Abstract
Organogenesis is a multistage process, but it has been difficult, by conventional analysis, to separate stages and identify points of transition in developmentally complex organs or define genetic pathways that regulate pattern formation. We performed a detailed time-series examination of global gene expression during kidney development and then represented the resulting data as self-organizing maps (SOMs), which reduced more than 30,000 genes to 650 metagenes. Further clustering of these maps identified potential stages of development and suggested points of stability and transition during kidney organogenesis that are not obvious from either standard morphological analyses or conventional microarray clustering algorithms. We also performed entropy calculations of SOMs generated for each day of development and found correlations with morphometric parameters and expression of candidate genes that may help in orchestrating the transitions between stages of kidney development, as well as macro- and micropatterning of the organ.
INTRODUCTION
In vitro and knockout studies, along with classical morphologic approaches, have provided a wealth of information about the genes and encoded proteins regulating mammalian organogenesis. Expression profiling has implicated additional genes, which, when analyzed in light of current understanding of biochemistry and gene regulation, suggest key pathways in organogenesis. However, an overall perspective of the multistage process of organ development and patterning is often lost in the details.
We present a broader view of organogenesis obtained through the comparison and analysis of visual representations of gene expression patterns in the developing kidney generated by unsupervised learning neural networks known as self-organizing maps (SOMs) (1). SOMs cluster genes by placing those with similar temporal gene expression profiles together into “metagenes” and creating images that can be termed metagene mosaics or portraits. SOMs have been used for the analysis of patterns of gene expression data (2–4). For example, SOMs were used to analyze hematopoietic differentiation on the basis of gene expression data (5), and this method was also applied to gene expression data from human lung cancers as a means of differential diagnosis (6). Here, we have applied this type of analytical approach to gene expression data derived from a time series of organogenesis. To obtain SOMs, we used microarray gene expression data and Gene Expression Dynamics Inspector (GEDI), a program for analysis of high-dimensional data (6–8). After doing a detailed global time-series analysis of SOMs reflecting metanephric, postnatal, and adult kidney gene expression, we performed entropy calculations for each SOM. Entropy is presumed to measure the variation in a series of states (in this case, the time course of kidney development) and could conceivably be an indicator of the amount of information and differences in organization carried by sets of genes during phenotypic changes (9, 10). We have considered the metagene mosaics as individual units, determined the entropy in each, and sought correlations of the entropies with a large set of quantitative data that parameterizes kidney development at the same time points from which the SOMs were derived. Together, the various approaches suggested distinct stages of kidney organogenesis. It also suggested genes potentially involved in transitions between stages of organogenesis and suggested pathways that may be linked to the morphological patterning of organs. A number of the genes have already been implicated in organogenesis by in vitro and knockout studies. Although much experimental validation is necessary, the approaches presented here not only suggest linkages of known pathways to specific stages of kidney development but also potentially to macropatterned events, such as definition of cortex and medulla, as well as more micropatterned events, such as the formation of glomeruli.
RESULTS
Metagene mosaics
The gene expression profiles of 15 time points (from biological triplicates) during rat pre- and postnatal kidney development, including just before the onset of metanephric kidney development [embryonic day (e) 12, through birth and adulthood] were visualized on 15 SOMs (Fig. 1). An SOM is based on an unsupervised learning approach for clustering large amounts of data. During the organization of its internal neural network, an SOM assigns large groups of input objects (here, genes) to smaller sets of “tiles” or through the transformation of N genes from gene microarray analyses to M gene clusters or metagenes (1). These metagenes are then distributed onto two-dimensional maps or mosaics, each representing the gene expression profile for a certain day (1). These mosaics, which in this case can be thought of as genetic portraits, contain a comparatively small number of metagenes in relation to the entire set of genes (fig. S4).
Fig. 1.
Metagene portraits of kidney development. (A) The top panels are the pseudocolored SOMs of the 650 metagene profiles generated from the gene expression data for the developing rat kidneys from e12 to adult (ad). Each tile represents a cluster of genes and the same clusters are positioned in the same space on the SOM allowing direct comparison of expression of clustered genes at the different time points. Red represents high expression, blue represents low expression. (B) Bottom panels are confocal fluorescent micrographs of coronal sections of a representative kidney from nearly each day of metanephric development. Sections were cut with a vibratome, fixed, and stained with fluorescently labeled lectins, which bind to either branching UBs (Dolichos biflorus lectin, green) or developing nephron orglomeruli (peanut agglutinin, red). nb, newborn; w1, 1week postpartum;w4, 4 weeks postpartum; ad, adult.
In our time-series analysis, each SOM represents a mosaic composed of 26 × 25 tiles, which contains 650 metagenes (see Materials and Methods for justification for analysis of 26 × 25 tiles). The number of genes in each metagene varies depending on the similarity distance of differential expression for the approximately 30,000 genes represented on the microarrays. The fixed coordinates of metagenes allowed for comparisons between the SOMs (Fig. 1). Put more simply, this means that the genes in the tile in the upper right corner of each image are the same and their expression pattern can be compared across each time point. The mosaics for each time point could also be grouped based on overall similarities in the patterns of metagene expression. By grouping and comparing the mosaics, we attempted to focus on key distinctions between different developmental stages.
Visual inspection of the patterns of the SOMs suggested that kidney organogenesis could be divided into six to eight distinct stages. The approximately similar mosaic patterns of the SOMs (and thus, patterns of gene expression) displayed in e13 through e16 suggest that these days of development represent a distinct stage in the organogenesis of the kidney. Similarly, additional groupings of developmental stages were identified based on their similar mosaic patterns: e17 through e18, e19 through e22, and 4-week-old (w4) and adult. In contrast, the mosaic patterns for e12, newborn (nb), and 1-week-old (w1) have little apparent similarity with the other mosaics and thus appear to correspond to distinct stages of development. Although certain stages (for example, e12 versus e13) are expected, based on readily apparent differences in morphology, to have different gene expression profiles, several other stages, despite having SOM portraits with clear transitions, are not obvious by conventional techniques.
Stages of kidney development
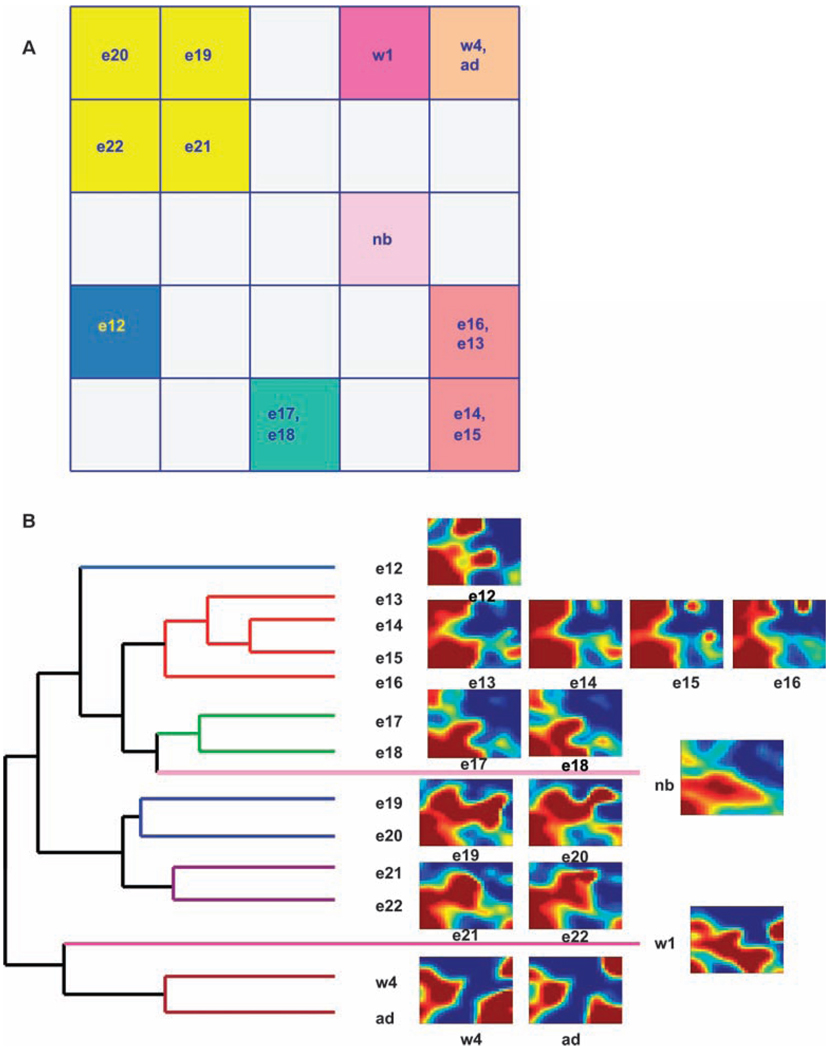
The grouping of the mosaic patterns into distinct stages was supported by two different techniques: (i) second-level SOM analysis (Fig 2A) and (ii) hierarchical clustering of the set of 15 metagene mosaics (Fig. 2B). For the second-level SOM analysis, each mosaic of 650 metagenes corresponding to the 15 days of development represents a “second-level metagene,” that is, a “metametagene.” The mosaics of the 650 metagenes from the 15 time points were then assigned to 5 × 5 SOM grids. The resulting second-level SOM grouped metagene mosaics into six to seven distinct stages of kidney development.
Fig. 2.
(A) Clustering of SOM metametagenes indicates stages of kidney organogenesis that group together and reveals underlying similarities in gene expression between and among stages. Each group is represented by a different color. Although some groups are adjacent, the chosen assignment into a different group seems more consistent with the overall data in B and A. Clustering of filtered data indicated a similar set of stages (see Materials and Methods and fig. S5A). (B) Dendogram showing clustering of metametagenes with the use of GeneSpring GX.
Hierarchical clustering of the metagene mosaics for each time point, represented as a hierarchical tree, or dendrogram, further supports the stages indicated by the dual methods of visual inspection and second-level SOM representation. As shown, the hierarchical tree is consistent with six to seven distinct stages and supports, but does not exactly coincide with, the prior grouping (compare Fig. 2, A and B). This grouping was not evident after conventional hierarchical clustering of the whole set of the transcripts.
Entropy analysis
The potential use of the SOMs rests in the fact that such an analysis transforms the gene expression data for many genes into a two-dimensional discrete map in a topologically ordered fashion. Nevertheless, it remains difficult to mathematically analyze the properties of SOMs. One intriguing possibility is entropy analysis. It has been shown that entropy-based analysis of images (and thus, potentially the SOMs) could represent a useful approach in considering the variation in a series of events, such as in a set of time-series data. By determining tile intensity and applying image histogram analysis of the SOMs as a probability distribution, a graphical determination of the variation contained in the developmental time series of SOMs was obtained (Fig. 3, A and B).
Fig. 3.
(A) Graph of Tsallis entropy of the metagenes corresponds to the stages of development (B). A similar profile was obtained for filtered data (see Materials and Methods and fig. S5B). Stage 1, e12; stage 2, e13 to e16; stage 3, e17 to e18; stage 4, e19 to e22; stage 5, nb (P0 to P1); stage 6, w1; stage 7, w4 to ad. Representative SOMs from each stage are shown in B. In B, purple arrows represent potential negative feedback loops that potentially stabilize the previous stage. In A, the blue downward arrow represents an extrapolation of the original slope of the entropy line after birth.
Entropy is conventionally viewed as a measure of the disorder in a system; thus, a higher entropy value might be associated with a lower state of organization, and a lower entropy value might be associated with a higher state of organization. That said, biological interpretations of entropy must be made with extreme caution. Nevertheless, a correlation emerged between the apparent visual “stability” of the images and the entropy values for the metagene mosaics. Periods of little change in entropy could conceivably correspond to periods of stable growth of the organ, whereas periods of changing entropy (either increasing or decreasing) may reflect transitional stages. Specifically, two regions of apparent stability (no change in entropy) correlated with days e13 through e16 and e19 through e22 (Fig. 3A), each of which represented a presumed stage. In addition, a period of stability is also suggested for week 4 after birth and later in adulthood (Fig. 3A). The periods of transition when gene expression patterns are highly dissimilar between adjacent stages appear to be reflected by changes in entropy and correspond to e12, e17 to e18, birth, and w1.
Correlation between “reverse” entropy and morphometric parameters during organogenesis
Kidney development involves two main morphogenetic processes: ureteric bud (UB) branching morphogenesis and nephron formation derived from the metanephric mesenchyme (MM). At e13, the UB has penetrated the MM, undergone its initial dichotomous branching event, and is now a T-shaped structure (Fig. 1B). Repetitive rounds of branching continue as development progresses with the tips of the UB extending toward the periphery of the organ. Rapprochement of the growing collecting ducts begins to occur around e17 to e18 (Fig. 1). Nephron formation as indicated by peanut agglutinin staining of structures that include glomeruli (the microscopic filtration apparatus of an individual nephron) becomes detectable from e17. Also at this stage, the kidney cortex and medulla become delineated; this corticomedullary definition is crucial for mature kidney function.
Although there is morphological support for the notion of developmental stages during organogenesis of the kidney and other tissues, the beginning and end of such stages are difficult to define morphologically. In fact, during kidney development many different basic morphogenetic processes occur simultaneously with differentiation proceeding from the interior of the kidney to the periphery. Nevertheless, the metagene and second-level analysis described, as well as the SOM entropy calculations, suggest that global gene expression can be used to separate these stages (even in the face of multiple morphogenetic processes occurring simultaneously). However, it is not known to what extent aggregate patterns of gene expression changes correspond to morphological changes. It is presumed that these gene expression changes reflect the processes that they drive, but it has been difficult, through profiling global gene expression patterns, to identify pathways involved in particular morphological processes evident by standard histological or marker analysis (11, 12). Therefore, to investigate potential correlations of the SOMs with morphological changes during development, quantitative morphometric analysis of the kidney sections was performed. Among the measures obtained for nearly each of the 15 time points were the following geometric parameters: kidney section area, kidney section perimeter, kidney section aspect ratio, kidney section major and minor axes, kidney roundness, cortex area, and medulla area. Among the structural-topological parameters measured were the number of tips, the tip density (number of tips per unit area), the number of glomeruli, and the glomerular density (glomeruli per unit area).
To obtain overlay plots of morphological parameters and entropy, we multiplied the previous entropy curve by (−1). For simplicity, the plot of reverse entropy is normalized. This type of reverse entropy is sometimes thought to be related to “organization” or “self-organization” of a system.
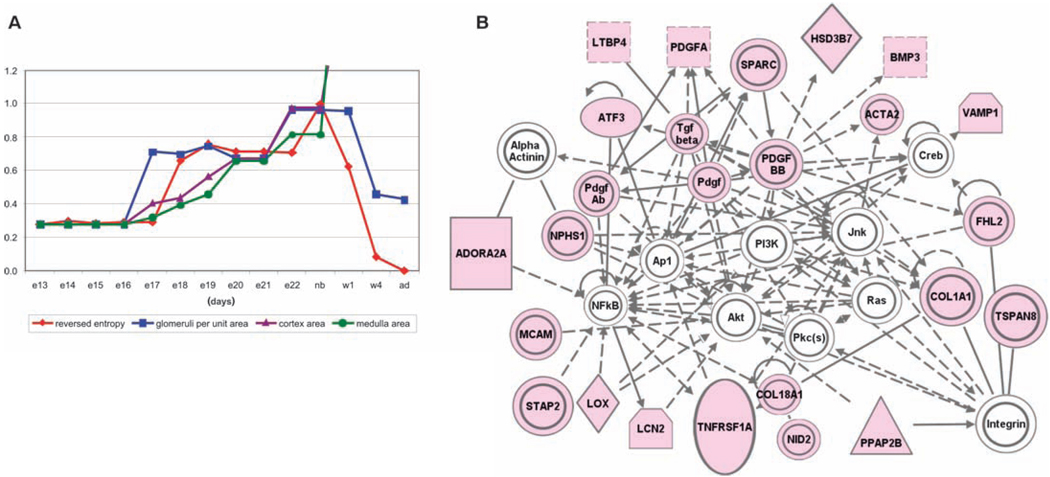
If reverse entropy is viewed as a measure of the organization of the system, an increase in reverse entropy could be interpreted as reflecting a period of change leading to increased organization. A decrease could conceivably reflect decreased organization. Correlation coefficients for reverse entropy were calculated for all measured parameters and the highest correlation of reverse entropy was with glomerular density (Fig. 4A). (In other words, the Tsallis entropy had a strong negative correlation with this morphological parameter.) Correlation analysis also showed a strong positive correlation (r ~ 0.85) between the profiles of glomerular density and a subset of all genes (table S1) (see Materials and Methods and fig. S4 for details of the correlation analysis.)
Fig. 4.
A potential gene network involved in glomerular development. (A) Correlation of SOM reverse entropy with morphological data of kidney development. The y axis is normalized reverse entropy and normalized morphological parameters; the x axis is time. Several morphometric parameters of kidney development correlate with the reversed entropy in the gene expression data. Glomerular density (glomeruli per unit area) shows the strongest correlation at all developmental times, whereas the growth parameters of cortex area and medulla area diverge with the SOM reverse entropy after e22. Spearman rank correlation between entropies for unfiltered and filtered data was 0.92 (fig. S5B). (B) Network representation of genes (by IPA) whose expression has a high correlation with glomerular density and thus the reverse entropy (table S1). Light magenta nodes include those genes previously implicated in glomerular development, UB branching, nephron number, or glomerular injury-repair response; white are additional genes suggested by the network analysis. Legends for network symbols (IPA) are presented in fig. S3. ADORA2A was excluded by filtering.
Network representations of candidate genes
Network representation (Fig. 4B) suggests that a group of genes exhibiting expression profiles that correlated with the glomerular density are part of a potential gene signaling network that includes genes believed to be responsible for organ development, in general, and kidney development, in particular. For example, nephrin (abbreviated NPHS1), collagen 18 (abbreviated COL18A1), and platelet-derived growth factors (abbreviated PDGF BB, PDGFA, and Pdgf)—which have been implicated in formation of the glomerulus—were among the identified genes (13–25). In addition, other genes that correlated with glomerular density, such as transforming growth factor β (abbreviated Tgf beta) and bone morphogenetic protein–3 (BMP-3), have been proposed to play a major role in branching morphogenesis and regulating nephron number (of which glomerular density is likely a reflection) (26, 27), or which function in the injury-repair response of the glomerulus, including diabetic nephropathy (28–30), or have been implicated in both processes.
Two aspects of organ development relevant to this type of analysis are (i) organ growth with homomorphic correspondence of each subsequent growth stage to the previous one and (ii) structural-topological development (patterning), wherein organizational changes in subsystems within the larger organ undergo distinct changes leading to patterning. It seems likely that different sets of genes will be responsible for these two aspects of organogenesis. A comparison of the properties of the reverse entropy with glomerular density, cortex area, and medullary area is shown in Fig. 4A. The reverse SOM entropic changes in the gene expression patterns for the whole organ between e13 and e20 correlate with these three morphometric parameters (but less so with the others that were measured). Changing glomerular density is a manifestation micropatterning of the kidney and approximately correlates with the reverse entropy of the SOMs during the entire period (e13 through adult). In addition, in the initial stages (up to e22), the profile of SOM reverse entropy correlates not only with glomerular density (structural-topological parameter) but also with the cortex and medulla average areas (geometric growth parameters). For a period after birth, these lines of development follow different destinations. The structural-topological parameter does not appear to undergo more change, suggesting that the self-organization of the organ is largely complete for this period of development, whereas the geometric parameters continue to increase. However, great caution must be exercised in the interpretation of these correlations between morphological or morphometric data and the calculated Tsallis entropies. Finer analysis of a much larger set of parameters will be required to further evaluate the broader possible utility of this approach for analyzing organogenesis and other normal and pathological states (31).
The differences between SOM portraits (and the corresponding SOM entropies) for different stages of organogenesis may help clarify the stages of kidney development, and these maps may also reflect signaling networks related to the profiles of gene expression seen in these stages. To evaluate this possibility, we examined the expressed genes found within a central region in the SOMs (outlined in Fig. 5). This region had been selected based on the genes listed in table S1 and their inclusion in specific metagene tiles. In other words, the size and shape of this region is a reflection of the candidate genes and the metagenes to which they map. Within this region, a set of genes was identified based on their changing pattern of expression, a decreasing expression from e13 to e16 followed by an increase in expression at e17 (Fig. 5A). This pattern of gene expression suggests an important role for this set of genes in morphogenetic processes critical for the transition from the relatively stable phase of development at e13 through e16 (as reflected by the metagene portraits and entropy calculations) to a new state at e17. Changes in expression for a selected set of genes at this time period were preliminarily supported by real-time reverse transcription polymerase chain reaction (RT-PCR) (table S2).
Fig. 5.
(A) Metagene profiles of days e13 to e17. For reasons explained in the text and fig. S4, genes found in the outlined region in each metagene mosaic were analyzed. (The small box in the center is excluded, as no specified genes fell into it.) (B and C) Putative networks of genes (and their expression: green, decreased expression at this time point; red, increased expression) found in the outlined region of the metagene profiles (A). (B) Expression at e13 to e16 and (C) expression at e17 of development. IBSP was excluded by filtering (see Materials and Methods). Legends for network symbols (IPA) are presented in fig. S3.
DISCUSSION
Among the candidate set of genes identified are several that regulate branching morphogenesis (members of the TGF-β superfamily, BMP1 and TGFβ1) (26, 27, 32), as well as those that are closely linked to genes thought to play key roles in other morphogenetic events. Network representation (Fig. 5, B and C) suggests links between several of these genes and those known to be involved in certain critical morphogenetic processes.
To speculate upon one example, the gene encoding FK-506 binding protein 8 (FKBP8), which is expressed in the kidney and in metastatic cancer cells (33–35), is expressed from e13 to e16 during kidney development and plays a role in dorsal-ventral patterning of the neural tube through regulation of apoptosis in this epithelial tissue (36–38). FKBP8, a transmembrane protein found in the mitochondria and endoplasmic reticulum, inhibits apoptosis in the neuroepithelium by recruiting the antiapoptotic protein, Bcl-2, to the mitochondria (37, 39). Taken together with the fact that mutational deletion of Bcl-2 results in renal dysplasia and the formation of renal cysts (40–43), this finding raises the possibility that FKBP8 is somehow involved in the patterning of the kidney through a mechanism similar to that in renal tubular epithelial cells. Pathway analysis implicated FKBP8 in a network with a few genes thought to play roles in epithelial biogenesis, polarization, or both (Fig. 5, B and C). For example, the Rho effector protein rhotekin (RTKN), which localizes to the Golgi apparatus [in association with PIST (a PDZ domain–containing protein)] in nonpolarized Madin-Darby canine kidney (MDCK) cells and at the adherens junction in polarized MDCK cells, was identified (44, 45). Rhotekin also binds to Lin-7B, another PDZ protein, which is thought to be involved in the polarization of neuronal cells (46). Thus, together with the fact that the e17 kidney appears to be active in the formation and development of the nephron (Fig. 1), one idea is that these genes might play roles in the development of the nascent tubular epithelium of the nephron, or in the maintenance and differentiation of the more mature epithelial UB, or both.
Our first- and second-level analyses of the SOMs for the kidney development time series revealed periods of instability between more stable periods (Figs. 1 and 2). This seems consistent with activation of previously unidentified pathways or an alteration of their configuration leading to major changes in gene expression. For example, certain genes expressed at e17 through e18 and visualized in the SOM portrait could conceivably function in the transition of gene expression seen at e19; the SOM portraits may thus be useful in identifying genetic pathways involved in regulating these transitions. A similar argument would apply to other transitions including the radical one at birth (Fig. 3A). Although negative feedback could potentially stabilize a stage of development, allowing the increase in size, volume, and number of structural elements (quantitative growth), positive feedback might generate changes, allowing transition to a different stage (qualitative jump). Thus, as indicated in Fig. 3B, we suggest that the organization of specific signaling networks that combine negative and positive feedback mechanisms create a situation where the metagene set remains relatively stable during e13 through e16 stages of development and then, perhaps through minimal alterations of a small group of genes in this metagene set, enable a transition to a different contour leading to previously unidentified structural-topological changes (patterning).
This view appears to be supported by the SOM entropy curve, which, after stabilization from e13 to e16, declines as the kidney develops (presumably somehow reflecting increasing organization in the developing organ) until the day of birth when there is an increase in the entropy of the system (Fig. 3A). Extrapolation of the original slope of the line after birth suggests that the entropy should continue to decrease (Fig. 3A). If it is reasonable to interpret the Tsallis entropy curve as we have, a continuing decrease in entropy would not be surprising, as the entropy decrease (as shown by the blue arrow) would presumably represent the developmentally regulated genes whose functions should be expected to be ending or, perhaps, switching to a stable maintenance function. One potential explanation is that increase in the entropy represents a burst in expression of those genes necessary for the proper renal function of the now independent organism, which has also gone from a wet environment (in utero) to a dry environment (11). The kidney, the key organ mediating fluid and electrolyte balance, is critical for dealing with the physiological burden imposed by this abrupt environmental shift. In this regard, it would be interesting to perform a similar analysis for lung development.
We have noted some intriguing correlations. If validated, the approach we have used here, in the context of kidney organogenesis, can, in principle, be used to analyze many types of time-series microarray data to define stages and transitions. It may also be used to narrow down sets of candidate pathways guiding these transitions, as well as those that might be responsible for particular morphological processes (for example, the developmental changes in glomerular density and the establishment of boundaries between cortex and medulla). As quantitative morphometric parameterization improves and is able to reliably measure specific events in a larger dynamic system involving dozens of cell types undergoing different kinds of morphogenetic processes (such as kidney organogenesis or regeneration after an acute injury), a finer molecular understanding of these complicated problems may be achieved with the type of approach we describe here. Nevertheless, the abstraction of time series of large data sets into entropy and comparison with morphological and, potentially, functional and other information requires much more exploration and validation.
MATERIALS AND METHODS
RNA isolation
Total RNA was isolated from the samples with the use of the Strataprep Total RNA isolation kit (Stratagene, La Jolla, CA). Triplicate samples for each condition consisting of 10 to 200 µg of total RNA were prepared, and IVT and hybridization to GeneChip Rat Genome 230 2.0 Arrays (Affymetrix Inc., 2007) was performed by the GeneChip Microarray Core at the UCSD Moores Cancer Center as previously described (11, 12, 47, 48).
Time points correspond to the initiation of metanephric kidney development (e12) through e22, as well as postgestational time points. For e12, presumptive metanephric tissue was isolated as the mesonephros. For the other stages, metanephroi composed of the UB and MM were isolated. For the early time points of metanephric development (e12, e13, e14, and e15), isolated tissues were pooled to obtain sufficient quantities of RNA for analysis. Later time points were unpooled samples. Newborn (nb) is P0 to P1.
Microarray analysis
The microarray data were obtained from the analysis of more than 31,000 probe sets representing more than 30,000 transcripts and variants from more than 28,000 well-substantiated rat genes found on the GeneChip Rat Genome 230 2.0Arrays (Affymetrix Inc., 2007). Primary data, as well as their initial processing (local background correction and removal of outlier spots), were obtained through the GeneChip Microarray Core at the UCSD Moore Cancer Center with proprietary software.
The data set is described as a matrix: X = {xs,k}, k = 1,…, 15; s = 1,…, 31,099, where k represents samples and s, spots or probes; each kth value is an average of three identical samples. The data set included 15 samples (45GeneChips) corresponding to the 15 days of Rattus norvegicus kidney development described in the text. To make outlier detection more reliable and to save expensive samples, we used three different methods relying on different principles: histogram comparison, Bland-Altman plots (49), and linear correlation analysis. The analysis was done for whole samples as well as for each gene in the time series.
The average values for each probe obtained from the different samples were calculated for every day of development. Gene expression data for the time series of each probe were normalized; also, the quantile normalization for each probe across the time series was used, with the formula NEi,j = Ei,j/Emax,j; i ∈ {1, …, 15}; j ∈ {1, …, 31,099}, where NEi,j and Ei,j are a normalized and ameasured expression for a specified sample for each probe, correspondingly, and Emax,j is the maximum expression for each probe. After normalization, all genes were expressed in the range 0 to 1 so as not to lose decreased-expression genes with significant differential expression.
We analyzed the data in both an unfiltered and filtered (fig. S5) fashion. Shown in the text are the unfiltered data because there are a number of filtering methods, each with its own potential for introducing some bias. Nevertheless, when we filtered the data by absent-call substitution with averages of remaining present-calls (for those genes with two or more present calls over the time course), similar clusterization of the time points, as well as the Tsallis entropy curves, was obtained (fig. S5).
Thus the data were prepared using two strategies whereby (i) the initial set of unfiltered expressions values were normalized; genes having absent calls were included in the metagene; (ii) the initial normalized set of genes was filtered, eliminating the genes having all absent calls and/or the genes having a single present call with the rest absent calls through the time series. In the remaining data, gene expression values with absent calls were substituted using the average value of the present calls for each particular gene. Figures S5A and S5B show that the SOM map is a robust method and the results for defining stages of development are similar for both data preparation strategies.
Representation of microarray data
Dynamic microarray data analysis was performed with the SOM algorithm and its implementation, GEDI (6–8), a program for the clustering of high-dimensional data (for example, cDNA microarrays), which provides integral visual presentation of large data sets (that is, SOMs). The genes found in each of the 650 metagene tiles are listed in table S3.
A metagenome has been constructed with GeneSpring GX (Agilent Technologies Inc, 2008). The second-level hierarchical dendograms shown in Fig. 2B were obtained with GeneSpring GX with the use of this metagenome. We created networks from genes selected by SOM analysis with Ingenuity Pathway Analysis (IPA) software, version 6.5 (Ingenuity Systems, Inc., 2008). The list of genes obtained from the SOM analysis was submitted to IPA. The examples of the resulting putative networks are shown in Figs. 4 and 5. See the Supplementary Materials for a brief outline of the approach to constructing and interpreting SOMs (fig. S4).
Calculation of entropies
We calculated the Tsallis entropy of metagene mosaics through the analysis of the tile intensity histograms derived from each SOM. The Tsallis entropy is a generalization of the standard Boltzmann-Gibbs entropy (50) and seems to have applicability to this type of problem (51). It is defined in discrete cases by the following formula: in which p denotes the probability distribution of interest and q is a real parameter (in our calculations, q = 0.25; when q is close to 1, the Tsallis entropy converts to the standard Boltzmann-Gibbs entropy).
Entropies were calculated for the SOMs having 16 × 15, 26 × 25, and 36 × 35 tiles (fig. S2). The general shapes of the entropy curves during kidney development are similar. The map of 26 × 25 tiles was chosen because it shows sharper differences between entropy values during the days of kidney development. For example, for development during the days e17, e18, and e19, three distinct stages are evident on the map of 26 × 25 tiles, whereas on the map of 36 × 35 tiles these differences are not as obvious. The 16 × 15 tile maps also have lower resolution at apparent transition points. More details about this analysis are presented in the Supplementary Materials (fig. S4).
Morphometric analysis
Samples were fixed overnight at 4°C in 4%paraformaldehyde in phosphate-buffered saline (PBS) and embedded in 4%agarose in PBS. Coronal serial sections (200 µm thick) cut with a vibratome were stored in PBS, and the middle coronal section of each kidney was selected and processed for lectin staining. Briefly, sections were incubated with 1%gelatin and 0.075%saponin in PBS for 30 min at 37°C and then washed (2 × 5 min) in neuroaminidase buffer [150 mM NaCl/50 mM sodium acetate, pH 5.5, in PBST (PBS with 0.1% Triton X-100)]. Sections were left overnight in neuraminidase (1 U/ml, Sigma) in neuraminidase buffer at 37°C before being transferred to fluorescently labeled peanut agglutinin (50 µg/ml, Sigma) and Dolichos biflorus agglutinin (50 µg/ml, Sigma) in PBST overnight at 4°C. Sections were washed three times for 1 hour each in PBST before mounting with Eukitt Mounting Medium (Electron Microscopy Sciences).
Images were captured with an Eclipse 80i microscope with D-Eclipse camera (Nikon) and EZ-C1 3.20 imaging software. Single optical sections were imaged with an open pinhole. Montage images were created for the older kidney sections with Image-Pro Plus, version 6.0 (Media Cybernetics). Images were processed by Image-Pro Plus, version 6.0 (Media Cybernetics), and the following parameters were measured. Geometric parameters included kidney section area; kidney section perimeter; kidney section aspect ratio; kidney section major and minor axes; kidney roundness; cortex area; medulla area; and Feret whole kidney, cortex, and medulla areas. Structural-topological parameters included number of tips, number of tips per area unit, number of glomeruli, and number of glomeruli per area unit (fig. S1).
As shown in Fig. 4A, Spearman rank correlation coefficients were calculated between reversed entropy and the measured morphological parameters (52). This calculation revealed that the parameter with the highest correlation was glomerular density. This result for glomerular density was then correlated in a similar fashion with the entire set of gene expression data. This second correlation revealed a set of genes potentially involved in glomerular development (table S1).
Preliminary analysis of gene expression by real-time RT-PCR
Expression of selected genes was analyzed by real-time RT-PCR. Metanephroi were obtained from e13.5, e15.5, and e17.5, and complementary DNAs were synthesized with SuperScript III first-strand synthesis systems for RT-PCR (Invitrogen, Carlsbad, CA). Primers for gene amplification were designed with Primer3 (www-genome.wi.mit.edu). Real-time PCR was carried out with an ABI 7500 Real-Time PCR system with an annealing temperature at 55°C. Increased or decreased expression was determined with reference to the expression of rat Gapdh.
Supplementary Material
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/1/49/ra16/DC1
Fig. S1. Process of obtaining measurements of glomeruli and tips from morphological images.
Fig. S2. Three entropy profiles calculated from SOMs with different map resolutions: 16 × 15, 26 × 25, and 36 × 35 tiles.
Fig. S3. Legend for network shapes and relationships presented in Figs. 4 and 5 (using IPA from Ingenuity Systems, www.ingenuity.com, with permission).
Fig. S4. Schematic of generation of SOMs, entropy calculations, correlations, and network analysis.
Fig. S5. Robustness of SOM clustering.
Table S1. List of genes with high correlation coefficients (>0.85) between expression profiles and glomerular density values during kidney development.
Table S2. Preliminary analysis of gene expression by RT-PCR.
Table S3. Affymetrix probe IDs of the genes found in each of the 650 metagene tiles.
REFERENCES AND NOTES
- 1.Kohonen T. Self-Organizing Maps. ed. 3. Heidelberg/New York: Springer; 2001. [Google Scholar]
- 2.Fernandez EA, Balzarini M. Improving cluster visualization in self-organizing maps: Application in gene expression data analysis. Comput. Biol. Med. 2007;37:1677–1689. doi: 10.1016/j.compbiomed.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Herrero J, Dopazo J. Combining hierarchical clustering and self-organizing maps for exploratory analysis of gene expression patterns. J. Proteome Res. 2002;1:467–470. doi: 10.1021/pr025521v. [DOI] [PubMed] [Google Scholar]
- 4.Papadimitriou S, Likothanassis SD. Kernel-based self-organized maps trained with supervised bias for gene expression data analysis. J. Bioinform. Comput. Biol. 2004;1:647–680. doi: 10.1142/s021972000400034x. [DOI] [PubMed] [Google Scholar]
- 5.Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: Methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Y, Eichler GS, Feng Y, Ingber DE, Huang S. Towards a holistic, yet gene-centered analysis of gene expression profiles: A case study of human lung cancers. J. Biomed. Biotechnol. 2006;2006:69141. doi: 10.1155/JBB/2006/69141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eichler GS, Huang S, Ingber DE. Gene Expression Dynamics Inspector (GEDI): For integrative analysis of expression profiles. Bioinformatics. 2003;19:2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- 8.Fujita A, Sato JR, Ferreira CE, Sogayar MC. GEDI: A user-friendly toolbox for analysis of large-scale gene expression data. BMC Bioinformatics. 2007;8:457. doi: 10.1186/1471-2105-8-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuhrman S, Cunningham MJ, Wen X, Zweiger G, Seilhamer JJ, Somogyi R. The application of Shannon entropy in the identification of putative drug targets. Biosystems. 2000;55:5–14. doi: 10.1016/s0303-2647(99)00077-5. [DOI] [PubMed] [Google Scholar]
- 10.Kouznetsova VL, Rakov MA. Self-Organization in Technical Systems. Kiev: Naukova Dumka; 1987. p. p. 17. (Russian) [Google Scholar]
- 11.Stuart RO, Bush KT, Nigam SK. Changes in global gene expression patterns during development and maturation of the rat kidney. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5649–5654. doi: 10.1073/pnas.091110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuart RO, Bush KT, Nigam SK. Changes in gene expression patterns in the ureteric bud and metanephric mesenchyme in models of kidney development. Kidney Int. 2003;64:1997–2008. doi: 10.1046/j.1523-1755.2003.00383.x. [DOI] [PubMed] [Google Scholar]
- 13.Betsholtz C, Karlsson L, Lindahl P. Developmental roles of platelet-derived growth factors. Bioessays. 2001;23:494–507. doi: 10.1002/bies.1069. [DOI] [PubMed] [Google Scholar]
- 14.Carroll TJ, McMahon AP. Secreted molecules in metanephric induction. J. Am. Soc. Nephrol. 2000;11 Suppl. 16:S116–S119. [PubMed] [Google Scholar]
- 15.Davies JA. Morphogenesis of the metanephric kidney. Scientific World Journal. 2002;2:1937–1950. doi: 10.1100/tsw.2002.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dressler GR. The cellular basis of kidney development. Annu. Rev. Cell Dev. Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- 17.Jalanko H, Patrakka J, Tryggvason K, Holmberg C. Genetic kidney diseases disclose the pathogenesis of proteinuria. Ann. Med. 2001;33:526–533. doi: 10.3109/07853890108995962. [DOI] [PubMed] [Google Scholar]
- 18.Levéen P, Pekny M, Gebre-Medhin S, Swolin B, Larsson E, Betsholtz C. Mice deficient for PDGF B show renal, cardiovascular, and hematological abnormalities. Genes Dev. 1994;8:1875–1887. doi: 10.1101/gad.8.16.1875. [DOI] [PubMed] [Google Scholar]
- 19.Lindahl P, Hellström M, Kalén M, Karlsson L, Pekny M, Pekna M, Soriano P, Betsholtz C. Paracrine PDGF-B/PDGF-Rβ signaling controls mesangial cell development in kidney glomeruli. Development. 1998;125:3313–3322. doi: 10.1242/dev.125.17.3313. [DOI] [PubMed] [Google Scholar]
- 20.Quaggin SE. Transcriptional regulation of podocyte specification and differentiation. Microsc. Res. Tech. 2002;57:208–211. doi: 10.1002/jemt.10076. [DOI] [PubMed] [Google Scholar]
- 21.Quaggin SE, Kreidberg JA. Development of the renal glomerulus: Good neighbors and good fences. Development. 2008;135:609–620. doi: 10.1242/dev.001081. [DOI] [PubMed] [Google Scholar]
- 22.Sampogna RV, Nigam SK. Implications of gene networks for understanding resilience and vulnerability in the kidney branching program. Physiology (Bethesda) 2004;19:339–347. doi: 10.1152/physiol.00025.2004. [DOI] [PubMed] [Google Scholar]
- 23.Schedl A, Hastie ND. Cross-talk in kidney development. Curr. Opin. Genet. Dev. 2000;10:543–549. doi: 10.1016/s0959-437x(00)00125-8. [DOI] [PubMed] [Google Scholar]
- 24.Soriano P. Abnormal kidney development and hematological disorders in PDGF beta-receptor mutant mice. Genes Dev. 1994;8:1888–1896. doi: 10.1101/gad.8.16.1888. [DOI] [PubMed] [Google Scholar]
- 25.Vainio S, Lin Y. Coordinating early kidney development: Lessons from gene targeting. Nat. Rev. Genet. 2002;3:533–543. doi: 10.1038/nrg842. [DOI] [PubMed] [Google Scholar]
- 26.Bush KT, Sakurai H, Steer DL, Leonard MO, Sampogna RV, Meyer TN, Schwesinger C, Qiao J, Nigam SK. TGF-β superfamily members modulate growth, branching, shaping, and patterning of the ureteric bud. Dev. Biol. 2004;266:285–298. doi: 10.1016/j.ydbio.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 27.Santos OF, Nigam SK. HGF-induced tubulogenesis and branching of epithelial cells is modulated by extracellular matrix and TGF-β. Dev. Biol. 1993;160:293–302. doi: 10.1006/dbio.1993.1308. [DOI] [PubMed] [Google Scholar]
- 28.Chen S, Jim B, Ziyadeh FN. Diabetic nephropathy and transforming growth factor-β: Transforming our view of glomerulosclerosis and fibrosis build-up. Semin. Nephrol. 2003;23:532–543. doi: 10.1053/s0270-9295(03)00132-3. [DOI] [PubMed] [Google Scholar]
- 29.Goldfarb S, Ziyadeh FN. TGF-β: A crucial component of the pathogenesis of diabetic nephropathy. Trans. Am. Clin. Climatol. Assoc. 2001;112:27–32. [PMC free article] [PubMed] [Google Scholar]
- 30.Ziyadeh FN. Mediators of diabetic renal disease: The case for TGF-β as the major mediator. J. Am. Soc. Nephrol. 2004;15 Suppl. 1:S55–S57. doi: 10.1097/01.asn.0000093460.24823.5b. [DOI] [PubMed] [Google Scholar]
- 31.Monte JC, Sakurai H, Bush KT, Nigam SK. The developmental nephrome: Systems biology in the developing kidney. Curr. Opin. Nephrol. Hypertens. 2007;16:3–9. doi: 10.1097/MNH.0b013e3280118a5a. [DOI] [PubMed] [Google Scholar]
- 32.Meyer TN, Schwesinger C, Bush KT, Stuart RO, Rose DW, Shah MM, Vaughn DA, Steer DL, Nigam SK. Spatiotemporal regulation of morphogenetic molecules during in vitro branching of the isolated ureteric bud: Toward a model of branching through budding in the developing kidney. Dev. Biol. 2004;275:44–67. doi: 10.1016/j.ydbio.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 33.Fong S, Mounkes L, Liu Y, Maibaum M, Alonzo E, Desprez PY, Thor AD, Kashani-Sabet M, Debs RJ. Functional identification of distinct sets of antitumor activities mediated by the FKBP gene family. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14253–14258. doi: 10.1073/pnas.2332307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam E, Martin M, Wiederrecht G. Isolation of a cDNA encoding a novel human FK506-binding protein homolog containing leucine zipper and tetratricopeptide repeat motifs. Gene. 1995;160:297–302. doi: 10.1016/0378-1119(95)00216-s. [DOI] [PubMed] [Google Scholar]
- 35.Pedersen KM, Finsen B, Celis JE, Jensen NA. muFKBp38: A novel murine immunophilin homolog differentially expressed in Schwannoma cells and central nervous system neurons in vivo. Electrophoresis. 1999;20:249–255. doi: 10.1002/(SICI)1522-2683(19990201)20:2<249::AID-ELPS249>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 36.Bulgakov OV, Eggenschwiler JT, Hong DH, Anderson KV, Li T. FKBP8 is a negative regulator of mouse sonic hedgehog signaling in neural tissues. Development. 2004;131:2149–2159. doi: 10.1242/dev.01122. [DOI] [PubMed] [Google Scholar]
- 37.Shirane M, Ogawa M, Motoyama J, Nakayama KI. Regulation of apoptosis and neurite extension by FKBP38 is required for neural tube formation in the mouse. Genes Cells. 2008;13:635–651. doi: 10.1111/j.1365-2443.2008.01194.x. [DOI] [PubMed] [Google Scholar]
- 38.Wong RL, Wlodarczyk BJ, Min KS, Scott ML, Kartiko S, Yu W, Merriweather MY, Vogel P, Zambrowicz BP, Finnell RH. Mouse Fkbp8 activity is required to inhibit cell death and establish dorso-ventral patterning in the posterior neural tube. Hum. Mol. Genet. 2008;17:587–601. doi: 10.1093/hmg/ddm333. [DOI] [PubMed] [Google Scholar]
- 39.Shirane M, Nakayama KI. Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat. Cell Biol. 2003;5:28–37. doi: 10.1038/ncb894. [DOI] [PubMed] [Google Scholar]
- 40.Sorenson CM. Nuclear localization of β-catenin and loss of apical brush border actin in cystic tubules of bcl-2 −/− mice. Am. J. Physiol. 1999;276:F210–F217. doi: 10.1152/ajprenal.1999.276.2.F210. [DOI] [PubMed] [Google Scholar]
- 41.Sorenson CM, Padanilam BJ, Hammerman MR. Abnormal postpartum renal development and cystogenesis in the bcl-2 −/− mouse. Am. J. Physiol. 1996;271:F184–F193. doi: 10.1152/ajprenal.1996.271.1.F184. [DOI] [PubMed] [Google Scholar]
- 42.Sorenson CM, Sheibani N. Focal adhesion kinase, paxillin, and bcl-2: Analysis of expression, phosphorylation, and association during morphogenesis. Dev. Dyn. 1999;215:371–382. doi: 10.1002/(SICI)1097-0177(199908)215:4<371::AID-AJA8>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 43.Ziehr J, Sheibani N, Sorenson CM. Alterations in cell-adhesive and migratory properties of proximal tubule and collecting duct cells from bcl-2 −/− mice. Am. J. Physiol. Renal Physiol. 2004;287:F1154–F1163. doi: 10.1152/ajprenal.00129.2004. [DOI] [PubMed] [Google Scholar]
- 44.Ito H, Iwamoto I, Mizutani K, Morishita R, Deguchi T, Nozawa Y, Asano T, Nagata K. Possible interaction of a Rho effector, Rhotekin, with a PDZ-protein, PIST, at synapses of hippocampal neurons. Neurosci. Res. 2006;56:165–171. doi: 10.1016/j.neures.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 45.Ito H, Iwamoto I, Morishita R, Nozawa Y, Asano T, Nagata K. Identification of a PDZ protein, PIST, as a binding partner for Rho effector Rhotekin: Biochemical and cell-biological characterization of Rhotekin-PIST interaction. Biochem. J. 2006;397:389–398. doi: 10.1042/BJ20052015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sudo K, Ito H, Iwamoto I, Morishita R, Asano T, Nagata K. Identification of a cell polarity-related protein, Lin-7B, as a binding partner for a Rho effector, Rhotekin, and their possible interaction in neurons. Neurosci. Res. 2006;56:347–355. doi: 10.1016/j.neures.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 47.Rosines E, Sampogna RV, Johkura K, Vaughn DA, Choi Y, Sakurai H, Shah MM, Nigam SK. Staged in vitro reconstitution and implantation of engineered rat kidney tissue. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20938–20943. doi: 10.1073/pnas.0710428105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakurai H, Bush KT, Nigam SK. Heregulin induces glial cell line-derived neurotrophic growth factor-independent, non-branching growth and differentiation of ureteric bud epithelia. J. Biol. Chem. 2005;280:42181–42187. doi: 10.1074/jbc.M507962200. [DOI] [PubMed] [Google Scholar]
- 49.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- 50.Tsallis C. Possible generalization of Boltzmann-Gibbs statistics. J. Stat. Phys. 1988;52:479–487. [Google Scholar]
- 51.Maszczyk T, Duch W. Artificial Intelligence and Soft Computing: Lecture Notes in Computer Science. vol. 5097/2008. Berlin/Heidelberg: Springer; 2008. pp. 643–651. [Google Scholar]
- 52.Spearman C. The proof and measurement of association between two things. Am. J. Psychol. 1904;15:72–101. [PubMed] [Google Scholar]
- 53.We thank E. Rosines for help with image processing and the Ghosh Laboratory at University of California, San Diego, for use of their vibratome. We also thank J. Monte, A. Dnyanmote, and T. F. Gallegos for helpful comments and M. Bettilyon for help with manuscript preparation. This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases grants R01-DK065831, R01-DK057286, and HL035018. This paper is dedicated to the memory of Robert O. Stuart.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.