PAIR3, an axis-associated protein, is essential for bouquet formation, initial homologous pairing and normal recombination, and SC assembly in rice.
Abstract
During meiosis, the paired homologous chromosomes are tightly held together by the synaptonemal complex (SC). This complex consists of two parallel axial/lateral elements (AEs/LEs) and one central element. Here, we observed that PAIR3 localized to the chromosome core during prophase I and associated with both unsynapsed AEs and synapsed LEs. Analyses of the severe pair3 mutant demonstrated that PAIR3 was essential for bouquet formation, homologous pairing and normal recombination, and SC assembly. In addition, we showed that although PAIR3 was not required for the initial recruitment of PAIR2, it was required for the proper association of PAIR2 with chromosomes. Dual immunostaining revealed that PAIR3 highly colocalized with REC8. Moreover, studies using a rec8 mutant indicated that PAIR3 localized to chromosomes in a REC8-dependent manner.
INTRODUCTION
Meiosis is a highly conserved process in all sexually reproducing eukaryotic organisms. During meiosis, pairing and recombination between homologous chromosomes are essential for the production of genetically distinct haploid sperm and eggs. The close association between homologues is enhanced by a meiotic-specific ladderlike proteinaceous structure, the synaptonemal complex (SC). Ultrastructural analysis of the SC reveals a tripartite structure with two parallel lateral elements (LEs) and one central element (CE). SC assembly starts during the premeiotic S-phase and early meiotic prophase with the formation of a single proteinaceous axis, the axial element (AE), along the two sister chromatids of each chromosome (the LE is called AE before synapsis of the homologous chromosomes; de Boer and Heyting, 2006). From zygotene to pachytene, the AEs of homologues are connected to each other along their entire length by the transverse filaments (TFs), and the CEs are formed in the middle, during a process called synapsis. At the diplotene stage, the SC disassembles and homologous chromosomes are separated, except at the chiasmata sites.
TF proteins have been characterized in several model organisms, including budding yeast (Zip1; Sym et al., 1993), mammals (Sycp1; Meuwissen et al., 1992), Caenorhabditis (Syp-1 and Syp-2; MacQueen et al., 2002; Colaiacovo et al., 2003), Drosophila (C(3)G; Page and Hawley, 2001), and Arabidopsis (ZYP1a and ZYP1b; Higgins et al., 2005). Although these TF proteins exhibit similar structures, they show a high degree of sequence divergence (Page and Hawley, 2004). A recent study revealed that ZEP1, the TF protein in rice, showed significant similarity with ZYP1 (Wang et al., 2010), suggesting that TF proteins are probably conserved in the plant kingdom.
AE assembly is one of the earliest known events of meiosis and is essential for the formation of cytologically characteristic leptotene chromosomes (Zickler and Kleckner, 1999; Wang et al., 2009a). The AE was initially defined by electron microscopy as a structure that forms between sister chromatids, along their entire length, before synapsis. Genetic and biochemical studies have identified AE components and showed that AEs play important roles in chromosome condensation, pairing, recombination, TF assembly, and prohibiting double-strand breaks from entering into recombination pathways that involve sister chromatids (Zickler and Kleckner, 1999; Page and Hawley, 2004; Severson et al., 2009).
Several AE components have been described in yeast, mammals, and Caenorhabditis elegans. The budding yeast Red1 and Hop1 are meiosis-specific proteins that localize to chromosomes. Mutants of red1 fail to make any discernible AE or SC structures and exhibit normal chromosome compaction, while hop1 mutants form long fragments of AEs but without any SCs and are defective in axial chromosome compaction (Rockmill and Roeder, 1990; Loidl et al., 1994; Smith and Roeder, 1997; Zickler and Kleckner, 1998). Synaptonemal complex proteins (SYCP) SYCP2 and SYCP3 are two known mammalian AE proteins that contain predicted coiled coils near the C terminus. They localize to both unsynapsed AEs and synapsed LEs. In the absence of either SYCP3 or SYCP2, AE/LE structures are disrupted (Dobson et al., 1994; Lammers et al., 1994; Offenberg et al., 1998; Yuan et al., 2000; Yang et al., 2006). The C. elegans HIM-3 protein localizes to the chromosome core and associates with both unsynapsed and synapsed chromosomes. In addition, HIM-3 plays important roles in chromosome pairing, synapsis, and segregation. Hence, HIM-3 protein is considered to be a meiotic chromosome core component (Zetka et al., 1999). HTP-3 in C. elegans also colocalizes with HIM-3 throughout the meiotic prophase, indicating that HTP-3 is also a component of meiotic chromosome axes (Goodyer et al., 2008; Severson et al., 2009). Thus far, although several AE-associated proteins have been reported in maize, Arabidopsis, and rice, no AE core component has yet been identified in plants (Mercier and Grelon, 2008).
A previous study characterized the PAIR3 gene and found that insertional mutations in its intron led to the elimination of synapsis in both male and female gametes (Yuan et al., 2009). Here, we also identified two pair3 mutants showing more severe phenotypes and demonstrated that PAIR3 is required for bouquet formation, homologous pairing, normal recombination, and SC installation. In addition, it was also revealed that PAIR3 localizes to the chromosome axis and is required for normal binding of PAIR2 to chromosome axes.
RESULTS
Characterization of pair3 mutants
We identified a tissue culture–derived mutant exhibiting complete sterility from the progeny of a japonica rice variety, Wuxiangjing 9. The mutant exhibited normal vegetative growth but showed complete sterility. Genetic analysis indicated that the mutant phenotype is controlled by a single recessive gene. The mutant failed to produce any viable pollen. In addition, the mutant could not set any seeds when pollinated with the wild-type pollens, indicating that the megagametogenesis was also affected by the mutation.
Molecular cloning of the pair3 gene
To isolate the mutated gene controlling the sterile phenotype, a large F2 mapping population was generated by crossing the heterozygous plants with indica rice variety 9311. F2 and F3 segregates (1100 in total) showing the sterile phenotype were used for gene mapping. Linkage analysis mapped the gene on the long arm of rice chromosome 10, which was further confined to a 30-kb region. By sequencing, one candidate gene, Os10g0405500, was found to be altered in the mutant. This gene was isolated recently and named PAIR3. A single nucleotide deletion at position 1044 in the coding region was found in the pair3-1 mutant (Supplemental Figure S1), which resulted in a frameshift mutation.
We also obtained one other mutant showing the same defects as pair3-1 from Nipponbare. Immunostaining with an antibody against PAIR3 failed to find any PAIR3 protein in the mutant. Thus, sequencing analysis was carried out and revealed that the mutant also contains a single nucleotide deletion at position 1538 in the coding region, and it was named pair3-2 (Supplemental Figure S1). Both mutations resulted in the loss of predicted coiled-coil motifs. Additionally, we utilized an RNA interference (RNAi) approach to down-regulate PAIR3 in rice variety Yandao 8. Twenty transgenic plants were obtained, and 13 plants exhibited severe reduction of fertility. The results confirmed that the loss of function of the PAIR3 gene led to the sterility phenotype.
Chromosome behavior in pair3 mutants
To understand the cause of the complete sterility in pair3 mutants, the meiotic chromosomes in pollen mother cells (PMCs) of both the wild type and pair3-1 mutant were investigated. In the wild type, chromosomes began to condense and form thin lines at leptotene (Supplemental Figure S2A). In zygotene, pairing and synapsis occurred between homologous chromosomes (Supplemental Figure S2B). The synapsis was completed, and 12 fully synapsed chromosomes were clearly visible at pachytene (Supplemental Figure S2C). With further chromosome condensation occurring in diplotene and diakinesis, SCs were gradually disassembled and the paired homologous chromosomes were held together by chiasmata (Supplemental Figure S2D). At metaphase I, all bivalents were aligned along the equatorial plate (Supplemental Figure S2E). Subsequently, homologous chromosomes were separated from each other and moved to the opposite poles of the cell at anaphase I (Supplemental Figure S2F). The second meiotic division was very similar to mitosis and finally produced tetrad spores (Supplemental Figure S2, G–I).
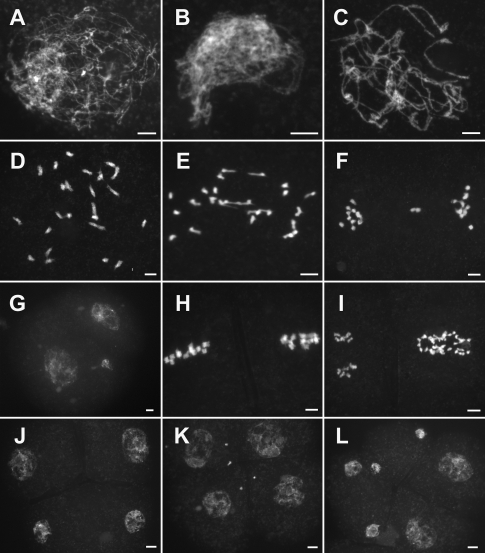
Compared with the wild type, there was no obvious difference in pair3-1 during leptotene (Figure 1A). However, pairing between homologous chromosomes was not observed from zygotene to pachytene (Figure 1, B and C). During diakinesis and metaphase I, many randomly distributed univalents were found (Figure 1D). We further quantified the chiasma frequency and showed that the pair3-1 mutant formed an average of 3.25 chiasmata corresponding to 3 bivalents per PMC (n = 20). In anaphase I, those univalents separated randomly toward two poles of the dyad (Figure 1E). In telophase I and prophase II, an uneven number of chromosomes could be seen in the two related cells (Figure 1, F and G). After the second division, tetrads and polyads with different numbers of chromosomes were formed (Figure 1, H–L). In addition, multiple micronuclei were frequently observed. These results implied that correct chromosome disjunction and cytokinesis were also affected by the mutation in the PAIR3 gene. Such abnormality was also found in the pair3-2 mutants but not in the previously reported pair3 mutants (Yuan et al., 2009). We suspected that the mutants identified in the present study contain alleles of pair3 that result in more severe defects.
FIGURE 1:
Meiosis in the pair3 mutant. (A) Leptotene. (B) Zygotene. (C) Pachytene. (D) Diakinesis. (E) Anaphase I. (F) Telephase I. (G) Prophase II. (H) Metaphase II. (I) Anaphase II. (J) Telophase II. (K) Telophase II. Tetrads with several randomly distributed chromosomes. (L) Hexayads. Scale bars, 5 μm.
Bouquet is abolished in pair3
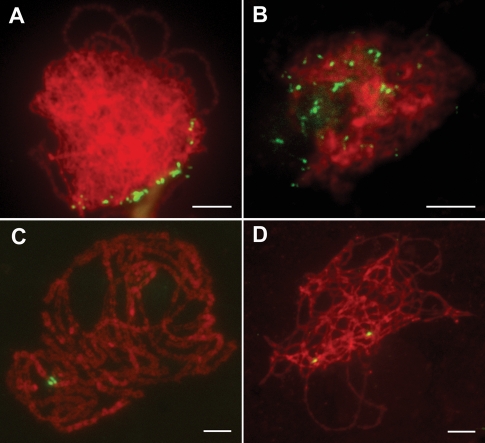
The clustering of telomeres into a “bouquet” is one of the mechanisms thought to facilitate these initial contacts leading to homologous chromosome alignment and subsequent pairing (Dawe, 1998; Harper et al., 2004; Hamant et al., 2006). To assess whether mutation of PAIR3 affects telomere bouquet formation, we probed meiotic chromosomes at the leptotene stage with the clone pAtT4 that contains telomeric repeats in both the wild type and the pair3-1 mutant. In the wild type, almost all telomeres attached to the nuclear envelope and clustered to a confined region, representing the typical bouquet configuration (Figure 2A). In the pair3-1 PMCs (n = 50), telomeres remained scattered throughout the nuclei and never formed bouquets (Figure 2B), implying that PAIR3 is required for normal bouquet formation in rice.
FIGURE 2:
PAIR3 is required for bouquet formation and homologous pairing. (A and B) Telomere bouquet formation analysis revealed by FISH using the telomere sequence as a probe (green) in both wild type (A) and the pair3 (B) nuclei. Chromosomes (red) are stained with DAPI. (C and D) Immunofluorescence micrographs showing paired and unpaired 5S rDNA signals (green) in wild type (C) and pair3 (D) mid-zygotene-stage nuclei, respectively. Bars, 5 μm.
PAIR3 is required for homologous chromosome pairing
To further examine the defects of homologous pairing in pair3, we performed fluorescence in situ hybridization (FISH) analysis using 5S rDNA as a probe to monitor the pairing status. In the wild type, the two unpaired 5S signals were observed at leptotene and zygotene. With the pairing of homologous chromosomes, the two signals were well paired during pachytene (Figure 2C). By contrast, paired 5S signals were never observed in all pair3-1 cells (n = 61; Figure 2D), implying that the pairing was lost in pair3-1. Therefore, PAIR3 is absolutely required for homologous chromosome pairing.
REC8 displays discontinuous signals during zygotene in pair3 mutants
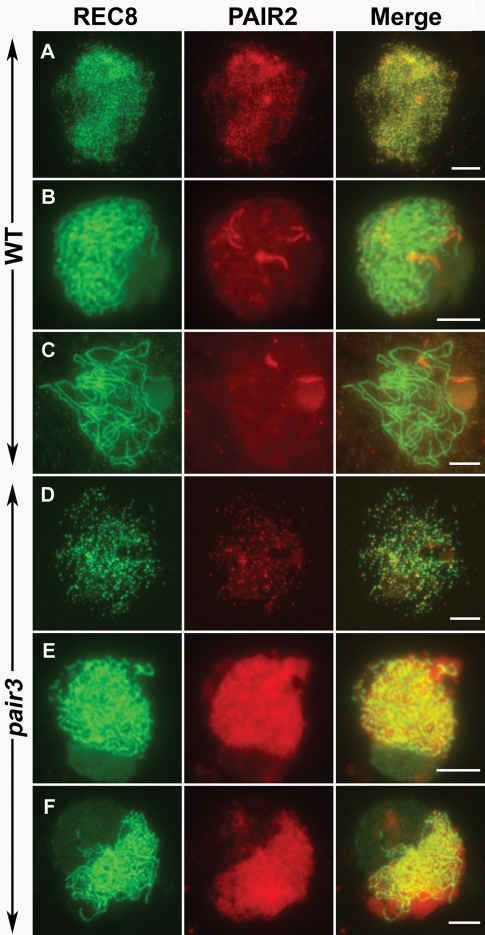
Meiosis-specific sister chromatid cohesion is essential for normal chromosome behavior in meiosis. The meiosis-specific cohesin REC8 mediates most of the cohesion (Watanabe and Nurse, 1999). An antibody against REC8 protein was used to discern whether the cohesion complex is normally installed on chromosomes in pair3 mutants. In the wild-type meiocytes, REC8 presented as punctate foci in the premeiotic interphase and leptotene (Figure 3A). In late leptotene, those foci assembled to form short linear signals on the chromosomes. In zygotene and pachytene, REC8 was frequently seen as continuous threads along the chromosomes (Figure 3, B and C). In pair3-1 meiocytes, REC8 proteins were also first visible as punctate foci (Figure 3D), which would also fuse and form short linear signals as those observed in the wild-type. Although REC8 linear signals were detected during zygotene, they were generally not continuous along the lengths of chromosomes, and frequent breaks/gaps were apparent (Figure 3E). At a stage corresponding to pachytene, when chromosomes were relatively condensed, REC8 signals became continuous and gaps/breaks were hardly detectable (Figure 3F). Thus, the loss of PAIR3 function may have only a minor effect on the normal progression of chromosome compaction or AE maturation.
FIGURE 3:
Dual immunolocalization of REC8 and PAIR2 in meiocytes of the wild type (A–C) and pair3 plant (D–E). (A) Leptotene. (B) Zygotene. (C) Pachytene. (D) Leptotene, REC8 and PAIR2 recruit normally. (E) Zygotene, REC8 shows discontinuous signals along the chromosomes, while PAIR2 exhibits diffused signals. (F) Pachytene, REC8 shows continuous signals along the chromosomes, while PAIR2 still appears as diffused signals. Bars, 5 μm.
PAIR2 requires PAIR3 to localize properly
PAIR2, a protein that associates with AEs in rice, is the homologue of yeast HOP1 and Arabidopsis ASY1 (Nonomura et al., 2006). The signals of HOP1/ASY1 in Arabidopsis and maize are frequently used to detect the assembly of AEs (Chelysheva et al., 2005; Golubovskaya et al., 2006; Pawlowski et al., 2009). In rice, PAIR2 proteins were visible as foci during interphase and leptotene in the wild type (Figure 3A). At zygotene, PAIR2 appeared as continuous lines on the chromosomes (Figure 3B). In pachytene, only a few residual PAIR2 signals could be observed on chromosomes. In pair3-1 meiocytes, PAIR2 also appeared as foci in interphase and early leptotene, but those foci rarely elongated to form short linear signals at leptotene. Instead, PAIR2 signals became diffused in the nucleoplasm in cells later than leptotene (Figure 3, B and C). These results suggest that normal PAIR2 association with chromosomes depended on PAIR3.
Normal chromosome structures are formed in a pair3 mutant
A previous study demonstrated that PAIR2 associates with AE in rice (Nonomura et al., 2006). Here, the abnormal PAIR2 signals imply that the AEs in pair3-1 might be severely disrupted; alternatively, it remains possible that AEs are normally formed but the association of PAIR2 with AEs is severely affected. With the aim of distinguishing between these two possibilities, we utilized a silver staining method to observe the chromosome axes in both the wild-type and pair3-1 mutant. In the wild type, synapsed LEs were obviously observed during pachytene (Figure 4A). In the pair3-1 mutant, apparently normal AEs were also observed (Figure 4B), suggesting that PAIR3 may not be required for the formation of chromosome axes.
FIGURE 4:
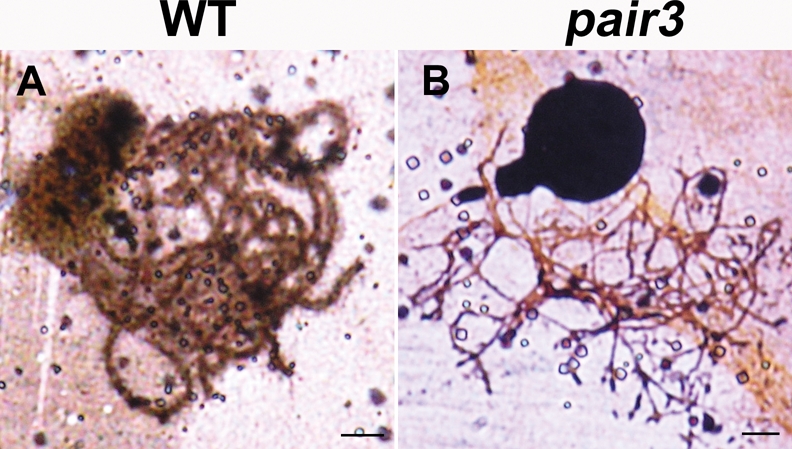
Silver staining of spread nuclei at pachytene stage in wild type and pair3 mutant. AE is formed in both wild type (A) and pair3 mutant (B). Bars, 5 μm.
PAIR3 is required for normal recombination and SC installation
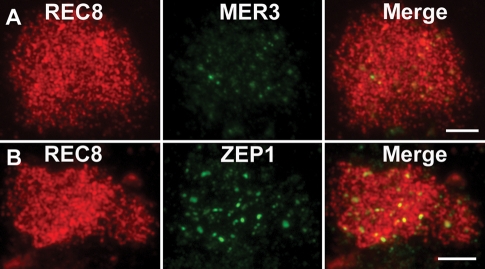
In the wild-type rice, MER3 foci form on the chromosomes at early leptotene, and those foci can be used as a marker to monitor most recombination events in rice (Wang et al., 2009b). To investigate whether normal recombination was also affected in the pair3-1 mutant, we observed the distribution of MER3 by immunostaining. In the wild type, MER3 appeared as foci from leptotene to zygotene. By contrast, only a few faint MER3 foci were detected on the chromosomes in the pair3-1 mutant (Figure 5A). This observation is consistent with a severe defect in recombination in pair3-1 mutants.
FIGURE 5:
The localization of MER3 and ZEP1 in pair3 mutant. (A) Only a few faint MER3 foci are detected in the pair3 mutant. (B) Only a few ZEP1 aggregates are observed in the pair3 mutant. Bars, 5 μm.
Lack of bivalent formation implies that chromosome synapsis was abolished in pair3-1 meiocytes. To determine whether initial assembly of SCs was affected in pair3-1 mutants, we performed dual-label immunostaining using antibodies against ZEP1 and REC8 raised in mouse and rabbit, respectively. ZEP1 is a major component of the CE in rice SCs. In the wild-type PMCs, ZEP1 first appeared as punctate foci at early zygotene and then elongated quickly to form short linear signals along the chromosomes. At pachytene, ZEP1 signals were located along the entire chromosomes. In most pair3-1 PMCs, only a few ZEP foci were observed (Figure 5B). However, short/fragmented ZEP1 signals were also found in a small number of nuclei. Together, the findings suggest that PAIR3 is important for proper ZEP1 assembly.
PAIR3 localizes along meiotic chromosomes during prophase I
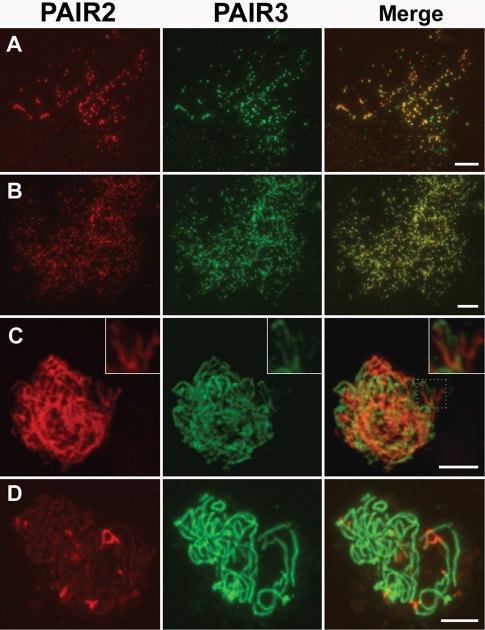
To investigate the spatial and temporal pattern of PAIR3 in wild-type microsporocytes, a polyclonal antibody against PAIR3 protein was raised in mouse. PAIR3 signals were first seen as foci in preleptotene. Those foci then elongated and formed filamentous structures on the chromosomes at leptotene (Supplemental Figure S3A). During zygotene, pachytene, and diplotene (Supplemental Figure S3, B–D), continuous PAIR3 signals were distributed along the entire length of the chromosomes. PAIR3 signals began to decrease at diakinesis (Supplemental Figure S3, E and F), and no signals were visible in subsequent stages.
To compare the PAIR3 and PAIR2 localization patterns, specific antibodies were used to double label meiotic chromosomes of wild-type PMCs at different stages. During interphase, discrete foci of PAIR2 and PAIR3 were extensively colocalized (Figure 6A). The total number of PAIR2 foci was slightly fewer than that of PAIR3 foci. That is, some PAIR3 foci failed to colocalize with a PAIR2 signal, while all PAIR2 signals also corresponded with PAIR3 staining (Figure 6A). At leptotene, all PAIR2 and PAIR3 stainings showed complete colocalization (Figure 6B). During zygotene, with the synapses of homologous chromosomes, PAIR2 proteins began to release from the chromosomes. Interestingly, remnant PAIR2 proteins often showed complementary staining patterns with PAIR3 proteins at this time (Figure 6C). Usually, one protein stains prominently, while the other is faint. Therefore, the merged images often exhibited red or green rather than yellow coloring. During pachytene, only a few PAIR2 fragments were observed, but PAIR3s were still distributed along the entire length of the chromosomes (Figure 6D).
FIGURE 6:
Dual immunolocalization of PAIR2 and PAIR3 in the wild-type prophase I. PAIR3 shows high colocalization with PAIR2 during interphase (A) and leptotene (B). (C) Zygotene, the magnified inset shows the complementary staining pattern between PAIR2 and PAIR3 protein. (D) Pachytene. Bars, 5 μm.
As the localization of PAIR3 was similar to that of REC8, we also conducted dual immunostaining experiments using antibodies against PAIR3 and REC8. The results clearly showed that PAIR3 signals completely overlapped with REC8 signals from leptotene to diplotene (Supplemental Figure S3, A–D). The only difference observed was that the disappearance of PAIR3 was slightly earlier than that of REC8. That is, at diakinesis, REC8 signals were visible in all cells, while PAIR3 signals could be observed in some but not all cells (Supplemental Figure S3, E and F). However, no PAIR3 signals were found in any of the pair3 PMCs observed, indicating that the mutants identified in this study may carry severe alleles.
PAIR3 localizes normally in pair2, mer3, and zep1 mutants
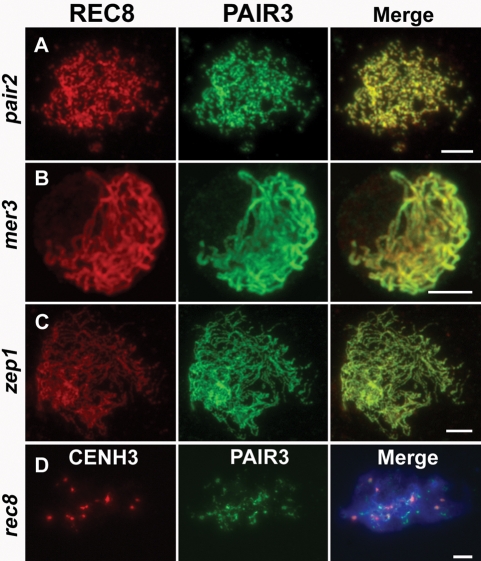
To determine whether PAIR3 localization was affected by meiotic recombination, the distribution of PAIR3 was also investigated in two recombination mutants, pair2 and mer3. In both mutants, PAIR3 localized on chromosomes normally and overlapped well with REC8 (Figure 7, A and B). Thus, PAIR3 localization is not affected by the recombination defects in the pair2 and mer3 mutants.
FIGURE 7:
The localization of PAIR3 in the pair2, mer3, zep1, and rec8 mutant. PAIR3 localizes on chromosomes normally in pair2 (A), mer3 (B), and zep1 (C). (D) PAIR3 localization is defective in rec8 mutant. Chromosomes are stained with DAPI (blue). Bars, 5 μm.
To determine whether the PAIR3 localization depended on the SC, we evaluated zep1 mutant cells and found that PAIR3 still associated with the chromosome axis in the absence of SC (Figure 7C), indicating that ZEP1 is not required for the normal PAIR3 localization.
PAIR3 localization is defective in rec8 mutants
In plants, sister chromatid cohesion has been shown to be required for the formation of meiotic chromosome axes. We next investigated whether PAIR3 localization was affected in rec8 mutants using an anti-CenH3 antibody to identify the PMCs. In the rec8 mutant, PAIR3 localization was severely disrupted, and only a few discrete PAIR3 signals were observed on the chromosomes (Figure 7D). This result indicated that the structural platform provided by cohesins may be essential for the localization of PAIR3.
DISCUSSION
PAIR3 localizes on chromosome axes during meiosis in rice
AEs play important roles in chromosome pairing and SC assembly. In addition, the recruitment of AEs is essential for chromosome condensation, pairing, and recombination. Although the identification of meiotic chromosome core proteins still remains elusive in plants, three axis-associated proteins have been described so far: ASY1/PAIR2/HOP1, REC8/SYN1/AFD1, and SCC3 (Mercier and Grelon, 2008). ASY1 is first observed as punctate foci in meiotic preleptotene and is maintained during pachytene, suggesting that ASY1 signals in Arabidopsis can indicate the structure of both AEs and LEs. PAIR2, the rice homologue of ASY1, also associates with AEs during preleptotene and leptotene. During zygotene, PAIR2 is released from the chromosomes during SC installation, suggesting that PAIR2 signals mainly reflect the structure of AEs, but not LEs (Nonomura et al., 2006). Additionally, normal AEs are observed in pair2-null mutant meiocytes, indicating that PAIR2 is not directly required for AE formation in meiosis (Nonomura et al., 2006). The cohesion proteins REC8 and SCC3 are components of the complex responsible for sister chromatid cohesion (Chelysheva et al., 2005; Golubovskaya et al., 2006; Zhang et al., 2006). Cohesins act as frameworks for the assembly of AEs/LEs, so cohesion signals during prophase I can reflect the location of both these elements. Mutations of cohesin genes always lead to chromosome condensation defects, formation of univalents, and chromosome fragmentation.
Here, the analyses of pair3 mutants indicated that PAIR3 is essential for bouquet formation, homologous pairing, and normal recombination as well as SC assembly. In addition, abnormal chromosome segregation and cytokinesis, which are universal defects of asynaptic mutants, were also observed in the pair3 mutants. The immunocytological experiments showed that PAIR3 appeared as foci in preleptotene. Those foci then fused to form short linear signals at leptotene. From zygotene to pachytene, the signals appeared as continuous signals. Dual immunostaining results showed that PAIR3 displayed extensive overlapping staining patterns with REC8 from preleptotene to pachytene, suggesting that PAIR3 localized to the axial cores of both synapsed and unsynapsed chromosomes, corresponding to LE and AE, respectively.
In the pair3 mutant, the ZEP1 protein failed to assemble along the chromosomes, suggesting that PAIR3 was required for SC assembly. Furthermore, PAIR3 localized normally to the unsynapsed chromosomes in the zep1 mutant. Therefore, this observation further confirmed that PAIR3 assembles before and independently of synapsis.
PAIR3 is required for PAIR2 localization to chromosomes
In the pair3 mutant, REC8 associated with chromosomes normally, suggesting that REC8 localization to the chromosome is independent of PAIR3. In the rec8 mutant, PAIR3 localization was severely disrupted, suggesting that REC8 is required for PAIR3 loading onto chromosomes. Similar dependence was also found in budding yeast, where the loading of Pds5 onto chromosomes requires the function of Rec8, but the association of Rec8 with chromosomes is independent of Pds5 (Zhang et al., 2005b; Jin et al., 2009). Both results are consistent with the fact that REC8 is required for cohesion and for formation of AEs/LEs.
A previous study demonstrated that PAIR2 associated with chromosome axes, and mutation of the PAIR2 gene caused the elimination of homologue pairing and synapsis and resulted in 24 completely unpaired univalents in the pair2 mutant during pachytene and diakinesis (Nonomura et al., 2004). In this study, the pair3 mutant displayed almost the same phenotype as the pair2 mutant, and it is likely that the PAIR2 and PAIR3 proteins work coordinately in the same pathway or process.
In the pair2 mutant, PAIR3 localized normally as in the wild type, so PAIR2 is not required for the assembly of PAIR3. In the pair3 mutant, PAIR2 signals appeared in leptotene as in the wild type, suggesting that PAIR3 is not required for initial recruitment of PAIR2. However, from late leptotene to pachytene, PAIR2 in the pair3 mutant did not localize onto chromosomes normally and its foci did not extend to form long-stretched signals. These results imply that PAIR3 recruitment to meiotic chromosome axes is a prerequisite for subsequent PAIR2 association. Thus, the loss of normal recombination and synapsis in the pair3 mutant may be due to the loss of normal anchoring of PAIR2 during meiosis.
Both PAIR2 and PAIR3 localize on the chromosome axes, but their duration is markedly different. Associated with synapsis during zygotene in the wild type, PAIR2 signals are released from the chromosome axes and are hardly detectable at pachytene (Nonomura et al., 2004). By contrast, PAIR3 signals can still be clearly observed on chromosomes during pachytene. The dependence of PAIR2 localization on PAIR3, as well as the longer persistence of PAIR3 signals on chromosomes, leads us to postulate that PAIR3 might function to provide a platform for PAIR2 proteins. Studies in Arabidopsis revealed that ASY1 acts as the interface between the developing chromosome axes and the recombination machinery, where it is required to ensure interhomologue recombination (Sanchez-Moran et al., 2007). Similarly, we propose that PAIR3 may work as the interface between the chromosome axes and PAIR2 protein, which is essential for normal functioning of the recombination machinery.
The dependence between AE-localized proteins has also been found in other species. For example, Red1 is essential for the recruitment of Hop1 to the chromosomes (Smith and Roeder, 1997), and SYCP3 is required for the normal binding of SYCP2 to the chromosomes (Pelttari et al., 2001). Meanwhile, HTP-3 is indispensable for the localization of HIM-3 to the chromosomes (Goodyer et al., 2008). In contrast to those AE mutants, however, the pair3 mutant in our study displayed normal assembly of chromosome axes. In addition, no chromosome condensation defect or chromosome fragmentation was detected. Those results indicated that the PAIR3 protein may not be a structural component of the chromosome axes but may belong to a structure that runs along the axes and associates with them.
Three identified axis-associated proteins in plants, namely, PAIR2/ASY1/HOP1, REC8/AFD1/SYN1, and SCC3, are all evolutionarily conserved among eukaryotes. However, PAIR3 shows little sequence similarity with other known proteins in Arabidopsis and other model organisms (Yuan et al., 2009), suggesting that PAIR3 may be a newly evolved meiotic protein in rice. Studies in other organisms have shown that, although the ultrastructural appearance of the SC is highly conserved, many of its components are ill conserved at the amino acid sequence level (de Boer and Heyting, 2006). Therefore, it is more likely that proteins with poor homology at the amino acid sequence level, but with similar structure, perform similar functions in other model organisms.
The PAIR3 protein contains putative coiled-coil motifs in its C-terminal domain. Similarly, the SYCP3/COR1 protein also has a predicted coiled-coil motif in the C-terminal half. In addition, both PAIR3 and SYCP3/COR1 assemble and disassemble in the same time as the SCs and label the AE/LE in a continuous manner. Mutation of either gene results in the defects in pairing, synapsis, and chiasma formation. However, SYCP3 is essential for the formation of LEs, while PAIR3 is not. Several lines of evidence suggest that the AE/LE seen by staining is only a small component of the axial structure by both morphological and functional criteria. In addition, it is likely that axial components may well occur in multiple (partially overlapping) layers (Zickler and Kleckner, 1999). Therefore, it is also possible that PAIR3 is an AE/LE component and localizes on one layer of the AE/LE.
In conclusion, our study strongly supports that the PAIR3 protein is associated with chromosome axes during meiosis and is required for the binding of PAIR2 to the chromosomes. Further investigation of the interaction between those meiotic proteins will promote the understanding of meiotic prophase chromosome cores in plants.
MATERIALS AND METHODS
Materials
The pair3-1 and pair3-2 mutants were identified from Wuxiangjing 9 (japonica) and Nipponbare (japonica), respectively. All plant materials were grown in individual lines in paddy fields. In summer, all materials were grown in Beijing, while in winter all materials were planted in Hainan Island in southern China.
Molecular cloning of PAIR3
To fine map PAIR3, InDel markers were developed based on sequence differences between indica variety 9311 and japonica variety Nipponbare according to the data published in http://www.ncbi.nlm.nih.gov/nucleotide. The primer sequences of InDel markers were as follows: P1 (5′-TACGTGTCCTTGTGCCTGA-3′ and 5′-ACAAATGAACTGAGAACGTG-3′), P2 (5′-CTTCCGATTAATTTCCTTTC-3′ and 5′-GGGTCTAGCTATTTGCTCT-3′), P3 (5′-TTCTACGAGCTTCCTACG-3′ and 5′-CTTAGACTCATTATAGTAC-3′), P4 (5′-CATATAGTGTCATATGCACC-3′ and 5′-GTCGTGTGTTTGCCAACAAT-3′), P5 (5′-CACTTCAGTGCTGGGTGTGC-3′ and 5′-TCAAAGGGCAAGTTAACGAC-3′), P6 (5′-GCGCATCCATGCATATCCAA-3′ and 5′-GACAAGGTGTTGCCCAAGAA-3′), P7 (5′-GTCAATGTGAATGGGAGACC-3′ and 5′-AGCAGCTCAAGGCAGCGA-3′), and P8 (5′-TTTCAGTGGCTCTGTTCGG-3′ and 5′-CAAAAGGAAATGAGTGAGGCT-3′).
Construction of RNAi and rice transformation
A 472-bp fragment of PAIR3 was amplified by PCR with primers P31F (5′-ACTCGAGATAATACGCCAGCACCT-3′) and P31R (5′-CAGATCTGTACGTCCTGGTCAATAC-3′); this fragment was cloned into the pGEM-T vector (Promega, Madison, WI) and sequentially cloned into the BamHI/SalI and BglII/XhoI sites of the pUC-RNAi vector. Subsequently, the stem loop fragment was cloned into the pCAMBIA2300-Actin vector. The resulting RNAi construct was transformed into an A. tumefaciens strain and then into mature embryo-derived callus of rice.
Antibody production
The antibodies against REC8, PAIR2, MER3, and ZEP1 were produced as described (Wang et al., 2009b, 2010). To generate the antibody against PAIR3, a 561-bp fragment of PAIR3 cDNA (amino acids 248 to 393) was amplified with primers P3GSTF (5′-GTGAATTCCCAGACCTATTACTCGTTCGCT-3′) and P3GSTR (5′-AGTCTCGAGTCATCGTGTTCACCCTTCTCT-3′). The fragments were digested with EcoRI/XhoI and ligated into the expression vector pGEX-4T-2. The expression vector was transformed into BL21 (DE3) and was induced by the addition of 0.3 mM isopropyl β-d-1-thiogalactopyranoside to the culture medium. The PAIR3-fusion peptides, mainly expressed in the soluble fraction, were purified using the MagneGST Protein Purification System (Promega). The specificity of PAIR3 antibody was checked by immunofluorescence.
Meiotic chromosome preparation
Young panicles of both wild type and pair3 were harvested and fixed in Carnoy’s solution (ethanol:glacial acetic = 3:1). Microsporocytes undergoing meiosis were squashed in an acetocarmine solution. Slides with chromosomes were frozen in liquid nitrogen. After removing the coverslips, the slides were dehydrated through an ethanol series (70%, 90%, and 100%). Chromosomes were counterstained with 4′,6-diamidino-phenylindole (DAPI) in an antifade solution (Vector Laboratories, Burlingame, CA). Chromosome images were captured under the Olympus BX61 fluorescence microscope with a micro-CCD camera. For silver staining, the slides were stained under coverslips in a moist chamber at 60° with a 2:1 (vol/vol) mixture of 50% aqueous silver nitrate to developer (2 g gelatin, l ml formic acid, 100 ml distilled water). The slides were washed off under running tap water, rinsed twice in distilled water, air dried, and photographed using an Olympus BX61 microscope. The chiasma frequency was quantified as described (Sanchez Moran et al., 2001; Wang et al., 2009b).
Fluorescence in situ hybridization
FISH analysis was conducted as described by Zhang et al. (2005a). Two repetitive DNA elements were used as the FISH probes: pTa794 has 5S ribosomal RNA genes from wheat (Cuadrado and Jouve, 1994), and pAtT4 contains telomeric repeats. Digoxigenin-11-dUTP-labeled probes were detected using an anti-digoxigenin conjugated with rhodamine antibody (Roche Diagnostics, Indianapolis, IN). Chromosomes were counterstained with DAPI in Vectashield anti-fading solution (Vector Laboratories). Original images were observed under Olympus BX61 fluorescence microscopy and captured with a DVC1412 CCD camera using IPLab software. The final images were merged using Adobe Photoshop CS2 software.
Immunofluorescence
Fresh young panicles were fixed in 4% (wt/vol) paraformaldehyde for 30 min at room temperature. Anthers in the proper stage were squashed on a slide with phosphate-buffered saline (PBS) solution and covered with a coverslip. After the slide was soaked in liquid nitrogen and the coverslip removed, the slide was dehydrated through an ethanol series (70%, 90%, and 100%) before being used in immunostaining. Slides were then incubated in a humid chamber at 37° for 4 h in different antibody combinations diluted 1:500 in TNB buffer (0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, and 0.5% blocking reagent). After three rounds of washing in PBS, Texas-red–conjugated goat anti-rabbit antibody and fluorescein isothiocyanate–conjugated sheep anti-mouse antibody (1:1000) were added to the slides. The chromosomes were counterstained with DAPI in an antifade solution (Vector Laboratories).
Supplementary Material
Acknowledgments
This work was supported by grants from the Ministry of Sciences and Technology of China (2011CB944602 and 2009ZX08009-036B) and the National Natural Science Foundation of China (30921061).
Abbreviations used:
- AE
axial element
- CE
central element
- LE
lateral element
- PMC
pollen mother cell
- SC
synaptonemal complex
- TF
transverse filament
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-08-0667) on November 30, 2010.
REFERENCES
- Chelysheva L, et al. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci. 2005;118:4621–4632. doi: 10.1242/jcs.02583. [DOI] [PubMed] [Google Scholar]
- Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Jouve N. Mapping and organization of highly-repeated DNA sequences by means of simultaneous and sequential FISH and C-banding in 6x-triticale. Chromosome Res. 1994;2:331–338. doi: 10.1007/BF01552727. [DOI] [PubMed] [Google Scholar]
- Dawe RK. Meiotic chromosome organization and segregation in plants. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:371–395. doi: 10.1146/annurev.arplant.49.1.371. [DOI] [PubMed] [Google Scholar]
- de Boer E, Heyting C. The diverse roles of transverse filaments of synaptonemal complexes in meiosis. Chromosoma. 2006;115:220–234. doi: 10.1007/s00412-006-0057-5. [DOI] [PubMed] [Google Scholar]
- Dobson MJ, Pearlman RE, Karaiskakis A, Spyropoulos B, Moens PB. Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J Cell Sci. 1994;107:2749–2760. doi: 10.1242/jcs.107.10.2749. [DOI] [PubMed] [Google Scholar]
- Golubovskaya IN, Hamant O, Timofejeva L, Wang CJ, Braun D, Meeley R, Cande WZ. Alleles of afd1 dissect REC8 functions during meiotic prophase I. J Cell Sci. 2006;119:3306–3315. doi: 10.1242/jcs.03054. [DOI] [PubMed] [Google Scholar]
- Goodyer W, Kaitna S, Couteau F, Ward JD, Boulton SJ, Zetka M. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev Cell. 2008;14:263–274. doi: 10.1016/j.devcel.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Hamant O, Ma H, Cande WZ. Genetics of meiotic prophase I in plants. Annu Rev Plant Biol. 2006;57:267–302. doi: 10.1146/annurev.arplant.57.032905.105255. [DOI] [PubMed] [Google Scholar]
- Harper L, Golubovskaya I, Cande WZ. A bouquet of chromosomes. J Cell Sci. 2004;117:4025–4032. doi: 10.1242/jcs.01363. [DOI] [PubMed] [Google Scholar]
- Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FC. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 2005;19:2488–2500. doi: 10.1101/gad.354705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Guacci V, Yu HG. Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis. J Cell Biol. 2009;186:713–725. doi: 10.1083/jcb.200810107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers JH, Offenberg HH, van Aalderen M, Vink AC, Dietrich AJ, Heyting C. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol Cell Biol. 1994;14:1137–1146. doi: 10.1128/mcb.14.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J, Scherthan H, Kaback DB. Physical association between nonhomologous chromosomes precedes distributive disjunction in yeast. Proc Natl Acad Sci USA. 1994;91:331–334. doi: 10.1073/pnas.91.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Colaiacovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Grelon M. Meiosis in plants: ten years of gene discovery. Cytogenet Genome Res. 2008;120:281–290. doi: 10.1159/000121077. [DOI] [PubMed] [Google Scholar]
- Meuwissen RL, Offenberg HH, Dietrich AJ, Riesewijk A, van Iersel M, Heyting C. A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. Embo J. 1992).;11:5091–5100. doi: 10.1002/j.1460-2075.1992.tb05616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Nakano M, Eiguchi M, Suzuki T, Kurata N. PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J Cell Sci. 2006;119:217–225. doi: 10.1242/jcs.02736. [DOI] [PubMed] [Google Scholar]
- Nonomura KI, Nakano M, Murata K, Miyoshi K, Eiguchi M, Miyao A, Hirochika H, Kurata N. An insertional mutation in the rice PAIR2 gene, the ortholog of Arabidopsis ASY1, results in a defect in homologous chromosome pairing during meiosis. Mol Genet Genomics. 2004;271:121–129. doi: 10.1007/s00438-003-0934-z. [DOI] [PubMed] [Google Scholar]
- Offenberg HH, Schalk JA, Meuwissen RL, van Aalderen M, Kester HA, Dietrich AJ, Heyting C. SCP2: a major protein component of the axial elements of synaptonemal complexes of the rat. Nucleic Acids Res. 1998;26:2572–2579. doi: 10.1093/nar/26.11.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 2001;15:3130–3143. doi: 10.1101/gad.935001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- Pawlowski WP, Wang CJ, Golubovskaya IN, Szymaniak JM, Shi L, Hamant O, Zhu T, Harper L, Sheridan WF, Cande WZ. Maize AMEIOTIC1 is essential for multiple early meiotic processes and likely required for the initiation of meiosis. Proc Natl Acad Sci USA. 2009;106:3603–3608. doi: 10.1073/pnas.0810115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelttari J, et al. A meiotic chromosomal core consisting of cohesin complex proteins recruits DNA recombination proteins and promotes synapsis in the absence of an axial element in mammalian meiotic cells. Mol Cell Biol. 2001).;21:5667–5677. doi: 10.1128/MCB.21.16.5667-5677.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS. Meiosis in asynaptic yeast. Genetics. 1990;126:563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Moran E, Armstrong SJ, Santos JL, Franklin FC, Jones GH. Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res. 2001;9:121–128. doi: 10.1023/a:1009278902994. [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E, Santos JL, Jones GH, Franklin FC. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007;21:2220–2233. doi: 10.1101/gad.439007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson AF, Ling L, van Zuylen V, Meyer BJ. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 2009;23:1763–1778. doi: 10.1101/gad.1808809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AV, Roeder GS. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M, Engebrecht JA, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Wang CJ, Carlton PM, Golubovskaya IN, Cande WZ. Interlock formation and coiling of meiotic chromosome axes during synapsis. Genetics. 2009a;183:905–915. doi: 10.1534/genetics.109.108688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, et al. MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J Cell Sci. 2009b;122:2055–2063. doi: 10.1242/jcs.049080. [DOI] [PubMed] [Google Scholar]
- Wang M, Wang K, Tang D, Wei C, Li M, Shen Y, Chi Z, Gu M, Cheng Z. The central element protein ZEP1 of synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell. 2010;22:417–430. doi: 10.1105/tpc.109.070789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Yang F, De La Fuente R, Leu NA, Baumann C, McLaughlin KJ, Wang PJ. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol. 2006;173:497–507. doi: 10.1083/jcb.200603063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Hoog C. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol Cell. 2000;5:73–83. doi: 10.1016/s1097-2765(00)80404-9. [DOI] [PubMed] [Google Scholar]
- Yuan W, Li X, Chang Y, Wen R, Chen G, Zhang Q, Wu C. Mutation of the rice gene PAIR3 results in lack of bivalent formation in meiosis. Plant J. 2009;59:303–315. doi: 10.1111/j.1365-313X.2009.03870.x. [DOI] [PubMed] [Google Scholar]
- Zetka MC, Kawasaki I, Strome S, Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Yang Q, Bao W, Zhang Y, Han B, Xue Y, Cheng Z. Molecular cytogenetic characterization of the Antirrhinum majus genome. Genetics. 2005a;169:325–335. doi: 10.1534/genetics.104.031146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tao J, Wang S, Chong K, Wang T. The rice OsRad21-4, an orthologue of yeast Rec8 protein, is required for efficient meiosis. Plant Mol Biol. 2006;60:533–554. doi: 10.1007/s11103-005-4922-z. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ren Q, Yang H, Conrad MN, Guacci V, Kateneva A, Dresser ME. Budding yeast PDS5 plays an important role in meiosis and is required for sister chromatid cohesion. Mol Microbiol. 2005b;56:670–680. doi: 10.1111/j.1365-2958.2005.04582.x. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.