Nitric oxide associated-1 (NOA1) is an evolutionarily conserved guanosine triphosphate binding protein that localizes predominantly to mitochondria in mammalian cells. Here we determine NOA1 function through generation of knock-out mice and in vitro assays.
Abstract
Nitric oxide associated-1 (NOA1) is an evolutionarily conserved guanosine triphosphate (GTP) binding protein that localizes predominantly to mitochondria in mammalian cells. On the basis of bioinformatic analysis, we predicted its possible involvement in ribosomal biogenesis, although this had not been supported by any experimental evidence. Here we determine NOA1 function through generation of knockout mice and in vitro assays. NOA1-deficient mice exhibit midgestation lethality associated with a severe developmental defect of the embryo and trophoblast. Primary embryonic fibroblasts isolated from NOA1 knockout embryos show deficient mitochondrial protein synthesis and a global defect of oxidative phosphorylation (OXPHOS). Additionally, Noa1–/– cells are impaired in staurosporine-induced apoptosis. The analysis of mitochondrial ribosomal subunits from Noa1–/– cells by sucrose gradient centrifugation and Western blotting showed anomalous sedimentation, consistent with a defect in mitochondrial ribosome assembly. Furthermore, in vitro experiments revealed that intrinsic NOA1 GTPase activity was stimulated by bacterial ribosomal constituents. Taken together, our data show that NOA1 is required for mitochondrial protein synthesis, likely due to its yet unidentified role in mitoribosomal biogenesis. Thus, NOA1 is required for such basal mitochondrial functions as adenosine triphosphate (ATP) synthesis and apoptosis.
INTRODUCTION
Mitochondria are the principal energy-producing organelles believed to have evolved from eubacteria that were engulfed by primordial eukaryotic cells (Gray et al., 2001). In the course of evolution mitochondria retained rudimentary genomes, whereas most of the other necessary genetic information was relocated to the nucleus of the host cell. As a result, in mammalian cells only 13 proteins (all of which are translated by mitochondrial and not cytosolic ribosomes) are encoded by mitochondrial DNA (mtDNA). Mitochondrial ribosomes (mitoribosomes) are assembled from more than 70 nuclearly encoded proteins (at least 29 in the 28S small subunit and 48 in the 39S large subunit) and 2 species of mitochondrially encoded rRNA (Koc et al., 2001). Relatively little is known about how these multiple components assemble into a catalytically active complex and what factors are required for completion of this process. We identified nitric oxide-associated-1 (NOA1) as a predominantly mitochondrially localized guanosine triphosphate (GTP) binding protein, homologous to the essential Bacillus subtilis GTPase, YqeH (Zemojtel et al., 2006b). YqeH defines a subfamily of circularly permutated GTPases conserved in some species of bacteria and all known eukaryotic organisms (Leipe et al., 2002). The protein belongs to a larger family of YlqF/YawG translation factors (TRAFAC), whose members, such as YlqF, have been implicated in ribosomal assembly in B. subtilis (Kim do et al., 2008). Independent of homology to bacterial YqeH, a possible involvement of NOA1 in mitoribosome function has been implicated by data from the yeast protein interactome (Zemojtel et al., 2006a). Here the NOA1 yeast homologue YOR205C was shown to be a part of a protein complex associated with the S5 protein of the small mitoribosomal subunit. YqeH has been suggested to participate in the biogenesis of the 30S ribosomal subunit and to assist in 50S ribosomal subunit assembly (Loh et al., 2007; Uicker et al., 2007). In a recent study, human NOA1 was shown to interact with complex I of the electron transport chain and with three mitoribosomal proteins, MRPL12, MRPS27, and MRPS29 (also known as death-associated protein 3 [DAP3]), indicating that NOA1 may interact with the mitoribosome (Tang et al., 2009). To date, however, no experimental data were available to support a role for NOA1 in mitochondrial protein synthesis. Similarly, a physiological role of the protein in mammalian development was also unknown. Here we present a comprehensive analysis of the cellular and developmental role of mammalian NOA1. We show that NOA1 inactivation impairs mitochondrial protein synthesis and causes global defect of oxidative phosphorylation (OXPHOS), defective apoptosis, and midgestation lethality of knockout mice.
RESULTS
Noa1 gene inactivation results in midgestation lethality
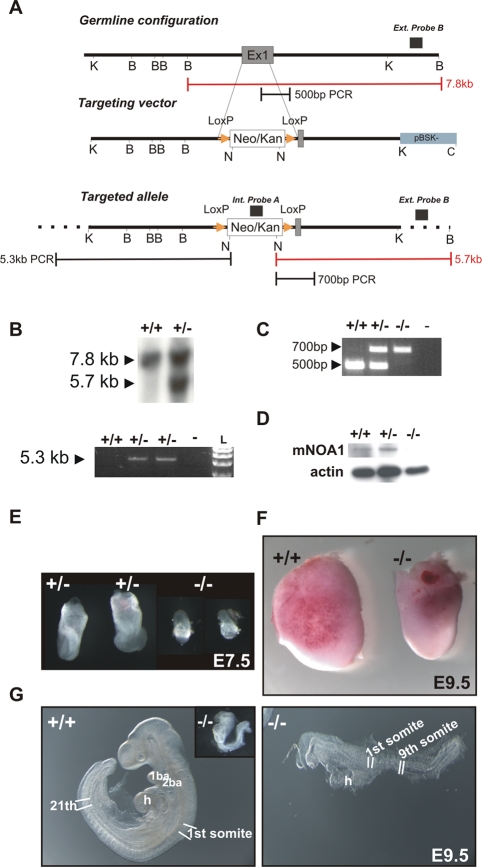
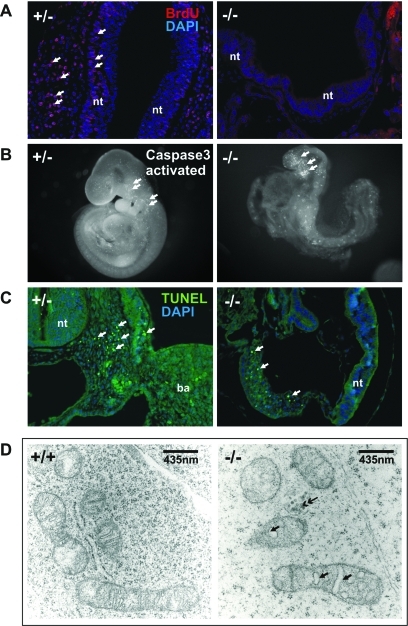
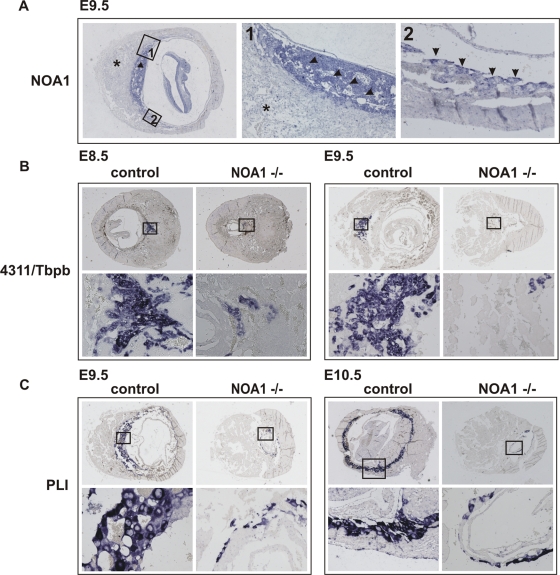
To experimentally address the function of NOA1 in a physiological in vivo context, we generated mice in which Noa1 was inactivated (Figure 1). Mutant embryos appeared growth retarded and at E9.5 a maximum of nine somites was observed (Figure 1, E–G, and Figure 2A). At E10.5 many embryos were necrotic, and no viable Noa1–/–embryos were detected thereafter, indicating midgestation lethality (Table 1). Cell proliferation as measured by bromodeoxyuridine (BrdU) incorporation was drastically reduced in E9.5 Noa1–/– embryos (Figure 2A), and few areas of apoptotic cell death were observed (Figure 2, B and C), indicating that cell proliferation as well as apoptosis was severely impaired. Transmission electron microscopy (TEM) of Noa1–/– embryos revealed abnormal mitochondria with characteristically swollen cristae (Figure 2D) but no abnormalities in other organelles. In addition, we observed severe defects of the placenta with a reduction of all three trophoblast layers, including the trophoblast giant cell (TGC) layer (Figure 3, Supplemental Figure S1). This finding was in line with Noa1 being expressed in the trophoblast but not in the maternal part of the placenta (Figure 3A). Together these results indicated that Noa1 is indispensable for normal development of the embryo and the placenta possibly due to an important function in mitochondria.
FIGURE 1:
Generation of the Noa1 knockout mice. (A) Strategy of Noa1 gene targeting. The top bar indicates the enzymatic cleavage sites at the Noa1 genomic locus (BamHI), K (KpnI), N (NotI), and C (ClaI). GTPase domain encoding Exon1 (dark grey segment) was targeted. Homologous recombination using this construct (middle bar) resulted in a targeted allele (bottom bar) where a large part of exon1 was replaced by the loxP flanked, PGK-gb2 promoter driven, neomycin/kanamycin resistance cassette (white segment – Neo/Kan; yellow triangles – loxP sites). Insertion of the selection cassette introduced a NotI endonuclease recognition site enabling Southern blot–based detection of homologous recombination with the internal and external probes (black boxes above the panels). Red rulers indicate expected BamHI/NotI fragment sizes for the wild-type locus as well as the altered fragment size in the homologously targeted locus. Black rulers indicate lengths of the long-range PCR products obtained from the wild type and homologously targeted locus. (B) Southern blot and long-range-PCR–based detection of the homologous recombination. (C) PCR-based genotyping strategy. (D) Western blot analyses of E9.5 wild type, Noa+/–, and Noa1–/– embryo lysates with β-actin as loading control. (E) Appearance of Noa1 knockout embryos at E7.5. (F) A comparison of the intact embryos at E9.5 is shown to illustrate the difference in size. (G) Noa1 knockout embryo development is arrested at prerotation stage (inset). E9.5 wild-type embryo displays 21 somites whereas NOA1-deficient littermate (magnified) reaches maximally a 9-somite stage. See also Supplemental Movies S1 and S2 available online.
FIGURE 2:
Phenotype of Noa1 knockout mice. (A) BrdU in-vivo labeling reveals arrest of proliferation in the E9.5 Noa–/– embryos. (B and C) Apoptosis in the E9.5 Noa1–/– embryos visualized by anti-activated caspase-3 immunolabeling in the whole mount preparations (B) and TUNEL staining on paraffin sections (C). Data are representative of at least three embryos analyzed for each genotype. (D) Electron microscopic analysis of the Noa1–/– embryos. Many mitochondria show aberrant morphology characterized by swollen intracristal spaces (single-headed arrows). Some appear collapsed into electron dense, shrunken remnants (double-headed arrow). Endoplasmic reticulum and other organelles appear normal in Noa1–/– embryos (data not shown). ba – branchial arche, h – heart, nt – neural tube.
Table 1.:
Intercross of Noa1+/− mice – offspring genotype distribution. Knockout embryos could not be detected beyond E10.5, indicating midgestation lethality. Heterozygous animals were born in the expected Mendelian ratio. No overt lethality of heterozygotes was observed in the postnatal period.
| Stage | Litters | Noa1+/+ | Noa1+/− | Noa1−/− | Total |
|---|---|---|---|---|---|
| E7.5 | 2 | 2 (10%) | 13 (65%) | 5 (25%) | 20 |
| E8.5 | 7 | 14 (25%) | 32 (58%) | 9 (16%) | 55 |
| E9.5 | 16 | 21 (17%) | 80 (66%) | 20 (17.0%) | 121 |
| E10.5 | 5 | 6 (17%) | 22 (61%) | 8 (22%) | 36 |
| E14.5 | 1 | 2 (33.3%) | 4 (66.6%) | 0 | 6 |
| E15.5 | 6 | 16 (31%) | 36 (69%) | 0 | 52 |
| E16.5 | 2 | 5 (33.3%) | 10 (66.6%) | 0 | 15 |
| P1 | 5 | 9 (25%) | 27 (75%) | 0 | 36 |
| P28 | 11 | 32 (38%) | 52 (62%) | 0 | 84 |
FIGURE 3:
Noa1 is indispensable for placental trophoblast development. In situ hybridization on the sections of E8.5, E9.5, and E10.5 wild type and mutant littermate embryos at the implantation sites. Boxed areas in the top panels are enlarged in bottom panels. (A) Noa1 expression is restricted to the placental trophoblast at E9.5 (arrow). Intense expression in the labyrinth and spongiotrophoblast (arrowheads – magnification 1×) and in the trophoblast giant cells (arrows – magnification 2×). Noa1 expression could not be detected in the maternal part of the placenta (star) or in the knockout embryos (data not shown). (B) Defect of the trophoblast progenitor cell differentiation in the Noa1–/– mice. 4311/Tbpb is expressed in the trophoblast progenitor cells which give rise to glycogen cells and secondary giant trophoblast cells. Diminished 4311/Tbpb expression in the E8.5, E9.5 ectoplacental cone. (C) Defect of trophoblast giant cell differentiation in E9.5 and 10.5 Noa1–/– embryos visualized by diminished expression of placental lactogen I (PLI).
Global OXPHOS defect in the Noa1–/– cells
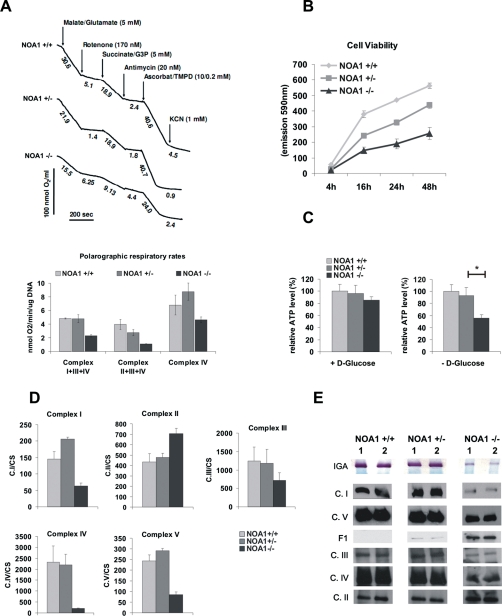
To gain further information on the biochemical consequences of Noa1 inactivation we isolated primary embryonic fibroblasts from E9.5 Noa1–/– embryos. Noa1-deficient cells grew in a medium known to compensate for mitochondrial respiratory deficiency and showed no overt abnormalities in the mitochondrial network (Supplemental Figure S3A). To test mitochondrial function we measured cellular respiration by polarographic assays in digitonin-permeabilized cells. Complex I–, III–, and IV–dependent oxygen consumption was severely reduced in mutant cells (Figure 4A), indicating a global OXPHOS defect. Consequently, Noa1 knockout cells showed reduced viability and cellular ATP content when grown under nutrient restriction (Figure 4, B and C). Similar results were obtained after NOA1 depletion in HeLa cells (Supplemental Figure S2).
FIGURE 4:
OXPHOS deficiency in Noa1–/– cells. (A) Cells were permeabilized with digitonin, and oxygen consumption was monitored polarographically in the presence of respiratory chain substrates and inhibitors. Note a marked decrease of the complex I–, III–, and IV–dependent oxygen consumption but not complex II–dependent oxygen consumption. Results represent mean ± SD of three independent experiments. (B) Reduced viability of the NOA1-deficient cells was determined with Alamarblue. (C) Noa1–/– cells show reduced ATP levels upon glucose deprivation. See also Supplemental Figure S6 for genetic complementation data. (D) Reduced enzymatic activities of the mitochondrial complexes I, III, IV, and V in the knockout cells. In contrast, complex II activity was increased. The enzymatic assay values are shown standardized to CS E3 activity. Results represent mean ± SD of three independent experiments. (E) In-gel activity (IGA) of complex I is decreased in knockout cells. Blue native electrophoresis (BN) showing reduction in quantities of the assembled complexes I, III, IV, and V and accumulation of the unassembled complex V intermediate (F1) in the homozygous Noa1 mutant cells.
Because the results of polarographic measurements are influenced by mitochondrial substrate import, we further addressed OXPHOS complex function in direct biochemical assays. Spectroscopic assays revealed that the activity of complexes I, III, IV, and V was strongly reduced in Noa1–/– cells, whereas the activity of complex II was increased (Figure 4D). Blue-native electrophoresis of isolated mitochondria showed a decreased amount of assembled complex I, III, IV, and V (Figure 4E) and an accumulation of the unassembled complex V (F1), whereas no free F1 subunit was detected in wild-type cells. Confirmatory to this, in-gel activity of complex I measured by the colorimetric enzymatic assay (immunoglobulin E [IgE]) was also strongly reduced (Figure 4E, top stripe). These findings suggested a general defect of mitochondrial protein synthesis in Noa1–/– cells, as complex I, III, IV, and V all include proteins encoded by the mtDNA, whereas all proteins of complex II are encoded in the nucleus and imported into the mitochondrium.
Compromised mitochondrial protein synthesis in Noa1–/– cells
To test this possibility, Noa1–/– cells were assayed for mitochondrial protein synthesis. Radioactive methionine labeling showed a deficiency in de novo mitochondrial protein synthesis (Figure 5A). Normal copy number and integrity of mtDNA were confirmed by quantitative PCR (qPCR) and long-range PCR, excluding defects of replication and/or mtDNA repair as a cause of compromised protein synthesis (Supplemental Figure S3, B and C). Expression of genes encoding proteins of the electron transport chain or mitoribosomal function was increased in the mutant cells, indicating that mtDNA transcription in the knockout cells was maintained, increased expression likely being compensatory to the protein synthesis defect (Figure 5B). Interestingly, 16S rRNA transcript level from the large ribosomal subunit was increased twofold in mutant cells whereas 12S rRNA remained at the control level indicative of a possible defect of mitoribosome biogenesis (Uicker et al., 2007). These results are consistent with NOA1 being required for mitochondrial protein synthesis.
FIGURE 5:
NOA1 deficiency impairs mitochondrial protein synthesis. (A) [35S]methionine labeling of mitochondrial protein synthesis in primary embryonic fibroblasts reveals deficient translation in NOA1 knockout cells. Concentrations of protein lysates were calculated by the bicinchoninic acid (BCA) method, and equal loading was confirmed by Coomassie staining following exposure (data not shown). (B) Real-time PCR quantification of the mitochondrial gene transcript levels. RNA species encoded by mitochondrial (underlined red) and nuclear (underlined blue) DNA were quantified. Data were normalized against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and actin expression. (C) Cell lysates were fractionated in the 10–30% (vol/vol) isokinetic sucrose gradients and analyzed by Western blots with antibodies against MRPL12 (39S mitoribosomal subunit) or MRPS18b (28S mitoribosomal subunit). Representative blots were analyzed densitometrically (bottom panel). See also Supplemental Figure S4 for genetic complementation data. comp. – knockout cells with retroviral complementation of NOA1 expression.
Altered mitoribosomal profile in the Noa1–/– cells
Protein synthesis within mitochondria is performed by 55S mitoribosomes, each of which consists of a small 28S subunit and a large 39S subunit. To determine whether the observed mitochondrial protein synthesis defect was due to a ribosomal dysfunction, we compared the mitoribosome sucrose density profiles of wild type or rescued cells and those lacking Noa1. The small subunit was traced with antibodies against the mitochondrial protein S18b (MRPS18b), which consistently revealed a similar profile between wild type (NOA1+/+) and knockout cells (NOA1–/–). Similar levels of the 28S small subunit were found in fractions 4 and 5 in the wild type, knockout (NOA1–/–), and complemented cells (NOA1–/– Comp.), but no 55S ribosomes could be visualized (Figure 5C, fractions 8–10). Antibodies against the mitochondrial protein L12 (MRPL12), which reacted with the large 39S subunit (fractions 6 and 7), revealed the decrease in 55S ribosomes in the NOA1–/– cell line (fractions 8–10). Furthermore, although Noa1 deficiency had only a minimal effect on the position or band intensity of the small subunit in the sucrose gradient (Figure 4C, bottom panels), the large subunits showed a marked shift to the slower migrating particles (lane 5, NOA1–/– cf. NOA1+/+); these effects were rescued by adding Noa1 in trans (NOA1–/– comp. in Figure 5C). Additionally, NOA1 complementation led to increased abundance of L12 protein in fractions 9 and 10. In line with the observed partial rescue of mitoribosomal assembly, retroviral complementation resulted in reactivation of mitochondrial protein synthesis, normalization of respiratory chain complex assembly, and improved viability of the knockout cells (Supplemental Figure S4). These data strongly suggest that inactivation of Noa1 resulted in an impaired assembly of the 55S mitoribosome.
The GTPase activities of mammalian NOA1 and its bacterial homologue YqeH are stimulated by ribosomal components from Escherichia coli
To gain further insight into NOA1 function, we purified mammalian NOA1 as well as its bacterial homologue YqeH from B. subtilis. Both mitochondrial NOA1 and bacterial YqeH showed a high and comparably strong intrinsic GTPase activity (Figure 6A). This observation was not made with other G-factors involved in translation, such as elongation factor G (EF-G), which can be stimulated exclusively by 70S ribosomes during protein synthesis and is boosted by empty 70S uncoupled from protein synthesis (Gordon, 1970; Qin et al., 2006). In line with a previously proposed role in 30S assembly, YqeH GTPase activity could be stimulated by E. coli 16S rRNA, more than twofold, whereas the addition of 23S rRNA had no effect (Figure 6B), similar to poly(U) addition (data not shown).
FIGURE 6:
GTPase activities of NOA1 and YqeH are differently stimulated by ribosomal components from E. coli. (A) Intrinsic GTPase activities of NOA1 and YqeH at 30°C. (B) Specific stimulation of YqeH GTPase activity by E. coli 16S rRNA at 30°C. (C) Specific stimulation of YqeH GTPase by equimolar amounts of E. coli 30S precursors (21S) in comparison to total reconstituted 30S subunits (21S+TP30), ribosomal subunits (30S, 50S), and 70S ribosomes at 20°C. (D) Stimulation of NOA1 GTPase by equimolar amounts of mitochondrial rRNA (mt12S, mt16S) and E. coli rRNA (Ec16S, Ec23S) at 20°C. (E) Stimulation of NOA1 GTPase by equimolar amounts of E. coli 30S precursors before (21S) and after total reconstitution of 30S particles (21S+TP30), ribosomal subunits (30S, 50S), and 70S ribosomes at 20°C.
In addition, 21S precursor particles of the 30S subunit stimulated the YqeH GTPase activity rather than mature 30S and 50S subunits or 70S ribosomes (Figure 6C). In contrast, NOA1 activity was stimulated by rRNAs of the small and large ribosomal subunits from both mammalian (human) mitochondria (12S and 16S rRNAs) and E. coli (16S and 23S rRNAs) by 40% (Figure 6D). Precursor of the 30S E. coli ribosome, the 21S particle prepared by total reconstitution (Nierhaus, 1990), stimulated NOA1 activity with approximately the same intensity (Figure 6E). Total reconstituted 30S particles as well as native 30S subunits induced a striking twofold increase of GTPase activity. Interestingly, the strongest effect was observed with 50S particles, whereas 70S ribosomes showed four times less stimulation underlining the specificity of the observed effects. These results suggest an involvement of both bacterial YqeH and mammalian NOA1 in ribosomal biogenesis.
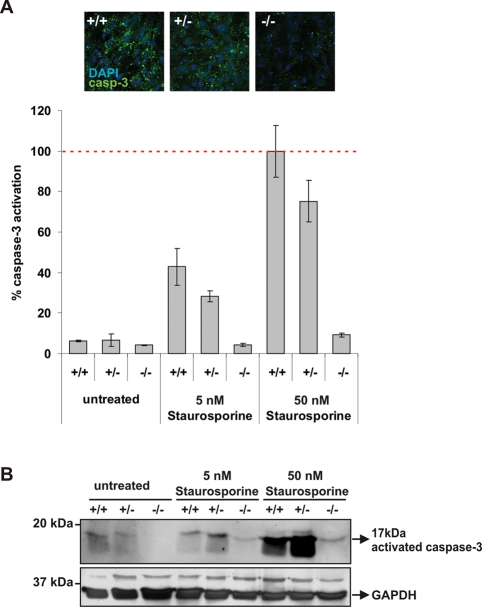
Noa1–/– cells are resistant to staurosporine-induced apoptosis
NOA1 was previously reported to be involved in regulation of apoptosis in HeLa cells (Tang et al., 2009). To verify whether the mitochondrial defects in NOA1-deficient cells would also be accompanied by the impairment of apoptosis, embryonic fibroblasts were treated with staurosporine and apoptosis was monitored by accumulation of the cleaved caspase-3. A robust caspase-3 activation was detected 24 h postinduction in the control cells but was entirely absent in the mutant cells (Figure 7). Retroviral reconstitution of NOA1 expression partially restored caspase-3 activation in Noa1–/– cells (Supplemental Figure S4E). Additionally, mitochondrial membrane potential determined with JC-1 staining was increased in knockout cells corroborating the observed apoptosis defect (Supplemental Figure S4F). Thus, NOA1 is necessary for activation of caspase-dependent apoptosis.
FIGURE 7:
Noa1-deficient cells are resistant to staurosporine-induced apoptosis. Embryonic fibroblasts were cultured on coverslips in the presence of increasing concentrations of staurosporine. Apoptosis was monitored by measuring activation of the caspase-3. (A) The caspase-3–positive area was quantified using ImageJ software and corrected for the cell number. Representative microscopic pictures are shown (data represent mean of at least 10 evaluated images with SEM as error bars). (B) Western blot with anti-activated caspase-3 antibodies. Blots were stripped and probed with the antibody against GAPDH for loading control.
DISCUSSION
In the current study we address both the cellular and the developmental function of NOA1 by inactivating the gene in mice. The vital role of Noa1 was underlined by a massively restricted growth and arrested proliferation at ∼E9.5 followed by the death of Noa1–/– mice at ∼E10.5 (Table 1 and Figures 1 and 2A). The mitochondria in these developing embryos showed aberrations of cristae morphology, whereas other organelles appeared not to be affected (Figure 2D). Thus, both the timing of the lethality as well as the results of electron microscopic analysis reflect the important mitochondrial function of NOA1. Interestingly, the developmental defect extended to the placental trophoblast, which constitutes the interface between maternal and embryonic circulation (Figure 3B, Supplemental Figure 1) (Red-Horse et al., 2004). The defect occurs at a relatively early stage of trophoblast differentiation as revealed by the diminished expression of 4311/Tbpb, demarcating the population of trophoblast progenitor cells at E8.5. The dependence of trophoblast development on NOA1 function exposes a crucial role of mitochondria in the development of the maternal–fetal interface. The importance of mitochondrial function in this process is further supported by the observation that knockouts of mitochondrial fusion factor Mtf2 as well as of mito-division factor Drp1 result in a deficiency of trophoblast giant cells (Chen et al., 2003; Wakabayashi et al., 2009).
The nature of NOA1 mitochondrial function was investigated in primary knockout mouse embryonic fibroblast (MEF) cells. Noa1–/– cells were shown to have impaired activities of the electron transfer chain complexes I, III, IV, and V but not of complex II, which lends strong support to NOA1 being required for expression of the mitochondrial genome. We therefore analyzed all stages of mtDNA expression and determined that NOA1 deficiency impairs protein synthesis but does not cause loss of mtDNA and/or transcription. Thus, NOA1 appears to be specifically required for mitochondrial protein synthesis. Sucrose gradient centrifugation and Western blot experiments revealed that knockout cells exhibit an aberrant migration of the large mitoribosomal subunit, indicating that NOA1 is required for correct mitoribosome assembly (Figure 5D). Involvement of NOA1 in the biogenesis of the small mitoribosomal subunit cannot be ruled out at present, however. In this study our choice of antibodies against mouse small mitoribosomal subunit was limited to anti-MRPS18b, which did not show any defect. Several other commercially available anti-MRP antibodies were tested, but none were capable of cross-reactivity with murine MRPs (data not shown).
Our knowledge about mitochondrial ribosome biogenesis and function is essentially based on the studies of bacterial and yeast model systems. Ribosome assembly comprises the processing and folding of pre-RNA and its concomitant assembly with ribosomal proteins. A large number of ribosomal constituents as well as nonribosomal factors are involved. Among these factors, the GTPases (a family of energy-consuming enzymes characterized by the so-called G-domain) have been shown to play key roles in the assembly of ribosomes in bacteria (Karbstein, 2007; Britton, 2009). As shown here, mitochondrial NOA1, similarly to its bacterial homologue YqeH, has GTPase activity. The intrinsic GTPase activity of the B. subtilis YqeH is strongly and specifically stimulated by 16S rRNA of the small subunit from E. coli as well as by the 21S reconstitution intermediate of the small subunit, which is highly similar to the native 21S precursor particle (for a review, see Nierhaus, 1991). The strong stimulation contrasts with the effects of 23S rRNA and the mature ribosomal subunits, which did not stimulate the intrinsic activity of YqeH. This finding supports the notion that the major activity of the bacterial YqeH is related to the early assembly phase of the small subunit. Interestingly, mammalian NOA1 was stimulated both by the bacterial 50S large ribosomal subunit and by the 30S small subunit. To a lesser degree, NOA1 was also stimulated by naked rRNAs and precursor 21S particles, whereas only residual stimulation was observed with full 70S ribosomes. These data show that NOA1, like its bacterial homologue YqeH, has GTPase activity that is stimulated by the bacterial ribosomal constituents. The apparent difference in NOA1 and YqeH specificity likely reflects their evolutionary divergence. Indeed, NOA1 contains an ∼100-amino-acid insertion in the GTP-binding domain, which is absent from bacterial YqeH and Arabidopsis NOA1 proteins. The latter could explain the differences in protein target recognition (Zemojtel et al., 2004). These data, together with the observed aberrant migration of the large ribosomal subunit in the Noa1 knockout cells, further suggest a possible involvement of NOA1 in the mitoribosome biogenesis.
Several proteins involved in mitochondrial protein synthesis have been shown to be involved in the process of apoptosis. Among them were mitoribosomal GTP-binding protein MRPS29/DAP3 (death-associated protein 3), MRPS30/PDCD9 or p52 (programmed cell death protein 9), and a protein with an apparent dual cell protective as well as pro-apoptotic function PDCD8/AIF (apoptosisinducing factor) (Cavdar Koc et al., 2001; O’Brien, 2002; Cheung et al., 2006). Interestingly, DAP3 was among several ribosomal proteins recently shown to interact with NOA1 (Tang et al., 2009).
Similarly to DAP3, NOA1 appears to be necessary for mitochondria-dependent apoptosis. The primary fibroblasts isolated from the knockout embryos were impaired in staurosporine-induced apoptosis (Figure 7A), and retroviral complementation partially reversed this phenotype (Supplemental Figure S4E). Importantly, NOA1 inactivation results in hyperpolarization of mitochondria, which likely contributes to the phenotype of defective apoptosis (Supplemental Figure S4F). In line with that, NOA1 knockdown in neuroblastoma cells was reported to result in mitochondrial hyperpolarization, preventing cytochrome c release (Parihar et al., 2008). Thus, although the exact molecular mechanism remains to be discovered, it is increasingly clear that a group of mitoribosome-associated proteins, including NOA1, is required for the execution of programmed cell death.
Interestingly, NOA1 appears to play a similar role in plants, as Arabidopsis NOA1 (formerly atNOS1 [Arabidopsis thaliana nitric oxide synthase 1]) was shown to localize to plastids, where it is necessary for protein synthesis (Flores-Perez et al., 2008). Similarly, plastidial NOA1 localization was shown in the phytoplankton diatom Phaeodactylum tricornutum (Vardi et al., 2008). Remarkably, deficient growth and the greening phenotype of atNOA1-deficient plants could be complemented by B. subtilis YqeH fused to the Arabidopsis NOA1 organelle localization sequence (Flores-Perez et al., 2008), clearly demonstrating that YqeH is a functional orthologue of plastid-localizing atNOA1. Like mitochondria, plastids are believed to have originated from eubacterial endosymbionts. Thus mammalian NOA1 is an evolutionarily extremely conserved GTPase, required for protein synthesis in prokaryote-derived organelles. Further investigation into NOA1 structure and function is needed to unravel the exact mechanisms of its function in relation to mitoribosome and apoptotic action. The Noa1 inactivation in mouse, however, demonstrates that this gene product plays a vital role in mitochondrial homeostasis and mammalian development.
MATERIALS AND METHODS
Tissue isolation and processing
BrdU solution (10 μl of 10 mM solution/1 g of mouse body weight) was injected intraperitoneally into pregnant mice 1 h before the mice were killed. Embryos were dissected from deciduas using an inverted microscope (Leica, Wetzlar, Germany). Whole embryos were photographed in phosphate-buffered saline (PBS) using a microscope-mounted digital camera (Leica, Germany). Embryonic stages were estimated by timed pregnancies and somite counts. The embryos were fixed in 4% paraformaldehyde (PFA) pH 7.4, dehydrated in ethanol, and paraffin embedded. The specimens were sectioned at 6 μm, stained with hematoxylin–eosin or processed for immunohistochemical staining.
MEF culture
Eviscerated embryos were collected in U-bottomed 96-well plates, cut into small fragments with dissection forceps, and digested in 25-μl of cell culture grade, 0.5% wt/vol trypsin at 37°C for 20 min. Subsequently, they were dispersed by pipetting and plated into one well of the 96-well, flat-bottomed plate in 200 μl of fresh DMEM containing glucose at 1 g/l, 10% fetal bovine serum (FBS), 2% penicillin/streptomycin, l-uridine at 50 μg/ml (Sigma, St. Louis, MO), 0.1 mM nonessential amino acids (Invitrogen, Carlsbad, CA), 1 mM sodium pyruvate (Sigma), and 10% Amniomax C-100 supplement (Invitrogen). The medium was changed every second day until cells reached confluence. Cells were subsequently expanded and continuously passaged until spontaneously immortalized colonies were derived. Cells were cultured at 37°C in 95% air / 5% CO2.
Retroviral infection
BOSC23 packaging cells were transiently transfected with a retroviral vector expressing mouse Noa1 (pQCXIP-mNOA1) using the CalPhos Mammalian Transfection Kit (Clontech Laboratories, Mountain View, CA). Retroviral stocks were collected 48 h post-transfection. Noa1-deficient MEFs were infected with the retrovirus in the presence of 6 μg of polybrene and selected for 4 days with puromycin at 2.5 μg/ml.
Immunofluorescence
For BrdU detection, sections were pretreated by 2N HCl treatment and incubated with a monoclonal antibody against BrdU (1:50; Roche Diagnostics, Indianapolis, IN). Apoptotic cell death was visualized in the whole mount specimens with antibodies against activated caspase-3 (Cell Signaling Technology, Danvers, MA). Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays on the paraffin sections were performed using the In Situ Cell Death Detection Kit (Roche Diagnostics) following the manufacturer’s protocol. Alexa-568 and/or Alexa-488 conjugated goat anti–rabbit secondary antibodies were used in all experiments.
Quantification of immunofluorescence staining
Embryonic fibroblasts were seeded on coverslips. After 12 h, the cells were treated with increasing concentrations of staurosporine (Sigma). Cells were processed 18 h later for immunofluorescence by fixing in 4% PFA and 1× PBS for 10 min and permeabilizing with 0.4% Triton X-100, 3% BSA, and 1× PBS for 10 min at 4°C. Incubation with an antibody against activated caspase-3 (Cell Signaling Technology) 1:400 was performed overnight in 3% BSA at 4°C. For visualization, an anti–rabbit immunoglobulin G (IgG) Alexa Fluor 488 (Invitrogen, Molecular Probes) conjugate was applied. Nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI), and cells were mounted in Fluoromount (Scientific Services, Sparrow Bush, NY). Cells were photographed using a fluorescence microscope (BX60; Olympus, Center Valley, PA) or scanning microscope (LSM510meta; Zeiss, Jena, Germany). For each treatment, at least 10 images were evaluated with ImageJ software. Experiments were repeated at least three times, and more than 1,000 cells per sample were counted.
Immunocytochemistry
Cells grown on gelatin-coated coverslips were fixed with 4% PFA and permeabilized in PBS/0.1% Triton X-100. Nonspecific binding was reduced by blocking with 5% bovine calf serum in PBS, and the cells were incubated with primary antibody in 1% BSA/PBS.
In situ hybridization
Embryos in deciduas were fixed overnight at 4°C in 4% PFA, dehydrated through an ethanol and xylene series, and embedded in paraffin blocks. Sections (6 μm) were cut in a transverse plane with respect to the placenta. Slides were processed for hematoxylin–eosin staining and in situ hybridization as previously described (Vortkamp et al., 1996). Riboprobe template vectors were provided by Malgorzata Gasperowicz and James Cross, University of Calgary, Canada. For genotyping, embryonic tissue was scraped off unstained slides. DNA was recovered and genotyped by PCR.
HeLa cell culture and small interfering RNA (siRNA) transfections
HeLa human cervical carcinoma cells were maintained in DMEM medium (4.5 g/l glucose) supplemented with 10% fetal calf serum (FCS), penicillin G at 100 IU/ml, streptomycin at 100 g/ml, and 2 mM l-glutamate and cultured at 37°C in a humidified atmosphere containing 5% CO2 and 95% air. The sequence of the sense strand of the NOA1 targeting siRNA was (GGUCAUACGUUACUCCAGA) dTdT. siRNA was transfected using DreamFect (Oz Biosciences, Marseille, France). 60% confluent cells were transfected with 1 μM siRNA (20 nmol of double-stranded RNA [dsRNA]) according to supplied protocol. Following transfection, cells were cultured in DMEM containing 10% FCS for 3 days. Cells were harvested 72 h post-transfection for analysis. HeLa cells transfected with scrambled siRNA were used as control.
Western blotting
Protein lysates were prepared in SDS radioimmunoprecipitation assay (SDS RIPA) buffer supplemented with a complete proteinase tablet (Roche), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM sodium vanadate, and 1 mM sodium fluoride. The lysate concentrations were determined using the Bradford assay (Bio-Rad, Hercules, CA). Lysates (40 μg of protein per well) were resolved in 10% SDS–PAGE and subsequently transferred to polyvinylidene fluoride (PVDF) membranes. The blots were probed with the rabbit anti-NOA1 polyclonal antibody generated against peptide: CVNVKGQRMKKSVAYK (1:100). Other antibodies used were: rabbit anti-actin (1:1,000; Sigma). Blots were developed with the Western Lightning® Plus–ECL system (PerkinElmer).
TEM
Whole E9.5 embryos were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer solution (pH 7.3) at 4°C, washed, and postfixed in 2% osmium tetroxide in the same buffer. Samples were dehydrated in ethanol, then embedded in Eponate 12 epoxy resin (Serva, Heidelberg, Germany) and sectioned on a Reichert-Jung Ultracut microtome (Leica, Wetzlar, Germany). Semithin sagittal sections of whole embryos were stained with methylene blue-azure to locate and mark the somite regions. The ultrathin sections were prepared, stained with uranyl acetate followed by lead citrate, and imaged using an EM906 Zeiss (Oberkochen, Germany) electron microscope at 80 kV.
Optical tomography (OPT)
The OPT method was used to visualize embryos in three dimensions (3D). The method accumulates projection images of an entire specimen from many different angles, and recalculates the original 3D structure by using a “back-projection” reconstruction algorithm (Sharpe et al., 2002). OPT scans were performed in UV light using a GFP1 filter for detection of the embryos’ autofluorescence using IPLab software from Scanalytics (Rockville, MD). Results were exported to QuickTime movies.
Polarography
MEFs and siRNA-treated HeLa cells were trypsinized, washed twice with PBS, and permeabilized with digitonin at 50 μg/ml. The optimum incubation time for permeabilization (50 s for MEFs; 120 s for HeLa) was determined in separate time series for each cell line and defined as the shortest duration after which 99% of the cells were trypan blue positive. Additionally, we verified adequate mitochondrial coupling by measuring the increase of respiration after decoupling with DNP (2,4-dinitrophenol) in comparison to the state IV respiration without ADP. The DNP / state IV respiration rates were 1.7 ± 0.1 in wild-type cells and 1.8 ± 0.2 in the knockout MEF cells. The cells (1 × 107) were resuspended in reaction buffer and introduced into the reaction chamber of a Clark-type electrode (Hansatech Instruments, Norfolk, England), and oxygen concentrations were measured in 1-ml volumes at 37°C with substrates and inhibitors according to standard protocols (Hofhaus et al., 1996). After completion of the polarographic measurement, cells were collected and DNA quantified using a Quant-iT PicoGreen DNA kit (Invitrogen) according to the supplied protocol. Oxygen consumption was given as mean ± SD nmol O2 per minute per μg of DNA (n ≥ 4).
Blue-native PAGE and complex I in-gel activity assay
Blue-native PAGE was used for separation of the OXPHOS complexes on 5–15% polyacrylamide-gradient gels as described before (Calvaruso et al., 2008). After electrophoresis of 40 μg of protein from MEFs, gels were further processed for in-gel activity assays and Western blotting. Assembly of the OXPHOS complexes was analyzed using monoclonal antibodies against subunits of complex I (NDUFA9, NDUFS3), complex III (Core2), complex V (α-ATPase) (Molecular Probes, Leiden, The Netherlands), and complexes II (SDHA) and IV (CoxVa) (MitoSciences, Eugene, OR).
OXPHOS enzyme complex measurements
The activities of the OXPHOS enzyme complexes were measured in MEFs as described previously (Lazarov and Cooperstein, 1951; Smeitink et al., 2001; Janssen et al., 2003, 2007). Citrate synthase (CS) activities were determined in cultured fibroblast as previously described (Srere, 1969).
Analysis of mitochondrial protein synthesis
In vitro pulse labeling of mitochondrial translation products was performed as described elsewhere (Boulet et al., 1992), with a few adaptations. In brief, cells were labeled for 60 min at 37°C in l-methionine–free and l-cysteine–free DMEM with 10% dialyzed FCS, [35S]methionine and [35S]cysteine at 200 μCi/ml (Tran35S-Label; MP Biomedicals, Eindhoven, The Netherlands), and emetine at 100 μg/ml, and were subsequently chased for 10 min in regular DMEM with 10% FCS. Total cellular protein was resuspended and incubated for 10 min in PBS containing 2% lauryl maltoside. Unsolubilized material was removed by centrifugation at 10,000 × g for 10 min. The protein concentration was determined (Micro BSA Protein Assay Kit; Pierce) and loading buffer was added to the supernatant 1:1 (Tricine Sample Buffer; Thermo Fisher Scientific, Rockford, IL). The samples were separated through 16% polyacrylamide gels and subsequently scanned on a FLA5100 (Fujifilm Life Science, Düsseldorf, Germany). Equal protein loading was confirmed using colloidal Coomassie Blue staining (data not shown).
Isokinetic sucrose gradient analysis of mitochondrial ribosomes
Cell lysis was performed in 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, + PI-Mix, 1 mM PMSF, and 10 mM MgCl2 for 30 min at 4°C. After a 10-min incubation, lysate was clarified by centrifugation at 12,000 × g at 4°C. Approximately 0.9 mg of supernatant was loaded onto a linear sucrose gradient (10–30% [vol/vol], 1 ml) in 50 mM Tris-HCl, pH 7.2, 10 mM Mg(OAc)2, 40 mM NH4Cl, 100 mM KCl, 1 mM PMSF, and chloramphenicol at 50 μg/ml and centrifuged for 2 h 15 min at 100,000 × g at 4°C. Fractions of 100 μl were collected, and 10 μl of each fraction was analyzed by Western blot. Antibodies used were anti-MRPS18b (ProteinTech Group, Chicago, IL) and anti-MRPL12 (Abcam, Cambridge, MA).
Quantitative RT–PCR
cDNAs were synthesized from 1 μg total RNAs with murine leukemia virus (MuLV) reverse transcriptase (Applied Biosystems, Carlsbad, CA). TaqMan universal PCR was then performed on an ABI PRISMs 7900 cycler (Applied Biosystems), using the SYBR green method according to the manufacturer’s instructions. Transcripts of the following genes were monitored: NOA – NOA1, 16S RNA, 12S RNA, ND1 – NADH dehydrogenase subunit 1; ND6 – NADH dehydrogenase subunit 6, CytOx1 – Cyt c oxidase subunit 1, CytOx2 – Cyt c oxidase subunit 2, cytB – cytochrome b, ATP6 – ATP synthase F0 subunit 6, EF-Tu – Elongation Factor Tu, L20 – mitochondrial ribosomal protein L20, AIF – apoptosis inducing factor (see also supplemental primer list).
mtDNA analysis
The mtDNA/nuclear DNA (mtDNA/nDNA) ratio was estimated using qPCR with primers specific for mtDNA-encoded Cox1 gene (subunit 1 of cytochrome c oxidase) and nuclearly encoded Ndufv1 gene (NADH:ubiquinone oxidoreductase) (see supplemental primer list). DNA quantification was repeated five times in independent qPCR reactions, and an average mtDNA/nDNA ratio was calculated. All values were expressed as means ± SD.
Mitochondrial genome integrity was tested using primers listed in the supplement and the Expand Long Template PCR System (Roche) according to the manufacturer’s protocol. PCR conditions were: 94°C for 3 min; 10 cycles: 94°C for 10 s, 65°C for 30 s, 68°C for 16 min; 20 cycles: 94°C for 10 s, 65°C for 30 s, 68°C for 16 min (+20 s/cycle). For detection of shorter products, elongation time was reduced to 8 min.
ATP assay
ATP levels were measured with the ATPlite Bioluminescence Assay kitSystem (Roche). Cells were trypsinized after a 36-h incubation time in culture medium with (4.5 g/l) or without glucose. The cells were washed once in PBS and resuspended in the kit’s dilution buffer.
Viability assay
Viability was measured with an alamarBlue assay according to supplied protocol (Invitrogen). Defined cell numbers were plated in 96-well formats, and the resazurin substrate reduction was monitored in the cell culture medium by measuring the accumulation of the fluorescent resorufin (excitation at 530 nm, emission at 590 nm).
Preparation of rRNA
16S rRNA and 23S+5S rRNA of E. coli were prepared by phenol extraction followed by ethanol precipitation starting with purified 30S and 50S ribosomal subunits, respectively (Lietzke and Nierhaus, 1988). The genes coding for the mitochondrial 12S and 16S rRNA were amplified from human mtDNA using the primers mt12S fw and mt12S rev or mt16S fw and mt16S rev, leading to the fusion of a T7 promoter sequence at the 5′-end of the genes. PCR fragments were digested with HindIII and EcoRI and cloned into pSP65 cleaved with the same enzymes. Transcription of the genes was performed in vitro using T7 RiboMAX Express (Promega, Madison, WI) with the new constructed plasmids (linearized with EcoRI) as template. Transcribed RNA was purified by gel filtration and ethanol precipitation.
Cloning, expression, and purification of B. subtilis YqeH and human NOA1
The gene coding for B. subtilis YqeH was amplified from genomic DNA by PCR and cloned into pET28a (Novagen, Merck KGaA, Darmstadt, Germany) via NdeI and XhoI, leading to an N-terminal His-tag fusion. The correct sequence was confirmed by sequencing. The gene was expressed in BL21(DE3) and the protein purified via Ni-NTA agarose (Qiagen, Valencia, CA, according to the manufacture’s manual) followed by anion exchange chromatography (MonoQ, GE Healthcare, Piscataway, NJ). The protein was finally dialyzed against 20 mM HEPES·KOH (pH 8.0 at 0°C), 6 mM Mg(acetate)2, 150 mM K(acetate), and 4 mM β-mercaptoethanol.
Full-length cDNAs for human NOA (IRALp962E057Q2) was obtained from the German Resource Center for Genome Research (RZPD, Berlin). The gene coding for ∆25a.a.-hNOA1 protein (hNOA1 without mitochondria targeting sequence – 25 amino acids of N terminus) was amplified by PCR and cloned into pGEX-6P (Amersham Pharmacia Biotech, Buckinghamshire, UK) via EcoRI and XhoI leading to an N-terminal GST-tag fusion. The gene was expressed in BL21-CodonPlus (DE3)-RP (Stratagene, Santa Clara, CA), and the protein was purified via Glutathione Sepharose 4B (Amersham Pharmacia Biotech, Piscataway, NJ) using PBS buffer pH 6.2 supplemented with 1% Triton X-100, according to the manufacture’s manual. The protein was finally dialyzed against 50 mM Tris, pH 7.0, at 4°C, 150 mM NaCl and concentrated using Centriprep YM-10 (Amicon, Bedford, MA).
30S reconstitution and poly(Phe) synthesis
0.125 A260 unit of 16S E. coli rRNA equivalent to 9 pmol was incubated with 0.05 equivalent units (e.u.) of total proteins of the 30S subunit (TP30) for 30 min at 40°C in 15 ml of Rec20 buffer: 20 mM HEPES-KOH (pH 7.6 at 0°C), 20 mM Mg(acetate)2, 400 mM NH4(acetate), and 4 mM β-mercaptoethanol (for details, see Nierhaus, 1990). Subsequently, poly(U)-dependent poly(Phe) synthesis was performed with 6 pmol of reconstituted particles, which were incubated at 37°C for 1 h in 40 μl with 6 pmol of 50S subunits, 13.4 μg of bulk transfer RNA (tRNAbulk), 33.4 μg of poly(U), 10 nmol of phenylalanine (23 dpm/pmol), 3.33 μl of S100 enzymes in the presence of 20 mM HEPES-KOH (pH 7.6 at 0°C), 6 mM Mg(acetate)2, 150 mM NH4(acetate), 4 mM β-mercaptoethanol, 2 mM spermidine, 0.05 mM spermine, 3 mM ATP, 1.5 mM GTP, and 5 mM acetyl phosphate. The reaction mix (35 μl) was subjected to hot trichloroacetic acid (TCA) precipitation and filtered through glass filters. Control reconstitutions were performed at 20°C for 1 h, but otherwise under identical conditions in the presence and absence of 9 pmol of YqeH. The B. subtilis YqeH factor did not accelerate the E. coli reconstitution process of the 30S subunit.
Preparation of 21S particle
600 A260 units of 16S rRNA and 240 e.u. of TP30 were reconstituted for 30 min in ice water. The incubated sample was centrifuged through a 10–30% sucrose gradient containing Rec20 in a Beckman Zonal rotor at 24,000 rpm for 19 h at 4°C. Fractions containing the heavy shoulder of the 21S peak were collected and the particles were pelleted and resuspended in Rec20 buffer. Protein content of the 21S particles was determined by two-dimensional electrophoresis, and the following proteins were detected: S4, S5, S7, S8, S9, S13, S14, S15, S16, and S18. The proteins S1 and S20 could not be resolved on the 30S control. The intactness of the rRNA was confirmed by RNA electrophoresis. Controls containing the 21S particles were subjected to a reconstitution incubation in the presence of TP30 and checked for poly(Phe) synthesis in the presence of mature 50S subunits. The results confirmed that the 21S particles were authentic intermediates, which could be processed to active 30S subunits.
GTPase assay
The assay was performed in a 50-μl reaction volume under optimized ion conditions (20 mM HEPES-KOH [pH 8.0 at 0°C], 4.5 mM Mg(acetate)2, 150 mM K(acetate), 2 mM spermidine, 0.05 mM spermine, and 4 mM β-mercaptoethanol). The incubation mixture contained 10 pmol of YqeH or NOA1, 2.5 nmol of [γ32P]GTP (20 dpm/pmol), and the indicated amounts of rRNA, 21S particles, or ribosomal subunits. Samples were incubated either 1 h at 30°C or 6 h at 20°C, and the reaction was stopped by the addition of 120 μl of 0.5 M H2SO4 and 1.5 mM NaH2PO4. The released γ-phosphate was extracted by mixing the samples with 30 μl of 200 mM MoNaO4 and 800 μl of water-saturated 2-butanol. Samples were vortexed for 1 min followed by centrifugation at 16,000 × g for 10 min at 4°C. The radioactivity obtained in the butanol phase was measured in a Wallac 1409 Liquid Scintillation Counter.
All animal experimental procedures were approved by the local animal ethics commission, State Office of Health and Welfare Berlin (LAGeSo).
Supplementary Material
Acknowledgments
We thank Monika Osswald, Petra Schrade, and Carola Dietrich for excellent technical assistance. We also thank Malgorzata Gasperowicz and James Cross for providing template vectors for placenta specific riboprobes. We also acknowledge Laurene Marchand for her contribution in establishing mitochondrial membrane potential assay. M. K. and N. K. were supported by a Young Investigator Award (Grant 2007–01-038) from the Children’s Tumor Fundation, New York, and by Bundesministerium für Bildung und Forschung Grant NF1–01GM0844. This work was also supported by the Sixth Framework of the European Commission, EuroGrow project LSHM-CT-2007–037471. M. P. was supported by a German Academic Exchange Service (DAAD, New York) grant and H. Y. by an Alexander-von-Humboldt stipendium. R.N.L. was supported by grant number BB/F011520/1 from the Biotechnology and Biological Sciences Research Council. I. M. and P. M. are supported by grants 1 M 6837805002 and 0021620806 from the Ministry of Education, Youth and Sports and by grant 252021 102107 from GAUK. M. S. was supported by the Deutsche Forschungsgemeinschaft, NeuroCure Exc 257 and is a member of the German network for mitochondrial disorders (mitoNET, 01GM0862), funded by the German Ministry of Education and Research (BMBF).
Abbreviations used:
- 3D
three dimensions
- DAP3
death-associated protein 3
- DAPI
4′,6-diamidino-2-phenylindole
- DNP
2,4-dinitrophenol
- dsRNA
double-stranded RNA
- FBS
fetal bovine serum
- GTP
guanosine triphosphate
- MEF
mouse embryonic fibroblast
- mtDNA
mitochondrial DNA
- MuLV
murine leukemia virus
- nDNA
nuclear DNA
- NOA
nitric oxide associated
- OPT
optical tomography
- OXPHOS
oxidative phosphorylation
- PBS
phosphate-buffered saline
- PFA
paraformaldehyde
- PMSF
phenylmethylsulfonyl fluoride
- PVDF
polyvinylidene fluoride
- qPCR
quantitative PCR
- RIPA
radioimmunoprecipitation assay
- siRNA
small interfering RNA
- TCA
trichloroacetic acid
- TEM
transmission electron microscopy
- TGC
trophoblast giant cell
- TRAFAC
translation factors
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-07-0643) on November 30, 2010.
REFERENCES
- Boulet L, Karpati G, Shoubridge EA. Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged-red fibers (MERRF) Am J Hum Genet. 1992;51:1187–1200. [PMC free article] [PubMed] [Google Scholar]
- Britton RA. Role of GTPases in bacterial ribosome assembly. Annu Rev Microbiol. 2009;63:155–176. doi: 10.1146/annurev.micro.091208.073225. [DOI] [PubMed] [Google Scholar]
- Calvaruso MA, Smeitink J, Nijtmans L. Electrophoresis techniques to investigate defects in oxidative phosphorylation. Methods. 2008;46:281–287. doi: 10.1016/j.ymeth.2008.09.023. [DOI] [PubMed] [Google Scholar]
- Cavdar Koc E, Ranasinghe A, Burkhart W, Blackburn K, Koc H, Moseley A, Spremulli LL. A new face on apoptosis: death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Lett. 2001;492:166–170. doi: 10.1016/s0014-5793(01)02250-5. [DOI] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung EC, et al. Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. EMBO J. 2006;25:4061–4073. doi: 10.1038/sj.emboj.7601276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Perez U, Sauret-Gueto S, Gas E, Jarvis P, Rodriguez-Concepcion M. A mutant impaired in the production of plastome-encoded proteins uncovers a mechanism for the homeostasis of isoprenoid biosynthetic enzymes in Arabidopsis plastids. Plant Cell. 2008;20:1303–1315. doi: 10.1105/tpc.108.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J. Regulation of the in vivo synthesis of the polypeptide chain elongation factors in Escherichia coli. Biochemistry. 1970;9:912–917. doi: 10.1021/bi00806a028. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. The origin and early evolution of mitochondria. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-reviews1018. reviews1018.1–reviews1018.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofhaus G, Shakeley RM, Attardi G. Use of polarography to detect respiration defects in cell cultures. Methods Enzymol. 1996;264:476–483. doi: 10.1016/s0076-6879(96)64043-9. [DOI] [PubMed] [Google Scholar]
- Janssen AJ, Smeitink JA, Van Den Heuvel LP. Some practical aspects of providing a diagnostic service for respiratory chain defects. Ann Clin Biochem. 2003;40:3–8. doi: 10.1258/000456303321016114. [DOI] [PubMed] [Google Scholar]
- Janssen AJ, Trijbels FJ, Sengers RC, Smeitink JA, Van Den Heuvel LP, Wintjes LT, Stoltenborg-Hogenkamp BJ, Rodenburg RJ. Spectrophotometric assay for complex I of the respiratory chain in tissue samples and cultured fibroblasts. Clin Chem. 2007;53:729–734. doi: 10.1373/clinchem.2006.078873. [DOI] [PubMed] [Google Scholar]
- Karbstein K. Role of GTPases in ribosome assembly. Biopolymers. 2007;87:1–11. doi: 10.1002/bip.20762. [DOI] [PubMed] [Google Scholar]
- Kim do J, Jang JY, Yoon HJ, Suh SW. Crystal structure of YlqF, a circularly permuted GTPase: implications for its GTPase activation in 50 S ribosomal subunit assembly. Proteins. 2008;72:1363–1370. doi: 10.1002/prot.22112. [DOI] [PubMed] [Google Scholar]
- Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem. 2001;276:43958–43969. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- Lazarov A, Cooperstein SY. Studies on the isolated islet tissue of fish. I. The cytochrome oxidase and succinic dehydrogenase contents of normal toadfish (Opsonus tau) Biol Bull. 1951;100:191–198. doi: 10.2307/1538530. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- Lietzke R, Nierhaus KH. Total reconstitution of 70S ribosomes from Escherichia coli. Methods Enzymol. 1988;164:278–283. doi: 10.1016/s0076-6879(88)64049-3. [DOI] [PubMed] [Google Scholar]
- Loh PC, Morimoto T, Matsuo Y, Oshima T, Ogasawara N. The GTP-binding protein YqeH participates in biogenesis of the 30S ribosome subunit in Bacillus subtilis. Genes Genet Syst. 2007;82:281–289. doi: 10.1266/ggs.82.281. [DOI] [PubMed] [Google Scholar]
- Nierhaus K. Reconstitution of ribosomes. In: Ribosomes and Protein Synthesis A Practical Approach, Oxford, UK: IRL Press at Oxford University Press. In: Spedding G, editor. 1990. pp. 161–189. [Google Scholar]
- Nierhaus KH. The assembly of prokaryotic ribosomes. Biochimie. 1991;73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- O’Brien TW. Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene. 2002;286:73–79. doi: 10.1016/s0378-1119(01)00808-3. [DOI] [PubMed] [Google Scholar]
- Parihar MS, Parihar A, Chen Z, Nazarewicz R, Ghafourifar P. mAtNOS1 regulates mitochondrial functions and apoptosis of human neuroblastoma cells. Biochim Biophys Acta. 2008;1780:921–926. doi: 10.1016/j.bbagen.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Qin Y, Polacek N, Vesper O, Staub E, Einfeldt E, Wilson DN, Nierhaus KH. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127:721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- Red-Horse K, Zhou Y, Genbacev O, Prakobphol A, Foulk R, McMaster M, Fisher SJ. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J Clin Invest. 2004;114:744–754. doi: 10.1172/JCI22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J, Ahlgren U, Perry P, Hill B, Ross A, Hecksher-Sorensen J, Baldock R, Davidson D. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- Smeitink J, Sengers R, Trijbels F, Van Den Heuvel L. Human NADH:ubiquinone oxidoreductase. J Bioenerg Biomembr. 2001;33:259–266. doi: 10.1023/a:1010743321800. [DOI] [PubMed] [Google Scholar]
- Srere P. Citrate synthase, EC 4.1.3.7, citrate oxaloacetate lyase (CoA-acetylating) Methods Enzymol. 1969;13:3–11. [Google Scholar]
- Tang T, Zheng B, Chen SH, Murphy AN, Kudlicka K, Zhou H, Farquhar MG. hNOA1 interacts with complex I and DAP3 and regulates mitochondrial respiration and apoptosis. J Biol Chem. 2009;284:5414–5424. doi: 10.1074/jbc.M807797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uicker WC, Schaefer L, Koenigsknecht M, Britton RA. The essential GTPase YqeH is required for proper ribosome assembly in Bacillus subtilis. J Bacteriol. 2007;189:2926–2929. doi: 10.1128/JB.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardi A, Bidle KD, Kwityn C, Hirsh DJ, Thompson SM, Callow JA, Falkowski P, Bowler C. A diatom gene regulating nitric-oxide signaling and susceptibility to diatom-derived aldehydes. Curr Biol. 2008;18:895–899. doi: 10.1016/j.cub.2008.05.037. [DOI] [PubMed] [Google Scholar]
- Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel T, et al. Plant nitric oxide synthase: a never-ending story? Trends Plant Sci. 2006a;11:524–525. doi: 10.1016/j.tplants.2006.09.008. author reply 526–528. [DOI] [PubMed] [Google Scholar]
- Zemojtel T, Penzkofer T, Dandekar T, Schultz J. A novel conserved family of nitric oxide synthase? Trends Biochem Sci. 2004;29:224–226. doi: 10.1016/j.tibs.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Zemojtel T, et al. Mammalian mitochondrial nitric oxide synthase: characterization of a novel candidate. FEBS Lett. 2006b;580:455–462. doi: 10.1016/j.febslet.2005.12.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.