Abstract
Excretion of uric acid, a compound of considerable medical importance, is largely determined by the balance between renal secretion and reabsorption. The latter process has been suggested to be principally mediated by urate transporter 1 (URAT1; slc22a12), but the role of various putative urate transporters has been much debated. We have characterized urate handling in mice null for RST, the murine ortholog of URAT1, as well as in those null for the related organic anion transporters Oat1 and Oat3. Expression of mRNA of other putative urate transporters (UAT, MRP2, MRP4, Oatv1) was unaffected in the knockouts, as were general indexes of renal function (glomerular filtration rate, fractional excretion of fluid and electrolytes). While mass spectrometric analyses of urine and plasma revealed significantly diminished renal reabsorption of urate in RST-null mice, the bulk of reabsorption, surprisingly, was preserved. Oat1- and Oat3-null mice manifested decreased secretion rather than reabsorption, indicating that these related transporters transport urate in the “opposite” direction to RST. Moreover, metabolomic analyses revealed significant alteration in the concentration of several molecules in the plasma and urine of RST knockouts, some of which may represent additional substrates of RST. The results suggest that RST, Oat1, and Oat3 each contribute to urate handling, but, at least in mice, the bulk of reabsorption is mediated by a transporter(s) that remains to be identified. We discuss the data in the context of recent human genetic studies that suggest that the magnitude of the contribution of URAT1 to urate reabsorption might vary with ethnic background.
Keywords: urate, organic anion transport, renal reabsorption, renal secretion, metabolomics
Uric acid is generated by the degradation of purines and cleared by further degradation to allantoin, catalyzed by hepatic uricase, or by excretion, which occurs largely in the kidney. Humans, unlike most mammals, lack uricase (68), and therefore renal excretion is a critical determinant of systemic urate levels (22, 46, 57). Elevation in these levels, as reflected by hyperuricemia (increased plasma urate), contributes to the development of gout and kidney stones and has also been linked to the metabolic syndrome (insulin resistance, hypertension, dyslipidemia) and cardiovascular disease (reviewed in Refs. 18, 43, 45, 58). Thus mechanisms of urate transport in the kidney might have considerable clinical implications.
Net renal excretion of urate appears to be dependent on the balance between filtration, secretion, and reabsorption, with the latter two processes (though acting in opposition) largely occurring in overlapping segments of the proximal tubule of the kidney and, indeed, potentially in the same proximal tubular cell (22, 43, 46). Recent studies (reviewed in Ref. 22) suggest that in humans the reabsorptive component of this overall process is principally mediated by urate transporter 1 (URAT1; slc22a12), a transmembrane protein on the apical surface of the proximal tubule. Specifically, in vitro-expressed URAT1 manifests functional properties consistent with those of proximal tubular urate reabsorption, including anion exchange and inhibition (in cis) by uricosuric agents (compounds that increase urinary urate) such as probenecid or benzbromarone (13); moreover, patients with congenital lack of renal urate reabsorption (idiopathic renal hypouricemia) have been found to carry loss-of-function mutations in URAT1 (13).
URAT1 is a member of a large family of non-primary active transport proteins, organic anion transporters (OATs; slc22), which are mostly expressed in the barrier epithelia of the major excretory organs of the body, the kidney and the liver (reviewed in Refs. 9, 14, 67). Four of the OATs have been definitively localized to the proximal tubule, Oat1 [slc22a6; originally identified as NKT (36 –39)] and Oat3 [slc22a8; originally identified as ROCT (5)] to its basolateral aspect (25, 31, 42, 60), and URAT1, as noted above, and OAT4 (slc22a11) to its apical (luminal) surface (3, 13). Although there were initially some contradictory data, each of these proximal tubular OATs is now believed to function via anion exchange (12, 13, 47, 52, 55). Accordingly, their direction of transport is largely determined by the concentration gradients of the transported and countertransported substrates. In other words, provided the appropriate concentration gradients, either basolateral or apical OATs can mediate either cellular entry or efflux of their substrates, which translates at the level of the whole kidney into the potential ability to participate in either renal secretion or reabsorption (67). Notably each of Oat1, Oat3, and OAT4, in addition to URAT1, have been shown to mediate urate transport in vitro (4, 20, 29), which, in accordance with the molecular physiology outlined above, has been used to support a role for these OATs in both the renal secretion and reabsorption of urate (reviewed in Refs. 22, 43, 46).
The renal-specific transporter RST (40), identified several years before URAT1, is the murine ortholog of URAT1 as determined by sequence homology and conservation of synteny (14, 16, 24). Moreover, RST, like URAT1, is localized predominantly (if not exclusively) at the apical surface of the proximal tubule (13, 15, 24, 40). When expressed in oocytes, RST did manifest transport of urate [albeit with significantly lower affinity—the affinity constant (Km) was 1.2 mM (24), compared with 371 µM for URAT1 (13)]. In addition, RST-mediated transport was cis-inhibited by uricosuric agents. However, when RST was expressed in mammalian cells, uptake of a tracer organic anion was not significantly inhibited by 500 µM urate, implying even lower affinity than that measured in oocytes (30). Nevertheless, collectively, these data have led to the belief that RST might subserve proximal tubular reabsorption of urate in mice (24).
Because of the ambiguities in the aforementioned data, we generated a colony of RST knockout mice so as to analyze RST function in vivo. We find that the knockout mice do manifest significantly increased urinary urate, indicating the involvement of RST in the renal reabsorption of urate. Surprisingly, however, the bulk of reabsorption is preserved, suggesting the contribution of other, independent reabsorptive mechanisms. Given this, we have also analyzed urate handling in Oat1 and Oat3 knockouts—these mice manifest slightly decreased secretion, rather than reabsorption. Thus net renal excretion of urate appears to be determined by multiple transporters, some of which may act in opposition in the same nephron segment. In addition, we have performed metabolomic analyses of the plasma and urine of the RST knockouts, revealing alteration in the concentration of several molecules, potentially representing additional endogenous substrates of RST. The results have implications for clinical studies aimed at identifying polymorphisms predisposing to gout and other diseases associated with altered uric acid levels, and for development of therapeutics for these conditions.
MATERIALS AND METHODS
Animals and genotyping
Derivation of RST knockout mice was performed as previously described for Oat1 knockout mice (17). Briefly, homologous recombination (performed at Deltagen, Redwood City, CA) was used to replace a 70-base segment in exon 3 of RST with a LacZ-Neo cassette (encoding β-galactosidase and a neomycin-selectable marker) containing multiple transcription and translation stop signals, resulting in the formation of a null allele. RST-null founders were back-crossed to C57BL/6J for seven generations to yield experimental animals. Control (wild type) mice were either descended from siblings of the progenitors of knockout mice (bred/raised in house along with the knockouts) or were commercially purchased C57BL/6J mice that were housed with the knockouts for 3–4 wk before use in experiments. Comparable results were obtained with either source of control animals.
Mice were group-housed under a 12:12-h light-dark cycle with ad libitum access to food and water, and experiments were performed on male mice [since expression of RST is markedly higher in males than in females (24)] that were age matched (within 2 wk) and between 12 and 20 wk of age. Experimental protocols were approved by the Institutional Animal Care and Use Committee and were in accordance with the Guide for the Care and Use of Laboratory Animals [National Institutes of Health, Bethesda, MD (Pub. No. 865-23)].
Mice were genotyped by PCR analysis of genomic DNA as previously described (17). Briefly, genomic DNA samples prepared from tail snips were used as templates in multiplex PCR reactions using a single reverse primer recognizing a sequence downstream of the site of recombination (5′-CAC TGC AAG GAG CAA GAA AGG GGT C-3′) and two forward primers recognizing sequences within the recombined region, one targeting the endogenous sequence and the other the inserted transgenic sequence (5′-AGA AGG GTG CTG ACC TGG AGC TAT C-3′ and 5′-GGG CCA GCT CAT TCC TCC CAC TCA T-3′, respectively); these primers were predicted to amplify a 224-bp PCR product from the native allele and a 366-bp product from the null allele. Thirty-five cycles of 20 s of 94°C denaturation, 20 s of 60°C annealing, and 20 s of 72°C extension were performed, and reactions were visualized by electrophoresis through 1.3% agarose gels stained with ethidium bromide.
The derivation and genotyping of Oat1 and Oat3 knockout mice was described previously (17, 54). For each of Oat1, Oat3, and RST, gene expression appears to be greater in males than in females (6–8, 24, 35). Therefore, all analyses were performed in male mice only.
Materials
Bovine serum albumin (BSA), inactin, oxonate, and urate (>98% pure) were obtained from Sigma-Aldrich (St. Louis, MO). [3H]inulin (~50 Ci/mmol) was obtained from Perkin-Elmer Life Sciences (Wellesley, MA).
Northern analysis and histology
Northern analysis and histology were performed as previously described (17). Briefly, 5 µg of total kidney RNA from three each of wild-type and RST knockout male mice was electrophoresed through a denaturing gel, transferred to a charged nylon membrane, and then hybridized to a 32P-labeled RST-specific probe [derived by random priming from the full-length cDNA sequence (Image clone BC035927)]. Tissues for histological analysis were collected from these same mice and analyzed at the University of California-San Diego (UCSD) Cancer Center Histology Shared Resource facility by hematoxylin and eosin and β-galactosidase staining.
Microarray and real-time PCR analyses
Microarray and quantitative real-time RT-PCR (qPCR) were performed as previously described (50, 51, 53). Briefly, RNA prepared from wild-type and knockout kidneys and livers was purified on RNeasy columns (Qiagen, Valencia, CA) and then subjected either to linear amplification (reverse transcription followed by in vitro transcription) for microarray analysis or to reverse transcription alone for real-time PCR. In the former case, the amplified RNA was labeled by incorporation of biotinylated nucleotides during in vitro transcription and then hybridized to Affymetrix microarrays, washed, and scanned per the standard Affymetrix protocol. [Hybridization and scanning were carried out at the UCSD/Department of Veterans Affairs Medical Center (VAMC) Genechip core laboratory (http://www.vmrf.org/research-websites/gcf), and microarray data analysis was performed as described in Statistics and calculations.] In the latter case, each cDNA sample was subjected to duplicate real-time PCR reactions with the Amplifluor Detection System (Serologicals, Clarkston, GA) in an Applied Bio-systems ABI Prism 7700 Sequence Detector. [Reactions were performed at the UCSD/VAMC Center for AIDS Research Genomics Core laboratory (http://cfar.ucsd.edu/genomics).] Gene expression values in each case were normalized to that of an internal control housekeeping gene, GAPDH, in the corresponding tissue cDNA sample. Gene-specific primer sequences (5′ to 3′) were as follows [note that the first 18 bases (ACT GAA CCT GAC CGT ACA) on each forward primer correspond to the “Z sequence” that is complementary to the “uniprimer” used in the Amplifluor system]: mOat1 (mSlc22a6; NM_008766), ACT GAA CCT GAC CGT ACA GCA TGA CTG CCG AGT TCT ACC (forward) and CAG CGC CGA AGA TGA AGA G (reverse); mOat3 (mSlc22a8; NM_031194), ACT GAA CCT GAC CGT ACA GCA GCC CTT CAT CCC TAA TG (forward) and CCT CCC AGT AGA GTC ATG GTC AC (reverse); RST (mSlc22a12; NM_009203), ACT GAA CCT GAC CGT ACA CCA TGC TAG GGC CTT TGG TA (forward) and GCA TCC AGG AGC CAT AGA CAC (reverse); Uox (urate oxidase; NM_009474), ACT GAA CCT GAC CGT ACA CAA GAA GGA TTA CCT ACA CGG TGA TA (forward) and CCA GGA CGT GCA CAG TGT TC (reverse).
General phenotypic analysis
General metabolic parameters, including oxygen consumption, carbon dioxide production, locomotor activity, and food and water intake were determined in indirect, open-circuit calorimeter chambers (Columbus Instruments, Columbus OH) as previously described (17). General chemical analyses were performed by the Clinical Laboratory of UCSD Medical Center (San Diego, CA).
Analysis of basal renal function
Determination of basal renal function and urinary clearance was performed as previously described (17). Briefly, mice were anesthetized for terminal clearance experiments with inactin and ketamine (each 100 mg/kg body wt) and placed on an operating table with a servo-controlled heating plate to maintain body temperature at 37.5°C. The trachea was cannulated with polyethylene tubing, and 100% oxygen was blown toward the tracheal tube throughout the experiment. The jugular vein was catheterized for continuous maintenance infusion of 2.25% BSA in 0.85% NaCl at a rate of 0.5 ml·h−1 ·30 g body wt−1. Kidney glomerular filtration rate (GFR) was assessed by [3H]inulin clearance via addition of [3H]inulin to the infusate to deliver 5 µCi·h−1 ·30 g body wt−1. The femoral artery was catheterized for blood pressure measurements and blood sample withdrawal. A polyethylene catheter was placed in the bladder via a suprapubic incision and served for collection of urine. Mice were permitted to stabilize for 60 min; subsequently, a timed urine collection was performed for 45 min, with blood drawn midway in the period. Urinary flow rate was determined gravimetrically. Concentrations of [3H]inulin were determined by liquid scintillation counting. Concentrations of Na+ and K+ in plasma and urine were also determined with a flame photometer (Cole-Parmer Instrument, Vernon Hills, IL).
Urine and plasma samples
Urine samples for mass spectrometric analysis of the levels of urate and other endogenous organic anions were obtained either by handling the mice, which elicited reflex urination, or by collections in metabolic cages (Nalgene, Rochester, NY). In either case, the values for organic anion concentration were normalized to the values for creatinine concentration in the same samples. Plasma samples were obtained immediately after urine sample collections by puncture of the retrobulbar plexus.
Quantification of urate, allantoin, and creatinine
Urine and plasma uric acid, allantoin, and creatinine concentrations were determined by liquid chromatography-electrospray tandem mass spectrometry (MS/MS). Urate measurement was based on a published method (19) with extensive modifications. The mass spectrometer (PE Sciex API 4000, Applied Biosystems, Foster City, CA) source was at 650°C, and nitrogen was used for desolvation and collision-induced dissociation. Data were collected with unit resolution and dwell times of 150 ms. Transitions monitored in the multiple reaction mode (MRM) were mass-to-charge ratio (m/z) 166.8→42.1, 168.8→43.2, and 156.8→97.0 in negative ionization mode (−4,500 V) for uric acid, [1,3-15N]uric acid, and allantoin, respectively, and m/z 114→44 and 117→47 with positive ionization (5,500 V) for creatinine and [methyl-2H3]creatinine, respectively.
Urine samples were diluted 50 times with 50 µM[1,3-15N]uric acid (Isomet, Palisades Park, NJ) or 50 µM [methyl-2H3]creatinine (CDN Isotopes, Pointe-Claire, QC, Canada) in the mobile phase solvent, briefly vortexed, centrifuged 5 min at 15,000 rpm, and diluted 10 times with the mobile phase. For plasma samples, 10 µl was deproteinized with 35 µl of methanol, vortexed, and centrifuged as above, and 15 µl of 0.1% formic acid in water and 10 µl of 50 µM [1,3-15N]uric acid or 50 µM [methyl-2H3]creatinine in the mobile phase were added. Aliquots of 20 µl were injected with a reverse-phase C18 HPLC column (3 × 150 mm; 5-µm Zorbax Eclipse XDB-C18, Agilent Technologies) with an isocratic mobile phase, 70% aqueous methanol + 0.1% formic acid (by volume), at a flow rate of 0.5 ml/min. Run time was 2.2 min with a equilibration time of 0.5 min. Samples were quantified by interpolating the peak area response ratios in a calibration curve in the range 0.25–5 mM in urine and 5–125 µM in plasma.
Since poor uric acid solubility was a concern, we measured the influence of adding increasing concentrations of alkali to make sure that all the uric acid excreted in the uric acid load clearance experiment was dissolved. Urine samples were additionally diluted sixfold (by volume) with 1 N NaOH, and there was no effect on the response ratio corrected for dilution.
Quantification of other endogenous organic anions by mass spectrometry
Urine and plasma levels of other endogenous organic anions were determined by gas chromatography-mass spectrometry (GC-MS) as previously described (17, 23). Briefly, plasma samples of 100 µl or urine volumes equivalent to 0.25–1 µmol of creatinine were reacted with pentafluorobenzylhydroxylamine to form oximes of oxoacids, aldehydes, and ketones. The lyophilized reaction products were extracted in 42% t-amyl alcohol:chloroform over a column of silicic acid, and the dried eluate was reacted with BSTFA/TRISIL to form trimethylsilyl derivatives. These were injected (0.5–1 µl) on a bonded-phase (DB5) capillary column (30 m × 0.25 mm) in an Agilent 6890/5973 GC-MS. Electron impact mass spectra were obtained in scan mode (50–650 amu at 2.4 cycles/s) and species quantified with calibrated response curves of selected ions. 4-Nitrophenol and 2-oxocaproic acid were used as internal standards.
Compounds measured included 2-hydroxy 3-methylvaleric, 2-hydroxyisovaleric, 2-hydroxyglutaric, 2-oxoglutaric, 2 oxo-3 methylvaleric, 2-oxoisovaleric, 3-hydroxypropionic, 3-hydroxyisobutyric, 4-hydroxyphenyllactic, aconitic, benzoic, citric, ethylmalonic, fumaric, glutaric, glyceric, glycolic, isocitric, lactic, linoleic, malic, N-acetylaspartic, orotic, palmitic, pyruvic, and succinic acids, 5-oxoproline, and uracil.
Metabolomics
Urine and plasma samples were extracted by adding 4 vols of ice-cold methanol, vortexing, and incubating at −20°C for 60 min and then centrifuged for 15 min at 4°C and 10K RCF. The supernatant was collected, the centrifugation was repeated, and the final supernatant was evaporated to dryness in a lyophilizer.
The equivalent of 0.4–1.4 µl of extracted urine (after normalization to the previously measured concentrations of creatinine) or 1 µl of extracted plasma was applied to a capillary reverse-phase column with dimensions 150 mm × 0.3 mm (diameter) (Zorbax SB-300, Agilent) at a flow rate of 4 µl/min. Buffer A was H2O + 0.1% formic acid, and buffer B was acetonitrile with 0.1% formic acid. In the case of plasma, the samples were eluted with a gradient from 5% to 95% buffer B over 45 min; in the case of urine, the gradient was 5% buffer B from 0–10 min, then 5–95% from 10 to 55 min, and then held at 95% to 60 min. Some of the urine samples were measured in duplicate. The column was interfaced to an electrospray ionization time-of-flight mass spectrometer (ESI-TOF; Agilent), and data were collected from either 100–1,000 m/z (plasma) or 70–1,000 m/z (urine) in continuum mode. Data were converted from the instrument format (.wiff) to the common format (.cdf) and were then analyzed with the analysis and nonlinear alignment program XCMS (49), in which the integrated intensities of all observed ions were compared between the knockout and wild-type groups.
Statistics and calculations
Microarray data were analyzed by Vampire (http://genome.ucsd.edu/microarray; Refs. 26, 27), a statistical package designed to specifically account for the expression-dependent variability in microarray data. The significance threshold was set at P < 0.05 after Bonferroni correction for multiple comparisons. The statistical significance of other differences between knockout and wild-type mice was determined with Student’s unpaired t-test (2 tailed).
RESULTS
Knockout of RST
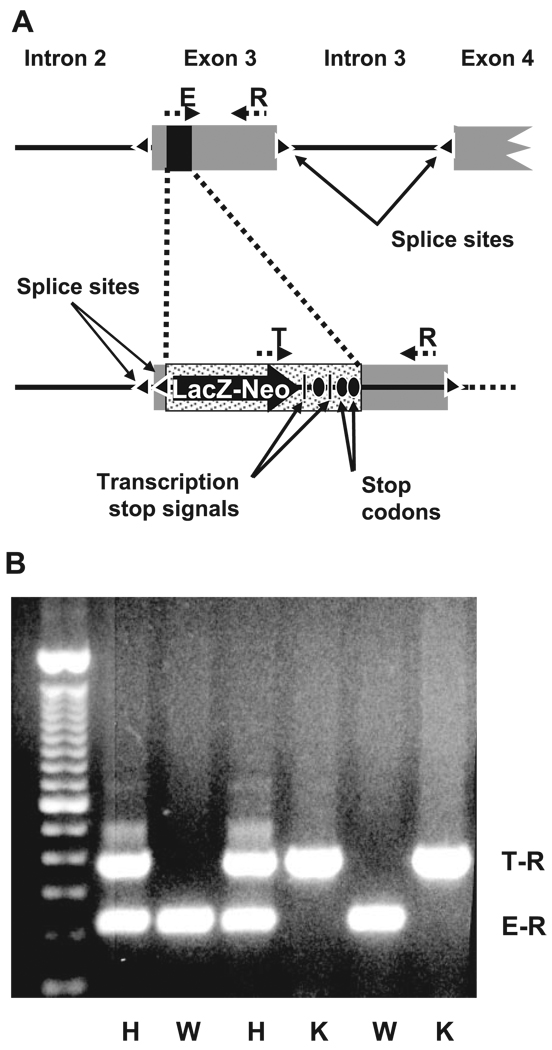
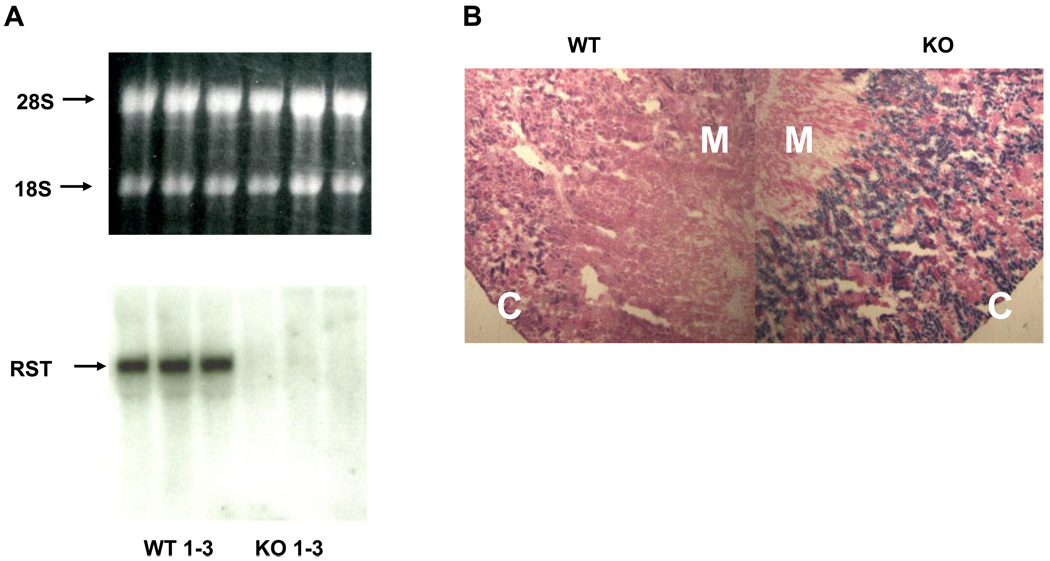
Homologous recombination was used to replace a segment of the third exon of RST with a β-galactosidase transgene flanked by upstream splice acceptor sites and downstream transcriptional as well as translational stop signals. This resulted in elimination of the ability to express the downstream eight (of 10 total) exons of RST and thus effectively generated a null allele (Fig. 1A). Mice heterozygous for this allele were bred to yield knockout and wild-type offspring, which were typed by PCR analysis of genomic DNA using primers specific for the native or transgenic sequence at the site of recombination (Fig. 1B; see materials and methods for details). Loss of RST gene expression in knockout mice was confirmed both by Northern analysis—the expected 2.3-kb hybridization signal was noted in RNA from wild-type mouse kidneys but not in RNA from knockouts (Fig. 2A)—and by qPCR as well as microarray analysis (see Fig. 4). In addition, histochemical analysis of kidney sections from knockout mice revealed a specific increase in galactosidase activity in the renal cortex (corresponding to the location of the proximal tubule), consistent with specific recombination of the transgene into the RST locus resulting in tissue-specific expression of β-galactosidase in the same location as usual RST gene expression (Fig. 2B).
Fig. 1.
Knockout of RST. A: schematic of targeted disruption of RST. Homologous recombination was performed to replace a 70-base segment (black rectangle) in exon 3 of RST with a LacZ-Neo cassette flanked by an upstream splice acceptor site (triangle) and multiple downstream transcription termination signals (vertical black bars) and in-frame stop codons (black ovals), resulting in formation of a null allele. The positions of the genotyping primers R, E, and T are indicated. B: genotyping of a typical litter from a cross between mice heterozygous for the RST null allele. A common “reverse” primer (R in A) and “forward” primers specific to either the endogenous or targeted (null) alleles (E and T, respectively, in A) were used in multiplex PCR on genomic DNA from individual offspring. Primers E and R amplify a 224-bp product (E-R) from the wild-type (WT) allele, while primers T and R amplify a 366-bp product (T-R) from the knockout (KO) allele; thus offspring forming only the E-R product are wild types (indicated by W), those forming only the T-R product are knockouts (K), and those forming both products are heterozygotes (H).
Fig. 2.
Northern analysis and β-galactosidase staining. A: kidney RNA from 3 WT (WT 1–3) and 3 RST KO (KO 1–3) mice was electrophoresed through a denaturing gel and then transferred to a nylon membrane and hybridized to a radiolabeled RST-specific probe. Top: the ethidium-stained RNA gel was visualized by UV transillumination to confirm sample integrity and uniformity of loading. Positions of the 28S and 18S rRNA bands are indicated. Bottom: an RST-specific hybridization signal was detected only in RNA from WT kidneys. B: micrographs of frozen kidney sections from WT and RST KO mice stained for β-galactosidase activity. Images depicted are representative of stains of sections from 3 different WT and KO animals and were acquired at the identical microscope and camera settings. Original magnification, ×4. Significant β-galactosidase staining is noted in KO renal cortex (C) (greater than the staining due to endogenous galactosidase-like activity that is noted in WT cortex), while staining is absent from the medulla (M), which does not contain proximal tubular tissue.
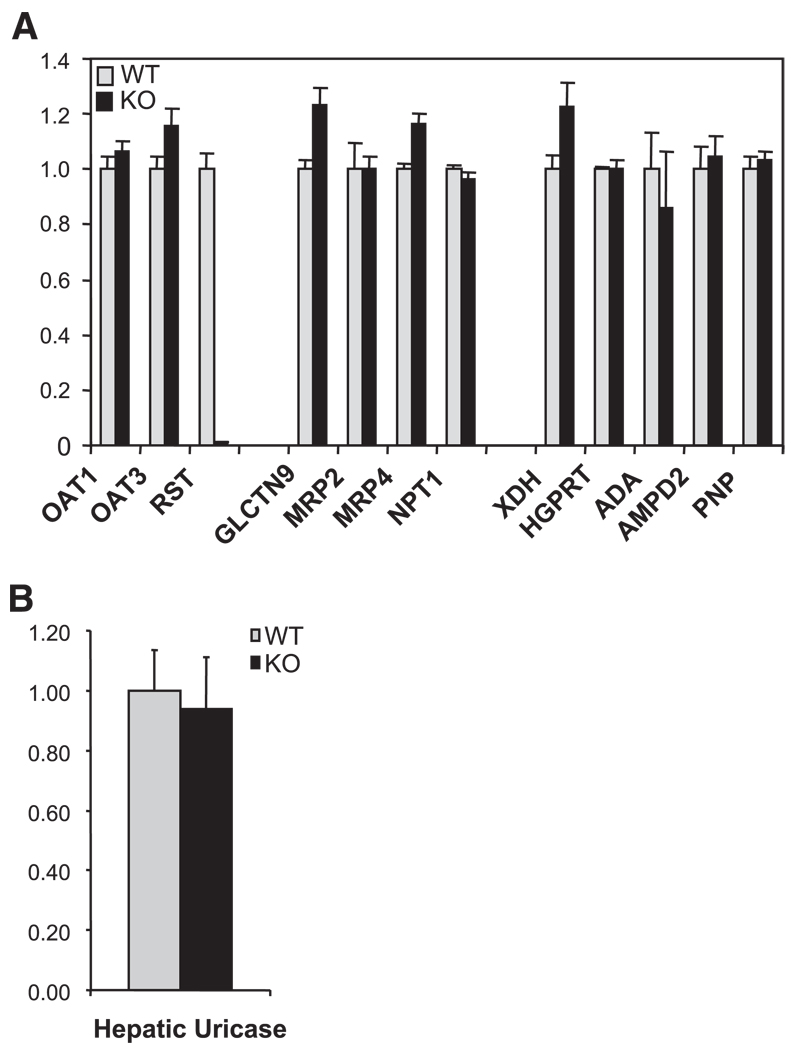
Fig. 4.
Expression of genes involved in urate transport and metabolism. A: kidney RNA samples from WT and RST KO mice were hybridized to high-density oligonucleotide arrays for global determination of gene expression (see also Table 2). Data are presented relative to expression in WT and represent means ± SE of measurements in 3 each of WT and KO mice. No significant differences between WT and KO were noted in the expression of multiple genes known or postulated to contribute to urate transport [Oat1, Oat3, Galectin 9 (UAT), Mrp2, Mrp4, and Npt1 (Oatv1)] or metabolism [xanthine dehydrogenase (XDH), hypoxanthine guanine phosphoribosyl transferase (HGPRT), adenosine deaminase (ADA), adenosine monophosphate deaminase 2 (AMPD2), and purinenucleoside phosphorylase (PNP)]. B: expression of uricase in liver RNA samples from WT and RST KO mice was determined by quantitative real-time PCR. Expression values in each sample were normalized to that of GAPDH and are presented relative to the level of expression in WT kidney. Data represent means ± SE of measurements in 4 each of WT and KO mice. No significant differences between the genotypes in expression of uricase mRNA were noted.
As observed with the previously described Oat1 and Oat3 knockout mice (17, 54), RST-null mice are viable, fertile, and without obvious histological or anatomic abnormalities; in addition, they are similar to their wild-type counterparts with respect to general metabolic parameters, including food and water intake, O2 consumption, CO2 production, and locomotor activity (Fig. 3), as well as other physiological metrics such as body weight, heart rate, blood pressure, plasma electrolytes, and hematocrit (Table 1). Thus the knockout mice appear grossly normal, permitting ready assessment of renal transport of urate and other organic anions as described below.
Fig. 3.
Analysis of metabolic parameters. Rates of oxygen consumption, carbon dioxide production, food and water intake, and locomotor activity were assessed in WT mice and RST KO mice by housing in calorimeter chambers. Data were collected over three 12-h dark cycles (D1–D3) and two 12-h light cycles (L1 and L2) and represent the means ± SE of measurements in 4 each of WT and KO mice. No significant differences between WT and KO mice were noted in any of the measured parameters.
Table 1.
General phenotypic analysis of wild-type and RST knockout mice
| Wild Type | RST Knockout | |
|---|---|---|
| Body weight, g | 29.3±0.5 | 31.4±1.1 |
| Mean arterial blood pressure, mmHg | 84±7 | 83±8 |
| Heart rate, 1/min | 501±23 | 522±17 |
| Hematocrit, % | 43±0.9 | 42.2±0.8 |
| Plasma [Na], mM | 145±2 | 143±2 |
| Plasma [K], mM | 4.2±0.2 | 4.3±0.4 |
Data represent means ± SE of measurements in 5 each of wild-type and knockout mice.
Gene expression in knockout mice
Before assessment of urate handling in the RST knockout mice, we investigated whether there was any alteration in expression of other genes potentially involved in the regulation of urate levels, which might compensate for, or otherwise modulate, any loss of urate transport in the knockouts. As noted in the introduction, within the OAT family of transporters of which RST is a member, the basolateral proximal tubular OATs, Oat1 and Oat3, have also been suggested to be involved in the renal transport of urate (on the basis of in vitro expression studies in Xenopus oocytes) (22, 43, 46). Moreover, it is also noteworthy that these two transporters are close phylogenetic relations of RST, and that the encoding genes of all three transporters are closely linked [within 2 Mb in both mouse (on chromosome 19) and human (on chromosome 11)] and may thus be coordinately regulated (15, 16). Accordingly, we initially determined renal expression of Oat1 and Oat3 in the knockout, as well as of RST as a control, by qPCR. We noted no significant alteration in expression of Oat1 and Oat3, while confirming the expected loss of RST mRNA (data not shown).
Of note, the human apical proximal tubular OAT, OAT4, has also been proposed to function as a urate transporter (20). However, the mouse genome (and also that of the rat) appears to lack an ortholog of OAT4. Human OAT4 is located on 11q13.1, adjacent to the locus of URAT1 [as we have previously described, the genes encoding OATs tend to occur in chromosomal pairs (15, 16)]. The syntenic mouse chromosomal segment on 19qA, although carrying RST and the mouse orthologs of other OAT4-neighboring genes, lacks evidence of the existence of a gene [such as the presence of gene predictions or of expressed sequence tags (ESTs)] at the locus expected for the murine ortholog of OAT4 (http://genome.ucsc.edu). Moreover, BLAST searches do not reveal any orthologous matches to OAT4 in mouse—the closest murine match to OAT4 (by BLAST) is RST (the orthologous relationship of which to URAT1 is established by the fact that each gene is the other’s best BLAST match in mouse and human).
We next performed a global survey of renal gene expression in the knockout via microarray hybridization. Lack of change in expression of Oat1 and Oat3 was confirmed; in addition, there were no major changes in mRNA levels of genes encoding additional proteins postulated to contribute to urate transport in the kidney—Mrp2 (11), Mrp4 (22, 62), UAT (46), and Oatv1 (Npt1) (43). Similarly, there were no major changes in mRNA levels of genes involved in urate metabolism—xanthine dehydrogenase (XDH), hypoxanthine guanine phosphoribosyl transferase (HGPRT), adenosine deaminase (ADA), adenosine monophosphate deaminase 2 (AMPD2), and purine-nucleoside phosphorylase (PNP) (41) (Fig. 4A). Uricase, which catalyzes the degradation of urate to allantoin, is expressed in murine liver, not kidney (41); as expected, expression of this gene was not detected on our microarrays. Accordingly, we performed qPCR to determine hepatic expression of uricase; once again, no differences were noted between wild-type and RST knockout mice (Fig. 4B).
The microarray data did, however, reveal significant changes in expression in several other genes whose products apparently do not directly participate in urate regulation (Table 2). Among these were genes annotated as being involved in metabolism (e.g., aldehyde dehydrogenase 1a1 and 1a7 were increased in the knockout relative to the wild type by ~9-fold and 6-fold, respectively), development/differentiation (e.g., Fos and Jun were decreased by ~8-fold and 3-fold, respectively), inorganic ion transport (e.g., the H+-K+-ATPase was decreased by ~3-fold), as well as biosynthesis, cell cycle progression, and apoptosis. However, caution is probably warranted in drawing functional inferences from these observations since it is possible that some of the alterations in gene expression are epiphenomenal. For example, the gene that immediately adjoins RST in the mouse genome (lying a mere 3 kb downstream; http://genome.ucsc.edu), Neurexin II, is among the genes manifesting downregulated expression in RST knockout kidney (by ~3-fold). Neurexin II [which is predominantly expressed in the nervous system (34) and to a lesser extent in the kidney (http://source.stanford.edu)] encodes a neural cell adhesion molecule with no obvious direct functional connection to urate transport or metabolism. It appears likely, therefore, that its repression is merely due to perturbation of local transcriptional control by the genomic rearrangement in the knockout.
Table 2.
Genes with significantly different renal expression in wild-type and RST knockout mice
| Locus | Description | Expression in WT, arbitrary units |
Ratio of Expression: KO/WT |
P | GO Molecular Function | GO Biological Process |
|---|---|---|---|---|---|---|
| Genes with decreased renal expression in KO compared with WT | ||||||
| Nr4a1 | Nuclear receptor subfamily 4, group A, member 1 | 7,585 | 0.23 | 4.60E-30 | Transcription factor | Apoptosis |
| Axud1 | AXIN1 upregulated 1 | 1,825 | 0.29 | 1.08E-09 | Apoptosis | |
| Gadd45g | Growth arrest and DNA-damage-inducible 45 gamma | 8,867 | 0.30 | 8.84E-26 | Protein binding | Apoptosis, development/differentiation |
| Gpaa1 | GPI anchor attachment protein 1 | 5,175 | 0.38 | 6.71E-17 | GPI anchor transamidase | Biosynthesis, GPI anchor |
| Ext1 | Exostoses (multiple) 1 | 2,235 | 0.45 | 8.58E-08 | Glycosyl transferase | Biosynthesis, proteoglycan |
| Gcnt1 | Glucosaminyl (N-acetyl) transferase 1, core 2 | 41,379 | 0.45 | 1.43E-16 | Glycosyl transferase | Biosynthesis, proteoglycan |
| Pkmyt1 | Protein kinase, membrane-associated tyrosine/threonine 1 | 1,651 | 0.37 | 5.27E-07 | Protein kinase | Cell cycle |
| Junb | Jun-B oncogene | 4,877 | 0.41 | 6.21E-15 | Transcription factor | Cell cycle |
| Fos | FBJ osteosarcoma oncogene | 5,386 | 0.13 | 1.33E-34 | Transcription factor | Cell cycle, development/differentiation |
| Jun | Jun oncogene | 10,829 | 0.38 | 2.97E-20 | Transcription factor | Cell cycle, development/differentiation |
| Per1 | Period homolog 1 (Drosophila) | 3,089 | 0.49 | 6.01E-09 | Transcription factor | Circadian rhythm |
| Plk3 | Polo-like kinase 3 (Drosophila) | 2,538 | 0.43 | 2.78E-09 | Protein kinase | Development/differentiation |
| Egr1 | Early growth response 1 | 4,0112 | 0.05 | 4.22E-53 | Transcription factor | Development/differentiation |
| Jarid2 | Jumonji, AT-rich interactive domain 2 | 2,749 | 0.49 | 4.62E-08 | Transcriptional repressor | Development/differentiation |
| Hbegf | Heparin-binding EGF-like growth factor | 3,615 | 0.45 | 2.89E-11 | Growth factor | Growth factor/cytokine signaling pathway |
| Tnfrsf9 | Tumor necrosis factor receptor superfamily, member 9 | 1,532 | 0.20 | 1.16E-09 | Receptor | Immune response |
| Pdk4 | Pyruvate dehydrogenase kinase, isoenzyme 4 | 4,895 | 0.49 | 4.34E-11 | Protein kinase | Metabolism, glucose |
| Zfp36 | Zinc finger protein 36 | 14,023 | 0.36 | 1.91E-22 | mRNA binding | mRNA catabolism |
| Sf1 | Splicing factor 1 | 2,165 | 0.45 | 1.53E-07 | mRNA binding | mRNA splicing |
| Fkbp5 | FK506 binding protein 5 | 2,563 | 0.25 | 2.83E-15 | Unfolded protein binding | Protein folding |
| Rabgap1 | RAB GTPase activating protein 1 | 2,136 | 0.31 | 6.96E-11 | GPCR activity | Signal transduction |
| Nrxn2 | Neurexin II | 2,680 | 0.34 | 9.24E-13 | Calcium channel regulator | Synaptic transmission |
| Slc22a12 | Solute carrier family 22 member 12 | 35,720 | 0.01 | 1.89E-57 | Transporter | Transport, organic anion |
| Atp4a | ATPase, H+/K+ exchanging, gastric, alpha polypeptide | 2,741 | 0.30 | 2.43E-14 | Transporter | Transport, inorganic cation transport |
| Nudt19 | Nudix-type motif 19 | 62,499 | 0.50 | 1.14E-13 | Hydrolase | |
| Ier2 | Immediate-early response 2 | 4,983 | 0.44 | 1.12E-13 | ||
| Malat 1 | Metastasis-associated lung adenocarcinoma transcript 1 | 5,497 | 0.49 | 1.57E-11 | ||
| Genes with increased renal expression in KO compared with WT | ||||||
| BC016495 | cDNA sequence BC016495 | 1,109 | 3.54 | 9.88E-21 | Protein kinase | Biosynthesis, pyrimidine nucleotide |
| Ccrk | Cell cycle-related kinase | 483 | 3.98 | 1.85E-11 | Protein kinase | Cell cycle |
| Rad9 | RAD9 homolog (S.pombe) | 1,130 | 2.09 | 1.61E-07 | Cell cycle | |
| Sema4b | Semaphorin 4B | 1,646 | 2.26 | 4.70E-12 | Receptor | Development/differentiation |
| Fen1 | Flap structure specific endonuclease 1 | 926 | 2.92 | 5.23E-13 | Endonuclease | DNA replication/repair |
| Cfd | Complement factor D (adipsin) | 940 | 5.99 | 2.67E-34 | Peptidase | Immune response |
| Ifi27 | Interferon, alpha-inducible protein 27 | 1,491 | 2.20 | 1.16E-10 | Immune response | |
| Car3 | Carbonic anhydrase 3 | 8,334 | 2.15 | 5.39E-16 | Carbonate dehydratase | Metabolism |
| Aldh1a7 | Aldehyde dehydrogenase family 1, subfamily A7 | 4,044 | 5.60 | 1.32E-41 | Oxidoreductase | Metabolism |
| Aldh1a1 | Aldehyde dehydrogenase family 1, subfamily A1 | 1,824 | 8.56 | 9.78E-48 | Oxidoreductase | Metabolism |
| Trf | Transferring | 727 | 2.59 | 6.47E-08 | Iron binding | Transport, iron |
| Pom121 | Nuclear pore membrane protein 121 | 4,845 | 2.06 | 4.49E-14 | Transporter | Transport, nucleocytoplasmic |
| Zdhhc12 | Zinc finger, DHHC domain containing 12 | 503 | 6.69 | 4.52E-26 | Acyltransferase | |
| Sdc4 | Syndecan 4 | 2,232 | 2.10 | 4.65E-12 | Cytoskeletal protein binding | |
| Col6a2 | Procollagen, type VI, alpha 2 | 1,419 | 2.05 | 1.26E-08 | Structural molecule | |
| Thrsp | Thyroid hormone responsive SPOT14 homolog | 660 | 3.59 | 1.60E-13 | ||
| cDNA sequence A230084D06 | 173 | 6.57 | 4.16E-07 | |||
A global comparison of renal gene expression in 3 each of wild-type (WT) and RST knockout (KO) mice was performed with high-density microarrays containing probes representing ~39,000 transcripts (Affymetrix MG 430 2.0 arrays). Genes with significantly different expression in WT and KO mice (as determined by Vampire; http://genome.ucsd.edu/microarray) and that were at least 2-fold increased or decreased are listed, along with their presumptive biological and molecular functions as annotated in the Gene Ontology (GO) database (www.geneontology.org). Genes with decreased expression in KO mice are presented first, followed by those with increased expression; within each group the lists are sorted by the GO biological process annotation.
Urate excretion in knockout mice
Before measurement of urinary excretion of urate in the knockout mice, we first assessed various basic indexes of renal function so as to ascertain that any changes in urate handling were not due to general defects in kidney function. Parameters measured included kidney weight, GFR (as determined by the clearance of labeled inulin), and absolute and fractional excretion of Na+, K+, and fluid (see materials and methods for details). In all cases, determinations in knockout and wild-type animals yielded similar values (Table 3).
Table 3.
Renal parameters in wild-type and RST knockout mice
| Wild Type | RST Knockout | |
|---|---|---|
| Kidney wt, mg/g bw | 5.8±0.2 | 6.5±0.5 |
| GFR, µl · min−1 · g bw−1 | 9.4±1.0 | 9.1±0.4 |
| Absolute renal fluid excretion, nl · min−1 · g bw−1 | 44±5 | 48±5 |
| Absolute renal Na excretion, nmol · min−1 · g bw−1 | 3.2±0.8 | 3.2±0.7 |
| Absolute renal K excretion, nmol · min−1 · g bw−1 | 7.4±0.9 | 9.3±1.0 |
| Fractional renal fluid excretion, % | 0.5±0.1 | 0.5±0.1 |
| Fractional renal Na excretion, % | 0.2±0.1 | 0.3±0.1 |
| Fractional renal K excretion, % | 20±4 | 24±2 |
Data are means ± SE of measurements in 5 each of wild-type and knockout mice. bw, Body weight; GFR, glomerular filtration rate.
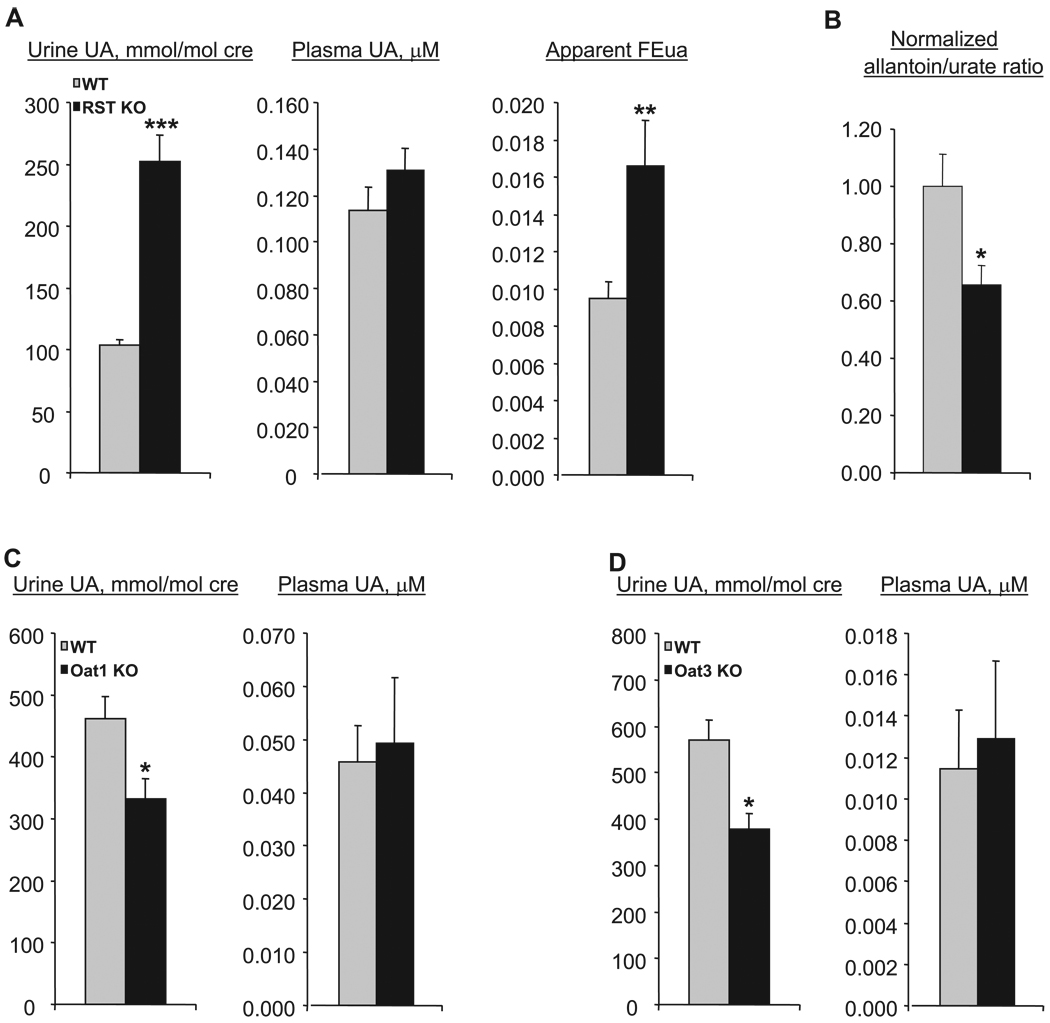
We then performed MS/MS to determine urate concentration in plasma and urine samples from a large set (26 each) of wild-type and knockout mice. (Urinary concentrations were normalized to creatinine in order to correct for variations in concentration due to differences in volume of urine produced.) Novel mass spectrometric methods were developed for these analyses as described in detail in materials and methods. Our measurements revealed that while plasma levels of urate were similar in the genotypes, there was a significant, approximately twofold, increase in urinary urate in the knockout mice (Fig. 5), indicating diminished renal reabsorption. Of note, doubling of the rate of urinary excretion would have been expected to result in decreased plasma levels. Thus the absence of change in plasma urate concentration suggests increased urate production or decreased degradation of urate to allantoin. While hepatic uricase mRNA expression was unaffected in the knockout mice (Fig. 4B), the plasma allantoin-to-urate ratio was significantly decreased, by about a third (Fig. 5B), indicating that decreased uricase-mediated degradation of urate accounts (at least in part) for the preserved plasma urate levels in the knockout mice.
Fig. 5.
Urinary and plasma levels of urate in WT and RST KO mice. A: urine and plasma samples were obtained from WT and RST KO mice, and urate (UA) levels were determined by tandem mass spectrometry. Urinary urate concentrations are presented normalized to that of creatinine (cre). The fractional excretion of urate (FEua) was calculated as the ratio of urine urate normalized to urine creatinine to plasma urate normalized to plasma creatinine (see text for details). Data represent means ± SE of measurements in samples derived from 18–26 different mice. KO mice manifest significantly increased urinary urate and fractional excretion of urate; **P < 0.01, ***P < 0.001. B: plasma allantoin and urate levels were determined in samples collected from WT and RST KO mice, and the allantoin-to-urate ratios were calculated. Data represent means ± SE of measurements in samples derived from 9 or 10 different mice of each genotype. KO mice manifest significantly decreased allantoin-to-urate ratios; *P < 0.05. C and D: urine and plasma urate were measured as in A in mice null for the RST-related transporters Oat1 (C) and Oat3 (D). Data are means ± SE of measurements in samples derived from 4 different mice each. Oat1 and Oat3 KO mice manifest decreased urinary urate compared with WT mice; *P < 0.05.
Surprisingly, despite the significant increase in urinary urate, calculation of fractional excretion of urate (FEua) indicates that the bulk of renal reabsorption was preserved in knockout mice, suggesting the independent contribution of other mechanisms to much of this transport process. FE of urate (or any compound) is equivalent to the ratio of the actual renal clearance of urate to the expected clearance if its excretion were due to filtration alone. FE is >1 in the presence of net secretion, <1 in the presence of net reabsorption, and equivalent to 1 in the absence of either. Since the FE of creatinine is ~1 (secretion and reabsorption contribute minimally to the renal excretion of creatinine), FEua can be approximated by the ratio of urine urate normalized to urine creatinine to plasma urate normalized to plasma creatinine. The apparent FEua was 0.009 in the wild type and 0.017 in the knockout (Fig. 5A), suggesting a net fractional reabsorption (1 − FEua) of ~98% in the knockout mice.
Comparable results were obtained in supplementary experiments (utilizing additional sets of mice) in which urate levels were measured with an uricase enzymatic method and creatinine was measured with the Jaffe colorimetric method (both assays were performed by the clinical laboratory of the UCSD Medical Center). Urinary urate levels (normalized to creatinine) were increased approximately twofold, plasma levels were essentially unchanged, and the fractional excretion increased from ~3% in the wild type to 5% in the knockout (data not shown), again indicating preservation of the bulk of reabsorption in the RST-null mice.
As noted in the introduction, Oat1 and Oat3 have been considered candidate urate transporters. Accordingly, we performed similar measurements of plasma and urine urate in Oat1 and Oat3 knockout mice (17, 54) to determine whether there were any changes in the renal excretion of urate. There was an ~30%, statistically significant decrease in urinary urate in both knockouts (Fig. 5, C and D), suggesting that Oat1 and Oat3 might participate in the renal secretion of urate. Thus neither transporter appears to contribute to the residual reabsorption revealed in the RST-null mice. We note also that the fractional excretion varied significantly in the different wild-type strains, each of which served as an appropriate control for experiments in comparison with the RST, Oat1, and Oat3 knockouts. Background effects may have a significant influence on urate handling for all three genes. Microarray analyses of gene expression in the kidneys of Oat1 and Oat3 knockout mice did not reveal any alterations in the expression of RST, or of the other putative urate transporters discussed above. However, it should be noted that the Oat1 knockout has somewhat diminished Oat3 gene expression and that, conversely, the Oat3 knockout has somewhat diminished Oat1 expression (61a). This is likely due to the pairing of the genes in the genome (15); however, the slight decrease of the paired OAT in each knockout does not appear to be functionally significant [transport of the Oat3-specific substrate estrone sulfate is unaffected in the Oat1 knockout (17), while that of the Oat1-specific substrate para-aminohippurate is unaffected in the Oat3 knockout (63)].
Metabolomics
All of the OATs are, to varying degrees, multispecific, suggesting that RST might mediate the transport of multiple endogenous compounds. In an attempt to identify such compounds, we initially performed GC-MS to determine the levels of ~30 of the most abundant small organic anions in the urine and plasma of wild-type and RST knockout mice (see materials and methods for a list of the measured compounds). We expected that any endogenous compounds normally transported (either secreted or reabsorbed) by RST might manifest altered concentration in either urine or plasma of the knockouts. Notably, previous measurements of largely the same set of compounds in Oat1 knockout mice identified several organic anions with significantly altered concentration consistent with their being endogenous Oat1 substrates (17). However, in two independent sets of experiments no significant differences were noted between RST knockout and wild-type mice in the urine or plasma concentrations of the assayed organic anions (not shown). Accordingly, this set of compounds, though containing likely endogenous Oat1 substrates, might not contain any substrates of RST. Likewise, none of these metabolites was significantly altered in the Oat3 knockouts (Eraly, Gangoiti, Barshop, and Nigam, unpublished observations). Thus, of all the specifically measured metabolites, the only organic anion altered in concentration in each of Oat1, Oat3, and RST knockout mice was urate. Of note, the organic acids measured included lactate and orotate, which had previously been identified as potential endogenous substrates or counterions for RST-mediated transport (13, 24). It remains possible that these compounds are substrates for RST in vivo but are not transported in sufficient degree as to impact overall urine or plasma levels.
Since targeted analyses did not lead to the identification of endogenous RST substrates, we next performed a global (metabolomic) survey of compounds in RST knockout mouse plasma and urine, utilizing a reverse-phase capillary HPLC column coupled to an ESI-TOF mass spectrometer (64). This resulted in the detection of 3,105 features in urine and 5,629 in plasma, which would include any metabolites ionizing under positive electrospray conditions and within the detection limit of the instrument. We then analyzed the data, using the recently developed software package XCMS to perform a nonlinear alignment of either urine chromatograms or plasma chromatograms, allowing for comparison of the intensities of each ion in the various samples (49). It should be noted, however, that, because of very large differences in matrix effects, metabolite composition, and relative metabolite concentrations, chromatograms are not comparable between plasma and urine, so that metabolites common to both fluids cannot be directly ascertained.
Of the features in urine (after normalization to creatinine), 48 (1.5%) were significantly different at a P value threshold of 1 × 10−3. Of these significant features, the vast majority, ~92%, were decreased in concentration in knockout compared with wild-type samples. Similar, though less pronounced, differences were noted in plasma, with 51 (0.9%) of detected features significantly different between the genotypes at a P value threshold of 10−3, of which 75% were decreased in the knockout. Since we were performing multiple comparisons it is expected that some of the above features represent “false positives.” However, under even the most conservative assumption of no overall difference in metabolite concentrations between knockout and wild-type mice, the expected numbers of false positives at the 10−3 significance thresholds would be only 3 and 6, respectively, in urine and plasma (3,105 × 10−3 = ~3; 5,629 × 10−3 = ~6). Thus a large majority of the apparently significantly different features are likely to represent compounds that are genuinely different in concentration in RST knockout and wild-type mice.
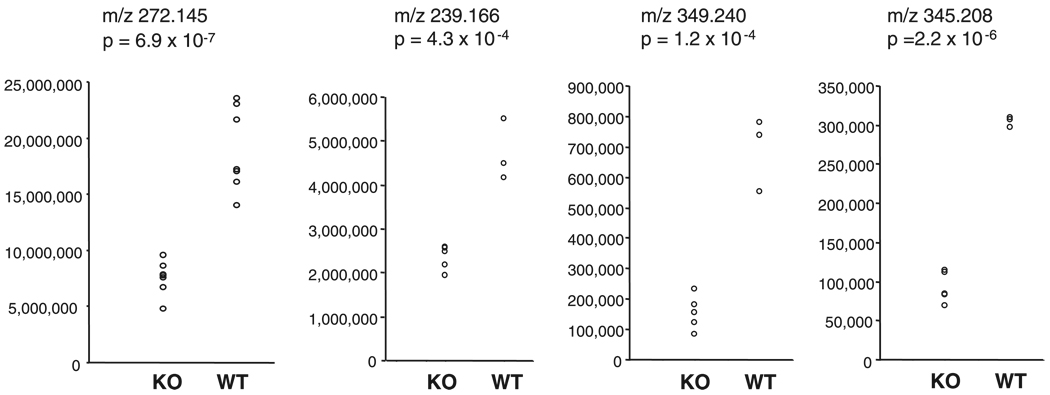
Overall, the metabolomics results suggest that there are significant differences in metabolite composition between the plasma and urine of RST knockout and wild-type mice. Data from four of the most significantly varying compounds, as selected not only on the basis of statistical significance but also by reference to the original chromatographic data, are shown in Fig. 6. However, although we determined the masses of these compounds to an accuracy of ~8 ppm, we were unable to identify any obvious matches among known metabolites, i.e., they correspond to entirely novel, previously undescribed molecules. [This was not an unexpected outcome since the identity of most of the metabolites present in biological matrixes and fluids is, as yet, undetermined (48).] One example for which there are a limited number of possible molecular formulas is the ion with m/z 239.166. There are two possible elemental compositions for an M+H ion within 8 ppm of this value, C14H23O3 and C13H24N2P; however, there are no known metabolites that correspond to either of these formulas. Identification of such unknowns cannot readily be accomplished via MS/MS since, unlike the case for peptides, the fragmentation patterns of small molecules are not predictable (44, 66). Thus an entirely separate research endeavor will be required to identify the unknowns from our study.
Fig. 6.
Significantly varying urine and plasma metabolites in WT and RST KO mice. Integrated intensities and mass-to-charge ratio (m/z) values for 4 of the most significantly varying compounds from the metabolomics analysis are shown. Far left panel corresponds to a compound significantly different in the urine of WT and KO mice, with the remaining panels depicting compounds that were significantly different in the plasma of WT and KO mice. Two-tailed t-test P values are indicated.
DISCUSSION
We report here the characterization of mice null for RST, the murine ortholog of the human urate transporter URAT1, and determination of renal urate excretion in these mice as well as in mice null for the related transporters Oat1 and Oat3. RST knockout mice, which appear otherwise physiologically normal, manifest increased urinary urate demonstrating the involvement of RST in the renal reabsorption of this compound, while Oat1 and Oat3 knockout mice manifest slightly decreased urinary urate consistent with a role for these transporters in the renal secretion of urate. Thus net urinary excretion of urate is influenced by multiple OATs, with some apparently acting in opposition to others, a finding of potential clinical significance if also found true in humans. Strikingly, in targeted measurements of urinary and plasma organic anions in the three knockouts, urate was the only compound altered in common. We also conducted untargeted mass spectrometric (metabolomic) analyses of knockout mouse plasma and urine, revealing alteration in concentration of several as yet unidentified molecules potentially representing novel endogenous compounds of RST. The most significantly varying of these substrates were “unknowns,” however, and their de novo chemical identification will require an entirely separate research endeavor outside of the scope of the present work.
While the loss of RST results in significantly increased urinary excretion of urate, the bulk of reabsorption was preserved, suggesting the presence of independent and/or redundant mechanisms contributing to this transport process of which RST is just one component. By contrast, humans with genetic defects in URAT1 appear to have markedly impaired urate reabsorption, resulting in hypouricemia (low plasma levels of urate) and uricosuria (high urinary levels of urate) (13). Conceivably, this discrepancy might be accounted for by the possibility that the compensating pathways for urate reabsorption that are operative in mice are overwhelmed in the presence of the higher systemic levels of urate that exist in humans (due to the loss of uricase), resulting in a greater relative contribution of URAT1 to reabsorption. In preliminary experiments, an increase in the filtered load of urate induced by administration of the uricase inhibitor oxonic acid did not lead to an increase in the difference in reabsorption between wild-type and RST knockout mice (data not shown), somewhat mitigating this hypothesis. Nevertheless, the possibility that a major contribution of RST to urate reabsorption might be uncovered in the absence of uricase would ultimately be best evaluated by performing crosses between RST knockout mice and those null for uricase.
An alternate possibility is that urate, though transported by RST in vitro, might not in fact be a significant substrate in vivo—physiological transport of urate might be a specific property of URAT1. Distinct lineage-specific functions of RST and URAT1 might be accounted for by evolutionary forces arising from the loss of uricase in the primate lineage. The resulting higher systemic urate levels would have predisposed to urate nephropathy (kidney disease due to blockage of tubules by urate crystals that form in the presence of high concentrations of urate). This could have resulted in selective pressure for acquisition of potent proximal tubular urate reabsorption (which would decrease levels in the downstream tubular lumen and thus decrease the risk of nephropathy), leading to the co-option of URAT1 to serve as a urate reabsorber specifically in the primate lineage.
This argument is supported by several lines of evidence. Functional data from in vitro studies provide circumstantial support—as noted earlier, the affinity of RST for urate is substantially lower that that of URAT1 [Km of ≥1.2 mM for RST vs. 371 µM for URAT1 (13, 24, 30)]. Moreover, the relative expression of RST in mouse kidney is lower than that of URAT1 in human kidney [while 29 of 101,446 total mouse kidney ESTs (0.03%) correspond to RST, 119 of 192,551 human kidney ESTs (0.06%) correspond to URAT1]. Importantly, genetic data provide direct support for the scenario described above. First, uricase knockout mice suffer early mortality from urate nephropathy (69), indicating the selective pressure that might have operated during primate evolution for development of protection against this disease. Second, human individuals with loss of URAT1 (i.e., patients with renal hypouricemia) have a clear propensity for (episodic) urate nephropathy (13), indicating that urate reabsorption might confer exactly this protection.
However, it is important to note that there is emerging evidence that URAT1 might not make as dominant a contribution to urate reabsorption in humans as had originally been supposed, and might even, depending on genetic background, account for only a minor component of total reabsorption. The nonsense mutation W258X in URAT1 results in truncation of approximately half the protein and abolishes transport function in vitro (13). In the initial report describing W258X it was noted that a Japanese individual homozygous for this mutation had an FEua of 0.95, whereas the usual FEua in humans is <0.1 (13). Thus reabsorption was almost totally abolished in this individual, suggesting a critical role for URAT1 in urate reuptake. However, in subsequent studies (28, 32, 56), the FEua of individuals homozygous for W258X was measured to be as low as 0.41 (32), with an average of 0.75 in the largest series of homozygotes (11 individuals; Ref. 28). Thus there was substantial residual renal reabsorption of urate in individuals presumably completely lacking in URAT1 function, which in at least one instance might have comprised the majority of maximal possible reabsorption. Moreover, a study of Greek Caucasians did not identify any URAT1 mutations among patients with loss of urate reabsorption resulting in renal hypouricemia (61). Finally, a striking recent case report from Mexico describes an individual homozygous for a frameshift mutation resulting in loss of over half the original sequence of URAT1 as well as in premature termination. Despite the almost certain loss of URAT1 function, this individual actually had hyperuricemia and gout, with an estimated FEua <5% (63a).
The transporter(s) underlying the residual reabsorption in the RST-null mice remains unclear. The present data indicate that Oat1 and Oat3 contribute to urate secretion rather than reabsorption. Other proteins postulated to participate in renal tubular transport of urate include, as mentioned earlier, Mrp2, Mrp4, UAT, and Oatv1 (Npt1) (reviewed in Refs. 22, 43, 46). None of the encoding genes (nor those encoding the lactate transporters discussed below) is clearly altered in mRNA expression in the RST knockout (Fig. 4B). Nevertheless, since any of these proteins might be under significant posttranscriptional control [as has previously been shown for UAT (33) as well as URAT1 (2, 10)], it remains possible that there might be compensatory alteration in their function in the knockout, e.g., due to changes in membrane localization. Of the above proteins, however, Mrp2, Mrp4, UAT, and Oatv1 have been postulated, on mechanistic grounds, to mediate urate efflux; thus, since each of these proteins is largely or exclusively confined to the apical membrane, they would be expected to function, like Oat1 and Oat3, in the tubular secretion rather than reabsorption of urate.
Interestingly, it was recently reported that mutant mice deficient in expression of renal Na+/lactate cotransporters (slc5a8 and slc5a12) manifest impaired reabsorption of urate with an approximately eightfold increase in urinary urate (59). These data were interpreted as follows: decreased tubular reabsorption of lactate (and possibly other RST-exchangeable organic anions) leads to diminished tubular cell availability of lactate, in turn resulting in decreased urate reabsorption via RST-mediated lactate/urate anion exchange. However, in light of the present results, it is also conceivable that one or both of these lactate transporters participates in an RST-independent mechanism of urate transport or even directly mediates urate uptake. Crosses between the lactate transporter-deficient and RST-null mice would help distinguish the above possibilities—a greater reabsorptive defect in the offspring of such crosses than in the RST knockouts would indicate a role for the lactate transporters in non-RST-mediated reabsorption.
Urate might confer certain physiological benefits explaining the existence of redundant mechanisms promoting its retention in the body. These include its antioxidant activity (1, 21) and probable contribution to the maintenance of blood pressure in salt-poor environments (65). Moreover, as noted above, urate in the tubular lumen can predispose to urate nephropathy, providing an additional rationale for a robust system for urate reabsorption. Nevertheless, in the context of the current human environment, excessive levels of systemic urate appear deleterious, clearly contributing to the development of gout and, while causal mechanisms are less clear here, correlating with cardiovascular disease and the metabolic syndrome (18, 43, 45, 58). Thus further characterization of the residual pathways for urate reabsorption in the RST knockout mice might open the door to development of therapeutics that are useful in a variety of pathophysiological contexts.
In conclusion, four or more genes (including RST, Oat1, and Oat3) regulate renal urate handling, at least in mice. While it remains to be determined whether any single gene is responsible for the bulk of urate transport, our results have potential medical significance. A number of studies are ongoing on polymorphisms influencing urate handling (in the context of gout, urate stones, and nephropathy, as well as metabolic syndrome). Our data suggest the need to consider searching for transporter genes other than RST, Oat1, and Oat3 that may contribute to the net renal excretion of urate.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the excellent technical assistance of Duke Vaughn, Kerstin Richter, and Shammi Closson and assistance from Dr. Jeff Long in the use of metabolism chambers.
GRANTS
S. A. Eraly was supported by NIH Grants DK-064839 and DK-075486, S. K. Nigam by Grant AI-057695, V. Vallon by Grant DK-56248 and by the Department of Veterans Affairs, and T. Rieg by a fellowship from the Deutsche Forschungsgemeinschaft (RI 1535/3-1). Quantitative real-time PCR was performed with the support of the Genomics Core at the Center for AIDS Research (AI-36214) and the Research Center for HIV and HCV Infection, San Diego Department of Veterans Affairs Healthcare System.
REFERENCES
- 1.Ames BN, Cathcart R, Schwiers E, Hochstein P. Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: a hypothesis. Proc Natl Acad Sci USA. 1981;78:6858–6862. doi: 10.1073/pnas.78.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anzai N, Miyazaki H, Noshiro R, Khamdang S, Chairoungdua A, Shin HJ, Enomoto A, Sakamoto S, Hirata T, Tomita K, Kanai Y, Endou H. The multivalent PDZ domain-containing protein PDZK1 regulates transport activity of renal urate-anion exchanger URAT1 via its C terminus. J Biol Chem. 2004;279:45942–45950. doi: 10.1074/jbc.M406724200. [DOI] [PubMed] [Google Scholar]
- 3.Babu E, Takeda M, Narikawa S, Kobayashi Y, Enomoto A, Tojo A, Cha SH, Sekine T, Sakthisekaran D, Endou H. Role of human organic anion transporter 4 in the transport of ochratoxin A. Biochim Biophys Acta. 2002;1590:64–75. doi: 10.1016/s0167-4889(02)00187-8. [DOI] [PubMed] [Google Scholar]
- 4.Bakhiya A, Bahn A, Burckhardt G, Wolff N. Human organic anion transporter 3 (hOAT3) can operate as an exchanger and mediate secretory urate flux. Cell Physiol Biochem. 2003;13:249–256. doi: 10.1159/000074539. [DOI] [PubMed] [Google Scholar]
- 5.Brady KP, Dushkin H, Fornzler D, Koike T, Magner F, Her H, Gullans S, Segre GV, Green RM, Beier DR. A novel putative transporter maps to the osteosclerosis (oc) mutation and is not expressed in the oc mutant mouse. Genomics. 1999;56:254–261. doi: 10.1006/geno.1998.5722. [DOI] [PubMed] [Google Scholar]
- 6.Buist SC, Cherrington NJ, Klaassen CD. Endocrine regulation of rat organic anion transporters. Drug Metab Dispos. 2003;31:559–564. doi: 10.1124/dmd.31.5.559. [DOI] [PubMed] [Google Scholar]
- 7.Buist SCN, Cherrington NJ, Choudhuri S, Hartley DP, Klaassen CD. Gender-specific and developmental influences on the expression of rat organic anion transporters. J Pharmacol Exp Ther. 2002;301:145–151. doi: 10.1124/jpet.301.1.145. [DOI] [PubMed] [Google Scholar]
- 8.Buist SCN, Klaassen CD. Rat and mouse differences in gender-predominant expression of organic anion transporter (OAT1-3; SLC22A6-8) mRNA levels. Drug Metab Dispos. 2004;32:620–625. doi: 10.1124/dmd.32.6.620. [DOI] [PubMed] [Google Scholar]
- 9.Burckhardt BC, Burckhardt G. Transport of organic anions across the basolateral membrane of proximal tubule cells. Rev Physiol Biochem Pharmacol. 2003;146:95–158. doi: 10.1007/s10254-002-0003-8. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham R, Brazie M, Kanumuru S, E X, Biswas R, Wang F, Steplock D, Wade JB, Anzai N, Endou H, Shenolikar S, Weinman EJ. Sodium-hydrogen exchanger regulatory factor-1 interacts with mouse urate transporter 1 to regulate renal proximal tubule uric acid transport. J Am Soc Nephrol. 2007;18:1419–1425. doi: 10.1681/ASN.2006090980. [DOI] [PubMed] [Google Scholar]
- 11.Dudas PL, Pelis RM, Braun EJ, Renfro JL. Transepithelial urate transport by avian renal proximal tubule epithelium in primary culture. J Exp Biol. 2005;208:4305–4315. doi: 10.1242/jeb.01879. [DOI] [PubMed] [Google Scholar]
- 12.Ekaratanawong S, Anzai N, Jutabha P, Miyazaki H, Noshiro R, Takeda M, Kanai Y, Sophasan S, Endou H. Human organic anion transporter 4 is a renal apical organic anion/dicarboxylate exchanger in the proximal tubules. J Pharmacol Sci. 2004;94:297–304. doi: 10.1254/jphs.94.297. [DOI] [PubMed] [Google Scholar]
- 13.Enomoto A, Kimura H, Chairoungdua A, Shigeta Y, Jutabha P, Cha SH, Hosoyamada M, Takeda M, Sekine T, Igarashi T, Matsuo H, Kikuchi Y, Oda T, Ichida K, Hosoya T, Shimokata K, Niwa T, Kanai Y, Endou H. Molecular identification of a renal urate anion exchanger that regulates blood urate levels. Nature. 2002;417:447–452. doi: 10.1038/nature742. [DOI] [PubMed] [Google Scholar]
- 14.Eraly SA, Bush KT, Sampogna RV, Bhatnagar V, Nigam SK. The molecular pharmacology of organic anion transporters: from DNA to FDA? Mol Pharmacol. 2004;65:479–487. doi: 10.1124/mol.65.3.479. [DOI] [PubMed] [Google Scholar]
- 15.Eraly SA, Hamilton BA, Nigam SK. Organic anion and cation transporters occur in pairs of similar and similarly expressed genes. Biochem Biophys Res Commun. 2003;300:333–342. doi: 10.1016/s0006-291x(02)02853-x. [DOI] [PubMed] [Google Scholar]
- 16.Eraly SA, Monte JC, Nigam SK. Novel slc22 transporter homologs in fly, worm, and human clarify the phylogeny of organic anion and cation transporters. Physiol Genomics. 2004;18:12–24. doi: 10.1152/physiolgenomics.00014.2004. [DOI] [PubMed] [Google Scholar]
- 17.Eraly SA, Vallon V, Vaughn DA, Gangoiti JA, Richter K, Nagle M, Monte JC, Rieg T, Truong DM, Long JM, Barshop BA, Kaler G, Nigam SK. Decreased renal organic anion secretion and plasma accumulation of endogenous organic anions in OAT1 knockout mice. J Biol Chem. 2006;281:5072–5083. doi: 10.1074/jbc.M508050200. [DOI] [PubMed] [Google Scholar]
- 18.Feig DI, Mazzali M, Kang DH, Nakagawa T, Price K, Kannelis J, Johnson RJ. Serum uric acid: a risk factor and a target for treatment? J Am Soc Nephrol. 2006;17:S69–S73. doi: 10.1681/ASN.2005121331. [DOI] [PubMed] [Google Scholar]
- 19.Gorman GS, Tamura T, Baggott JE. Mass spectrometric method for detecting carbon 13 enrichment introduced by folate coenzymes in uric acid. Anal Biochem. 2003;321:188–191. doi: 10.1016/s0003-2697(03)00398-1. [DOI] [PubMed] [Google Scholar]
- 20.Hagos Y, Stein D, Ugele B, Burckhardt G, Bahn A. Human renal organic anion transporter 4 operates as an asymmetric urate transporter. J Am Soc Nephrol. 2007;18:430–439. doi: 10.1681/ASN.2006040415. [DOI] [PubMed] [Google Scholar]
- 21.Hediger MA. Kidney function: gateway to a long life? Nature. 2002;417:393–395. doi: 10.1038/417393a. [DOI] [PubMed] [Google Scholar]
- 22.Hediger MA, Johnson RJ, Miyazaki H, Endou H. Molecular physiology of urate transport. Physiology (Bethesda) 2005;20:125–133. doi: 10.1152/physiol.00039.2004. [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann G, Aramaki S, Blum-Hoffmann E, Nyhan WL, Sweetman L. Quantitative analysis for organic acids in biological samples: batch isolation followed by gas chromatographic-mass spectrometric analysis. Clin Chem. 1989;35:587–595. [PubMed] [Google Scholar]
- 24.Hosoyamada M, Ichida K, Enomoto A, Hosoya T, Endou H. Function and localization of urate transporter 1 in mouse kidney. J Am Soc Nephrol. 2004;15:261–268. doi: 10.1097/01.asn.0000107560.80107.19. [DOI] [PubMed] [Google Scholar]
- 25.Hosoyamada M, Sekine T, Kanai Y, Endou H. Molecular cloning and functional expression of a multispecific organic anion transporter from human kidney. Am J Physiol Renal Physiol. 1999;276:F122–F128. doi: 10.1152/ajprenal.1999.276.1.F122. [DOI] [PubMed] [Google Scholar]
- 26.Hsiao A, Ideker T, Olefsky JM, Subramaniam S. VAMPIRE microarray suite: a web-based platform for the interpretation of gene expression data. Nucleic Acids Res. 2005;33:W627–W632. doi: 10.1093/nar/gki443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsiao A, Worrall DS, Olefsky JM, Subramaniam S. Variance-modeled posterior inference of microarray data: detecting gene-expression changes in 3T3-L1 adipocytes. Bioinformatics. 2004;20:3108–3127. doi: 10.1093/bioinformatics/bth371. [DOI] [PubMed] [Google Scholar]
- 28.Ichida K, Hosoyamada M, Hisatome I, Enomoto A, Hikita M, Endou H, Hosoya T. Clinical and molecular analysis of patients with renal hypouricemia in Japan: influence of URAT1 gene on urinary urate excretion. J Am Soc Nephrol. 2004;15:164–173. doi: 10.1097/01.asn.0000105320.04395.d0. [DOI] [PubMed] [Google Scholar]
- 29.Ichida K, Hosoyamada M, Kimura H, Takeda M, Utsunomiya Y, Hosoya T, Endou H. Urate transport via human PAH transporter hOAT1 and its gene structure. Kidney Int. 2003;63:143–155. doi: 10.1046/j.1523-1755.2003.00710.x. [DOI] [PubMed] [Google Scholar]
- 30.Imaoka T, Kusuhara H, Adachi-Akahane S, Hasegawa M, Morita N, Endou H, Sugiyama Y. The renal-specific transporter mediates facilitative transport of organic anions at the brush border membrane of mouse renal tubules. J Am Soc Nephrol. 2004;15:2012–2022. doi: 10.1097/01.ASN.0000135049.20420.E5. [DOI] [PubMed] [Google Scholar]
- 31.Kojima R, Sekine T, Kawachi M, Cha SH, Suzuki Y, Endou H. Immunolocalization of multispecific organic anion transporters, OAT1, OAT2, and OAT3, in rat kidney. J Am Soc Nephrol. 2002;13:848–857. doi: 10.1681/ASN.V134848. [DOI] [PubMed] [Google Scholar]
- 32.Komatsuda A, Iwamoto K, Wakui H, Sawada K, Yamaguchi A. Analysis of mutations in the urate transporter 1 (URAT1) gene of Japanese patients with hypouricemia in northern Japan and review of the literature. Ren Fail. 2006;28:223–227. doi: 10.1080/08860220600580365. [DOI] [PubMed] [Google Scholar]
- 33.Lipkowitz MS, Leal-Pinto E, Cohen BE, Abramson RG. Galectin 9 is the sugar-regulated urate transporter/channel UAT. Glycoconj J. 2004;19:491–498. doi: 10.1023/B:GLYC.0000014078.65610.2f. [DOI] [PubMed] [Google Scholar]
- 34.Lise MF, El-Husseini A. The neuroligin and neurexin families: from structure to function at the synapse. Cell Mol Life Sci. 2006;63:1833–1849. doi: 10.1007/s00018-006-6061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ljubojevic M, Herak-Kramberger CM, Hagos Y, Bahn A, Endou H, Burckhardt G, Sabolic I. Rat renal cortical OAT1 and OAT3 exhibit gender differences determined by both androgen stimulation and estrogen inhibition. Am J Physiol Renal Physiol. 2004;287:F124–F138. doi: 10.1152/ajprenal.00029.2004. [DOI] [PubMed] [Google Scholar]
- 36.Lopez-Nieto CE. NKT cDNA sequence, GenBank Accession Number MMU52842. 1996 March 27; Submitted. [Google Scholar]
- 37.Lopez-Nieto CE, Nigam SK. Selective amplification of protein-coding regions of large sets of genes using statistically designed primer sets. Nat Biotechnol. 1996;14:857–861. doi: 10.1038/nbt0796-857. [DOI] [PubMed] [Google Scholar]
- 38.Lopez-Nieto CE, You G, Barros EJG, Beier DR, Nigam SK. Molecular cloning and characterization of a novel transport protein with very high expression in the kidney. J Am Soc Nephrol. 1996;7:1301. (Abstract) [Google Scholar]
- 39.Lopez-Nieto CE, You G, Bush KT, Barros EJG, Beier DR, Nigam SK. Molecular cloning and characterization of NKT, a gene product related to the organic cation transporter family that is almost exclusively expressed in the kidney. J Biol Chem. 1997;272:6471–6478. doi: 10.1074/jbc.272.10.6471. [DOI] [PubMed] [Google Scholar]
- 40.Mori K, Ogawa Y, Ebihara K, Aoki T, Tamura N, Sugawara A, Kuwahara T, Ozaki S, Mukoyama M, Tashiro K. Kidney-specific expression of a novel mouse organic cation transporter-like protein. FEBS Lett. 1997;417:371–374. doi: 10.1016/s0014-5793(97)01325-2. [DOI] [PubMed] [Google Scholar]
- 41.Moriwaki Y, Yamamoto T, Higashino K. Enzymes involved in purine metabolism—a review of histochemical localization and functional implications. Histol Histopathol. 1999;14:1321–1340. doi: 10.14670/HH-14.1321. [DOI] [PubMed] [Google Scholar]
- 42.Motohashi H, Sakurai Y, Saito H, Masuda S, Urakami Y, Goto M, Fukatsu A, Ogawa O, Inui K. Gene expression levels and immunolocalization of organic ion transporters in the human kidney. J Am Soc Nephrol. 2002;13:866–874. doi: 10.1681/ASN.V134866. [DOI] [PubMed] [Google Scholar]
- 43.Mount DB. Molecular physiology and the four-component model of renal urate transport. Curr Opin Nephrol Hypertens. 2005;14:460–463. doi: 10.1097/01.mnh.0000170749.10785.04. [DOI] [PubMed] [Google Scholar]
- 44.Mutch DM, O’Maille G, Wikoff WR, Wiedmer T, Sims PJ, Siuzdak G. Mobilization of pro-inflammatory lipids in obese Plscr3-deficient mice. Genome Biol. 2007;8:R38. doi: 10.1186/gb-2007-8-3-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakagawa T, Kang DH, Feig D, Sanchez-Lozada LG, Srinivas TR, Sautin Y, Ejaz AA, Segal M, Johnson RJ. Unearthing uric acid: an ancient factor with recently found significance in renal and cardiovascular disease. Kidney Int. 2006;69:1722–1725. doi: 10.1038/sj.ki.5000391. [DOI] [PubMed] [Google Scholar]
- 46.Rafey MA, Lipkowitz MS, Leal-Pinto E, Abramson RG. Uric acid transport. Curr Opin Nephrol Hypertens. 2003;12:511–516. doi: 10.1097/00041552-200309000-00005. [DOI] [PubMed] [Google Scholar]
- 47.Sekine T, Watanabe N, Hosoyamada M, Kanai Y, Endou H. Expression cloning and characterization of a novel multispecific organic anion transporter. J Biol Chem. 1997;272:18526–18529. doi: 10.1074/jbc.272.30.18526. [DOI] [PubMed] [Google Scholar]
- 48.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 49.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 50.Stuart RO, Bush KT, Nigam SK. Changes in gene expression patterns in the ureteric bud and metanephric mesenchyme in models of kidney development. Kidney Int. 2003;64:1997–2008. doi: 10.1046/j.1523-1755.2003.00383.x. [DOI] [PubMed] [Google Scholar]
- 51.Stuart RO, Bush KT, Nigam SK. Changes in global gene expression patterns during development and maturation of the rat kidney. Proc Natl Acad Sci USA. 2001;98:5649–5654. doi: 10.1073/pnas.091110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sweet DH, Chan LM, Walden R, Yang XP, Miller DS, Pritchard JB. Organic anion transporter 3 (Slc22a8) is a dicarboxylate exchanger indirectly coupled to the Na+ gradient. Am J Physiol Renal Physiol. 2003;284:F763–F769. doi: 10.1152/ajprenal.00405.2002. [DOI] [PubMed] [Google Scholar]
- 53.Sweet DH, Eraly SA, Vaughn DA, Bush KT, Nigam SK. Organic anion and cation transporter expression and function during embryonic kidney development and in organ culture model systems. Kidney Int. 2006;69:837–845. doi: 10.1038/sj.ki.5000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sweet DH, Miller DS, Pritchard JB, Fujiwara Y, Beier DR, Nigam SK. Impaired organic anion transport in kidney and choroid plexus of organic anion transporter 3 (Oat3 [Slc22a8]) knockout mice. J Biol Chem. 2002;277:26934–26943. doi: 10.1074/jbc.M203803200. [DOI] [PubMed] [Google Scholar]
- 55.Sweet DH, Wolff NA, Pritchard JB. Expression cloning and characterization of ROAT1. The basolateral organic anion transporter in rat kidney. J Biol Chem. 1997;272:30088–30095. doi: 10.1074/jbc.272.48.30088. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka M, Itoh K, Matsushita K, Matsushita K, Wakita N, Adachi M, Nonoguchi H, Kitamura K, Hosoyamada M, Endou H, Tomita K. Two male siblings with hereditary renal hypouricemia and exercise-induced ARF. Am J Kidney Dis. 2003;42:1287–1292. doi: 10.1053/j.ajkd.2003.08.032. [DOI] [PubMed] [Google Scholar]
- 57.Terkeltaub R, Bushinsky DA, Becker MA. Recent developments in our understanding of the renal basis of hyperuricemia and the development of novel antihyperuricemic therapeutics. Arthritis Res Ther. 2006;8 Suppl 1:S4. doi: 10.1186/ar1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Terkeltaub RA. Clinical practice. Gout. N Engl J Med. 2003;349:1647–1655. doi: 10.1056/NEJMcp030733. [DOI] [PubMed] [Google Scholar]
- 59.Thangaraju M, Ananth S, Martin PM, Roon P, Smith SB, Sterneck E, Prasad PD, Ganapathy V. c/ebpδ Null mouse as a model for the double knock-out of slc5a8 and slc5a12 in kidney. J Biol Chem. 2006;281:26769–26773. doi: 10.1074/jbc.C600189200. [DOI] [PubMed] [Google Scholar]
- 60.Tojo A, Sekine T, Nakajima N, Hosoyamada M, Kanai Y, Kimura K, Endou H. Immunohistochemical localization of multispecific renal organic anion transporter 1 in rat kidney. J Am Soc Nephrol. 1999;10:464–471. doi: 10.1681/ASN.V103464. [DOI] [PubMed] [Google Scholar]
- 61.Tzovaras V, Chatzikyriakidou A, Bairaktari E, Liberopoulos EN, Georgiou I, Elisaf M. Absence of SLC22A12 gene mutations in Greek Caucasian patients with primary renal hypouricaemia. Scand J Clin Lab Invest. 2007;67:589–595. doi: 10.1080/00365510701222868. [DOI] [PubMed] [Google Scholar]
- 61a.Vallon V, Rieg T, Ahn S-Y, Wu W, Eraly SA, Nigam SK. Overlapping in vitro and in vivo specificities of the organic anion transporters OAT1 and OAT3 for loop and thiazide diuretics. Am J Physiol Renal Physiol. 2008 January 23; doi: 10.1152/ajprenal.00528.2007. doi:10.1152/ajprenal.00528.2007. [DOI] [PubMed] [Google Scholar]
- 62.Van Aubel RAMH, Smeets PHE, van den Heuvel JJMW, Russel FGM. Human organic anion transporter MRP4 (ABCC4) is an efflux pump for the purine end metabolite urate with multiple allosteric substrate binding sites. Am J Physiol Renal Physiol. 2005;288:F327–F333. doi: 10.1152/ajprenal.00133.2004. [DOI] [PubMed] [Google Scholar]
- 63.VanWert AL, Bailey RM, Sweet DH. Organic anion transporter 3 (Oat3/Slc22a8) knockout mice exhibit altered clearance and distribution of penicillin G. Am J Physiol Renal Physiol. 2007;293:F1332–F1341. doi: 10.1152/ajprenal.00319.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63a.Vazquez-Mellado J, Alvarado-Romano V, Burgos-Vargas R, Jimenez-Vaca AL, Pozo-Molina G, Cuevas-Covarrubias SA. Homozygous frameshift mutation in the SLC22A12 gene in a patient with primary gout and high levels of serum uric acid. J Clin Pathol. 2007;60:947–948. doi: 10.1136/jcp.2006.037473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Want EJ, Cravatt BF, Siuzdak G. The expanding role of mass spectrometry in metabolite profiling and characterization. Chembiochem. 2005;6:1941–1951. doi: 10.1002/cbic.200500151. [DOI] [PubMed] [Google Scholar]
- 65.Watanabe S, Kang DH, Feng L, Nakagawa T, Kanellis J, Lan H, Mazzali M, Johnson RJ. Uric acid, hominoid evolution, and the pathogenesis of salt-sensitivity. Hypertension. 2002;40:355–360. doi: 10.1161/01.hyp.0000028589.66335.aa. [DOI] [PubMed] [Google Scholar]
- 66.Wikoff WR, Gangoiti JA, Barshop BA, Siuzdak G. Metabolomics identifies perturbations in human disorders of propionate metabolism. Clin Chem. 2007;53:2169–2176. doi: 10.1373/clinchem.2007.089011. [DOI] [PubMed] [Google Scholar]
- 67.Wright SH, Dantzler WH. Molecular and cellular physiology of renal organic cation and anion transport. Physiol Rev. 2004;84:987–1049. doi: 10.1152/physrev.00040.2003. [DOI] [PubMed] [Google Scholar]
- 68.Wu X, Lee CC, Muzny DM, Caskey CT. Urate oxidase: primary structure and evolutionary implications. Proc Natl Acad Sci USA. 1989;86:9412–9416. doi: 10.1073/pnas.86.23.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu X, Wakamiya M, Vaishnav S, Geske R, Montgomery C, Jr, Jones P, Bradley A, Caskey C. Hyperuricemia and urate nephropathy in urate oxidase-deficient mice. Proc Natl Acad Sci USA. 1994;91:742–746. doi: 10.1073/pnas.91.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]