 contains the CMV/chicken beta-actin enhancer/promoter (gift from Dr J. Miyazaki), followed by a floxed selection-stop cassette, a downstream cDNA for hygromycin resistance (hyg), and, finally, the human DMPK 3′-UTR (nt 2096–2841, accession number NM_004409), modified with restriction sites for insertion of expanded CTG repeats (see below). Plasmids in this series also contain an attB element for integration in the host genome by PhiC31 integrase (55). The selection-stop cassette contains the cDNA encoding a puromycin resistance-eGFP fusion protein (56) followed by the ‘triple-stop' transcription terminator (a series of three SV40 polyadenylation signals, Dr C. Lobe) (57). Previously, we found that insertion of this triple-stop element between the CMV/chicken beta-actin enhancer/promoter and a downstream reporter caused >105-fold repression of luciferase activity (C. Thornton and C. Jiang, unpublished data).
Plasmid pLC15-F for constitutive transcription of CTG repeats in the forward direction (equivalent to the sense DMPK transcript, producing a CUG repeat RNA) was derived from pLC16 by removal of the selection-stop cassette and hygromycin resistance gene. A separate transcription unit encoding puromycin resistance (derived from pTREpuro, Clontech) was inserted at an upstream position (Fig. 3A). For constitutive transcription of (CTG)•(CAG) repeats in both directions, plasmid LC15-BD was derived from pLC15-A by inserting the ROSA26 promoter (gift from Dr E. Sandgren) downstream of DMPK 3′-UTR (Fig. 3A). Plasmid LC15-R for constitutive transcription in the reverse direction (equivalent to the antisense DMPK transcript, producing a CAG repeat RNA) was made from pLC15-BD by removing the CMV/chicken beta-actin enhancer/promoter (Fig. 3A).
The expanded CTG repeats were generated in the ‘repeat donor' plasmid, pDWD, by cell-free cloning using Phi29 polymerase and rolling circle amplification as previously described (21). Versions of pDWD were made with repeat tracts of 250 or 800 repeats. The length of the hyper-expanded repeat was too long for full sequence confirmation. However, repeats generated by this method are uninterrupted when sequenced from either end (21), and the method for expanding the repeat tract is not expected to generate sequence interruptions. The repeat tract was excised from pDWD by digestion with BstEII and BstXI and then directionally ligated into these same sites in the DMPK 3′-UTR of pLC plasmids. This procedure inserts the expanded repeat at its normal position in the DMPK 3′-UTR, except that the following sequence is included at the 5′-end of the repeat tract: GCCTTAGCCACGCCACTGGCTTAAGTCGGTAACCCACCCGGTG. The ligation product was directly used to transfect fibroblasts.
Cell culture and transfection
Human fetal lung fibroblasts (MRC-5 cells) were obtained from the Coriell Institute at passage 27. Cells were cultured in MEM (Gibco) supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 mg/ml streptomycin. 5 × 105 cells were co-transfected with 200 ng of repeat-containing plasmid and 1.8 µg of plasmid PhiC31o encoding the PhiC31 integrase (58). Transfection was performed using Nucleofector technology (Lonza) according to the manufacturer's program U-23. Stably transfected clones were selected with puromycin (0.5 µg/ml). To excise the selection-stop cassette, cells were nucleofected with pHSVCreWT encoding Cre recombinase (gift from Dr W. Bowers). Cells with recombination were selected using hygromycin B (50 µg/ml). Excision of the selection-stop cassette was confirmed by PCR using forward primer: 5′-TTCGGCTTCTGGCGTGTGAC-3′ and reverse primers 5′-TAGGTCAGGGTGGTCACGAG-3′ or 5′-GTTGGCGACCTCGTATTGGG-3′. For experiments on non-proliferating cells, fibroblasts were grown to confluence. Residual cell division in contact-inhibited cells was assessed using a BrdU Staining Kit (Invitrogen) according to the manufacturer's instructions.
Repeat length analysis
Genomic DNA was extracted from MRC-5 clones using the Gentra Puregene Cell Kit (Qiagen). Expanded CTG repeats were sized by small-pool PCR followed by Southern blot. Small-pool PCR was performed according to the method of Gomes-Pereira (59) with some modifications. Briefly, the transgene sequence containing the CTG repeat was amplified by Expand Long Template PCR System (Roche) using primers 5′-ACCCTAGAACTGTCTTCGACTCC-3′ and 5′-TTCCCGAGTAAGCAGGCAGAG-3′ through 24 rounds of PCR. The small-pool PCR products were resolved on 0.7% agarose gels buffered with 40 mm Tris–acetate, 1 mm EDTA. PCR products were transferred to nylon membranes (Roche) and then hybridized with digoxigenin (DIG)-labeled (CAG)7 LNA probe as described previously (14). At least 40 alleles were analyzed for each clone. Differences in the expansion length and heterogeneity were tested using Chi-square analysis. An estimate of mutation frequency was calculated under the conservative assumption that each variant allele resulted from a single expansion or contraction event.
RT–PCR
RNA was harvested with TRIZOL (Molecular Research Center) and treated with Turbo DNA-free (Ambion) to remove contaminating DNA. One microgram of total RNA was primed with oligo(dT) plus random hexamers and reverse transcribed with Superscript II (Invitrogen) followed by treatment with RNase H. For RT–PCR to examine expression of hygromycin resistance cassette, the hyg sequence was amplified for 28 cycles using primers 5′-CTACTTCGAGCGGAGGCATC-3′ and 5′-ACACAGCCATCGGTCCAGAC-3′. As an internal control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified as described previously (60). For strand-specific RT–PCR, cDNAs complementary to both sense and antisense strand RNA were synthesized with the tagged primers 5′-CGACTGGAGCACGAGGACACTGATCTAGAGAATAGGAACTTCGGAATAG-3′, and 5′-CGACTGGAGCACGAGGACACTGAGGCTGAGGCCCTGACGTGGATGGGCAAAC-3′, respectively. The first-round PCR was carried out with the tagged primer LK (CGACTGGAGCACGAGGACACTGA) and a sense-specific (5′-TGACTGACCGCGTTACTCCCACA-3′) or antisense-specific primer (5′- GGAGCAAAACAGAATGGCTGGC-3′) for 15 cycles. The second round of PCR used the LK primer and nested primers for the sense strand, 5′-GCAACGTGCTGGTTATTGTG-3′ or the antisense strand, 5′- TTCCCGAGTAAGCAGGCAGAG-3′ for 20 additional cycles. PCR products for both studies were analyzed on agarose gels using ImageQuant software (Molecular Dynamics).
mRNA quantification
Quantitative RT–PCR of transgene mRNA was performed using TaqMan Gene Expression assays on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). The probes and primers hybridized to identical sequence in the transgene and DMPK mRNA, in the 3′-UTR. The level of endogenous DMPK plus transgene-derived mRNA was normalized to the mean expression level of two housekeeping transcripts, 18S rRNA and RNA polymerase II polypeptide A (POLR2A). DMPK primer sequences were 5′-CTATCGTTGGTTCGCAAAGTG-3′ and 5′-GCAAATTTCCCGAGTAAGCAG-3′. The probe sequence was 5′-AAGCTTTCTTGTGCATGACGCCC-3′.
FISH
FISH was performed as previously described (8) except that cells were fixed for 15 min at RT with 3% paraformaldehyde and permeabilized with 0.5% Triton X-100.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the University of Rochester Paul D. Wellstone Muscular Dystrophy Cooperative Research Center (NIH/NS048843) with support from the National Institute of Health (AR049077, AR48143); the Muscular Dystrophy Association (to M.N.); the Saunders Family Research Fund; and postdoctoral fellowships (to M.N.) from the Cell Science Research Foundation and the Uehara Memorial Foundation.
ACKNOWLEDGEMENTS
The authors would like to thank Drs Junichi Miyazaki, Corinne Lobe, Eric Sandgren and William Bowers for providing plasmids, and members of the Thornton and Pearson laboratories for helpful suggestions and discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.La Spada A.R., Taylor J.P. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 2010;11:247–258. doi: 10.1038/nrg2748. doi:10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopez Castel A., Cleary J.D., Pearson C.E. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. doi:10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 3.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T., et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. doi:10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 4.Redman J.B., Fenwick R.G., Jr, Fu Y.H., Pizzuti A., Caskey C.T. Relationship between parental trinucleotide GCT repeat length and severity of myotonic dystrophy in offspring. JAMA. 1993;269:1960–1965. doi:10.1001/jama.269.15.1960. [PubMed] [Google Scholar]
- 5.Kovtun I.V., McMurray C.T. Features of trinucleotide repeat instability in vivo. Cell Res. 2008;18:198–213. doi: 10.1038/cr.2008.5. doi:10.1038/cr.2008.5. [DOI] [PubMed] [Google Scholar]
- 6.Dion V., Wilson J.H. Instability and chromatin structure of expanded trinucleotide repeats. Trends Genet. 2009;25:288–297. doi: 10.1016/j.tig.2009.04.007. doi:10.1016/j.tig.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krahe R., Ashizawa T., Abbruzzese C., Roeder E., Carango P., Giacanelli M., Funanage V.L., Siciliano M.J. Effect of myotonic dystrophy trinucleotide repeat expansion on DMPK transcription and processing. Genomics. 1995;28:1–14. doi: 10.1006/geno.1995.1099. doi:10.1006/geno.1995.1099. [DOI] [PubMed] [Google Scholar]
- 8.Jiang H., Mankodi A., Swanson M.S., Moxley R.T., Thornton C.A. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. doi:10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 9.Ashizawa T., Dubel J.R., Harati Y. Somatic instability of CTG repeat in myotonic dystrophy. Neurology. 1993;43:2674–2678. doi: 10.1212/wnl.43.12.2674. [DOI] [PubMed] [Google Scholar]
- 10.Thornton C.A., Johnson K., Moxley R.T. Myotonic dystrophy patients have larger CTG expansions in skeletal muscle than in leukocytes. Ann. Neurol. 1994;35:104–107. doi: 10.1002/ana.410350116. doi:10.1002/ana.410350116. [DOI] [PubMed] [Google Scholar]
- 11.Wong L.J., Ashizawa T., Monckton D.G., Caskey C.T., Richards C.S. Somatic heterogeneity of the CTG repeat in myotonic dystrophy is age and size dependent. Am. J. Hum. Genet. 1995;56:114–122. [PMC free article] [PubMed] [Google Scholar]
- 12.Zatz M., Passos-Bueno M.R., Cerqueira A., Marie S.K., Vainzof M., Pavanello R.C. Analysis of the CTG repeat in skeletal muscle of young and adult myotonic dystrophy patients: when does the expansion occur? Hum. Mol. Genet. 1995;4:401–406. doi: 10.1093/hmg/4.3.401. doi:10.1093/hmg/4.3.401. [DOI] [PubMed] [Google Scholar]
- 13.Martorell L., Monckton D.G., Gamez J., Johnson K.J., Gich I., de Munain A.L., Baiget M. Progression of somatic CTG repeat length heterogeneity in the blood cells of myotonic dystrophy patients. Hum. Mol. Genet. 1998;7:307–312. doi: 10.1093/hmg/7.2.307. doi:10.1093/hmg/7.2.307. [DOI] [PubMed] [Google Scholar]
- 14.Nakamori M., Sobczak K., Moxley R.T., 3rd, Thornton C.A. Scaled-down genetic analysis of myotonic dystrophy type 1 and type 2. Neuromuscul. Disord. 2009;19:759–762. doi: 10.1016/j.nmd.2009.07.012. doi:10.1016/j.nmd.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López Castel A., Nakamori M., Tomé S., Chitayat D., Gourdon G., Thornton C.A., Pearson C.E. Expanded CTG repeat demarcates a boundary for abnormal CpG methylation in myotonic dystrophy patient tissues. Hum. Mol. Genet. 2010 doi: 10.1093/hmg/ddq427. doi:10.1093/hmg/ddq427 (Online published on November 1, 2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho D.H., Thienes C.P., Mahoney S.E., Analau E., Filippova G.N., Tapscott S.J. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. doi:10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Bowater R.P., Jaworski A., Larson J.E., Parniewski P., Wells R.D. Transcription increases the deletion frequency of long CTG.CAG triplet repeats from plasmids in Escherichia coli. Nucleic Acids Res. 1997;25:2861–2868. doi: 10.1093/nar/25.14.2861. doi:10.1093/nar/25.14.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin Y., Dion V., Wilson J.H. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. doi:10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y., Wilson J.H. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. doi:10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y., Dent S.Y., Wilson J.H., Wells R.D., Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc. Natl Acad. Sci. USA. 2010;107:692–697. doi: 10.1073/pnas.0909740107. doi:10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osborne R.J., Thornton C.A. Cell-free cloning of highly expanded CTG repeats by amplification of dimerized expanded repeats. Nucleic Acids Res. 2008;36:e24. doi: 10.1093/nar/gkn025. doi:10.1093/nar/gkn025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chalberg T.W., Portlock J.L., Olivares E.C., Thyagarajan B., Kirby P.J., Hillman R.T., Hoelters J., Calos M.P. Integration specificity of phage phiC31 integrase in the human genome. J. Mol. Biol. 2006;357:28–48. doi: 10.1016/j.jmb.2005.11.098. doi:10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- 23.Sheerin A., Thompson K.S., Goyns M.H. Altered composition and DNA binding activity of the AP-1 transcription factor during the ageing of human fibroblasts. Mech. Ageing Dev. 2001;122:1813–1824. doi: 10.1016/s0047-6374(01)00319-0. doi:10.1016/S0047-6374(01)00319-0. [DOI] [PubMed] [Google Scholar]
- 24.Davis B.M., McCurrach M.E., Taneja K.L., Singer R.H., Housman D.E. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl Acad. Sci. USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. doi:10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamshere M.G., Newman E.E., Alwazzan M., Athwal B.S., Brook J.D. Transcriptional abnormality in myotonic dystrophy affects DMPK but not neighboring genes. Proc. Natl Acad. Sci. USA. 1997;94:7394–7399. doi: 10.1073/pnas.94.14.7394. doi:10.1073/pnas.94.14.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shelbourne P.F., Keller-McGandy C., Bi W.L., Yoon S.R., Dubeau L., Veitch N.J., Vonsattel J.P., Wexler N.S., Arnheim N., Augood S.J. Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum. Mol. Genet. 2007;16:1133–1142. doi: 10.1093/hmg/ddm054. doi:10.1093/hmg/ddm054. [DOI] [PubMed] [Google Scholar]
- 27.Jansen G., Willems P., Coerwinkel M., Nillesen W., Smeets H., Vits L., Howeler C., Brunner H., Wieringa B. Gonosomal mosaicism in myotonic dystrophy patients: involvement of mitotic events in (CTG)n repeat variation and selection against extreme expansion in sperm. Am. J. Hum. Genet. 1994;54:575–585. [PMC free article] [PubMed] [Google Scholar]
- 28.Martorell L., Gamez J., Cayuela M.L., Gould F.K., McAbney J.P., Ashizawa T., Monckton D.G., Baiget M. Germline mutational dynamics in myotonic dystrophy type 1 males: allele length and age effects. Neurology. 2004;62:269–274. doi: 10.1212/wnl.62.2.269. [DOI] [PubMed] [Google Scholar]
- 29.Martorell L., Johnson K., Boucher C.A., Baiget M. Somatic instability of the myotonic dystrophy (CTG)n repeat during human fetal development. Hum. Mol. Genet. 1997;6:877–880. doi: 10.1093/hmg/6.6.877. doi:10.1093/hmg/6.6.877. [DOI] [PubMed] [Google Scholar]
- 30.Pearson C.E., Nichol E.K., Cleary J.D. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. doi:10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 31.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. doi:10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 32.Manley K., Shirley T.L., Flaherty L., Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in untington disease transgenic mice. Nat. Genet. 1999;23:471–473. doi: 10.1038/70598. doi:10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 33.Wheeler V.C., Lebel L.A., Vrbanac V., Teed A., te Riele H., MacDonald M.E. Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum. Mol. Genet. 2003;12:273–281. doi: 10.1093/hmg/ddg056. doi:10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- 34.Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. doi:10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang S., Jaworski A., Ohshima K., Wells R.D. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. doi:10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- 36.Khajavi M., Tari A.M., Patel N.B., Tsuji K., Siwak D.R., Meistrich M.L., Terry N.H., Ashizawa T. “Mitotic drive” of expanded CTG repeats in myotonic dystrophy type 1 (DM1) Hum. Mol. Genet. 2001;10:855–863. doi: 10.1093/hmg/10.8.855. doi:10.1093/hmg/10.8.855. [DOI] [PubMed] [Google Scholar]
- 37.Yang Z., Lau R., Marcadier J.L., Chitayat D., Pearson C.E. Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type 1 disease locus in human cells. Am. J. Hum. Genet. 2003;73:1092–1105. doi: 10.1086/379523. doi:10.1086/379523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wohrle D., Kennerknecht I., Wolf M., Enders H., Schwemmle S., Steinbach P. Heterogeneity of DM kinase repeat expansion in different fetal tissues and further expansion during cell proliferation in vitro: evidence for a casual involvement of methyl-directed DNA mismatch repair in triplet repeat stability. Hum. Mol. Genet. 1995;4:1147–1153. doi: 10.1093/hmg/4.7.1147. doi:10.1093/hmg/4.7.1147. [DOI] [PubMed] [Google Scholar]
- 39.Peterlin B., Logar N., Zidar J. CTG repeat analysis in lymphocytes, muscles and fibroblasts in patients with myotonic dystrophy. Pflugers Arch. 1996;431:R199–R200. doi: 10.1007/BF02346337. doi:10.1007/BF02346337. [DOI] [PubMed] [Google Scholar]
- 40.Furling D., Coiffier L., Mouly V., Barbet J.P., St Guily J.L., Taneja K., Gourdon G., Junien C., Butler-Browne G.S. Defective satellite cells in congenital myotonic dystrophy. Hum. Mol. Genet. 2001;10:2079–2087. doi: 10.1093/hmg/10.19.2079. doi:10.1093/hmg/10.19.2079. [DOI] [PubMed] [Google Scholar]
- 41.Storbeck C.J., Sabourin L.A., Waring J.D., Korneluk R.G. Definition of regulatory sequence elements in the promoter region and the first intron of the myotonic dystrophy protein kinase gene. J. Biol. Chem. 1998;273:9139–9147. doi: 10.1074/jbc.273.15.9139. doi:10.1074/jbc.273.15.9139. [DOI] [PubMed] [Google Scholar]
- 42.Parniewski P., Bacolla A., Jaworski A., Wells R.D. Nucleotide excision repair affects the stability of long transcribed (CTG*CAG) tracts in an orientation-dependent manner in Escherichia coli. Nucleic Acids Res. 1999;27:616–623. doi: 10.1093/nar/27.2.616. doi:10.1093/nar/27.2.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cleary J.D., Tome S., Lopez Castel A., Panigrahi G.B., Foiry L., Hagerman K.A., Sroka H., Chitayat D., Gourdon G., Pearson C.E. Tissue- and age-specific DNA replication patterns at the CTG/CAG-expanded human myotonic dystrophy type 1 locus. Nat. Struct. Mol. Biol. 2010;17:1079–1087. doi: 10.1038/nsmb.1876. doi:10.1038/nsmb.1876. [DOI] [PubMed] [Google Scholar]
- 44.Libby R.T., Hagerman K.A., Pineda V.V., Lau R., Cho D.H., Baccam S.L., Axford M.M., Cleary J.D., Moore J.M., Sopher B.L., et al. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 2008;4:e1000257. doi: 10.1371/journal.pgen.1000257. doi:10.1371/journal.pgen.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moseley M.L., Zu T., Ikeda Y., Gao W., Mosemiller A.K., Daughters R.S., Chen G., Weatherspoon M.R., Clark H.B., Ebner T.J., et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet. 2006;38:758–769. doi: 10.1038/ng1827. doi:10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 46.Ladd P.D., Smith L.E., Rabaia N.A., Moore J.M., Georges S.A., Hansen R.S., Hagerman R.J., Tassone F., Tapscott S.J., Filippova G.N. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. doi:10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- 47.Lin Y., Leng M., Wan M., Wilson J.H. Convergent transcription through a long CAG tract destabilizes repeats and induces apoptosis. Mol. Cell Biol. 2010;30:4435–4451. doi: 10.1128/MCB.00332-10. doi:10.1128/MCB.00332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prescott E.M., Proudfoot N.J. Transcriptional collision between convergent genes in budding yeast. Proc. Natl Acad. Sci. USA. 2002;99:8796–8801. doi: 10.1073/pnas.132270899. doi:10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reddy K., Tam M., Bowater R., Barber M., Tomlinson M., Edamura K.N., Wang Y.H., Pearson C.E. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq935. doi:10.1093/nar/gkq935 (Online published on November 4, 2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolland D.J., Wood A.L., Johnston C.M., Bunting S.F., Morgan G., Chakalova L., Fraser P.J., Corcoran A.E. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol. 2004;5:630–637. doi: 10.1038/ni1068. doi:10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- 51.Slean M.M., Panigrahi G.B., Ranum L.P., Pearson C.E. Mutagenic roles of DNA “repair” proteins in antibody diversity and disease-associated trinucleotide repeat instability. DNA Repair (Amst.) 2008;7:1135–1154. doi: 10.1016/j.dnarep.2008.03.014. doi:10.1016/j.dnarep.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 52.Liu L.F., Wang J.C. Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. doi:10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Napierala M., Bacolla A., Wells R.D. Increased negative superhelical density in vivo enhances the genetic instability of triplet repeat sequences. J. Biol. Chem. 2005;280:37366–37376. doi: 10.1074/jbc.M508065200. doi:10.1074/jbc.M508065200. [DOI] [PubMed] [Google Scholar]
- 54.Wheeler T.M., Sobczak K., Lueck J.D., Osborne R.J., Lin X., Dirksen R.T., Thornton C.A. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. doi:10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thyagarajan B., Olivares E.C., Hollis R.P., Ginsburg D.S., Calos M.P. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol. Cell Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. doi:10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abbate J., Lacayo J.C., Prichard M., Pari G., McVoy M.A. Bifunctional protein conferring enhanced green fluorescence and puromycin resistance. Biotechniques. 2001;31:336–340. doi: 10.2144/01312st05. [DOI] [PubMed] [Google Scholar]
- 57.Lobe C.G., Koop K.E., Kreppner W., Lomeli H., Gertsenstein M., Nagy A. Z/AP, a double reporter for cre-mediated recombination. Dev. Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. doi:10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- 58.Raymond C.S., Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS ONE. 2007;2:e162. doi: 10.1371/journal.pone.0000162. doi:10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gomes-Pereira M., Bidichandani S.I., Monckton D.G. Analysis of unstable triplet repeats using small-pool polymerase chain reaction. Methods Mol. Biol. 2004;277:61–76. doi: 10.1385/1-59259-804-8:061. [DOI] [PubMed] [Google Scholar]
- 60.Kimura T., Nakamori M., Lueck J.D., Pouliquin P., Aoike F., Fujimura H., Dirksen R.T., Takahashi M.P., Dulhunty A.F., Sakoda S. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum. Mol. Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. doi:10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.