It is shown that RalA is regulated by a Ral GAP complex (RGC 1/2) in insulin action and links PI 3-kinase signaling to RalA activation. Akt phosphorylates the complex and inhibits its function, resulting in increased RalA activity and glucose uptake.
Abstract
Insulin stimulates glucose transport in muscle and adipose tissue by translocation of glucose transporter 4 (GLUT4) to the plasma membrane. We previously reported that activation of the small GTPase RalA downstream of PI 3-kinase plays a critical role in this process by mobilizing the exocyst complex for GLUT4 vesicle targeting in adipocytes. Here we report the identification and characterization of a Ral GAP complex (RGC) that mediates the activation of RalA downstream of the PI 3-kinase/Akt pathway. The complex is composed of an RGC1 regulatory subunit and an RGC2 catalytic subunit (previously identified as AS250) that directly stimulates the guanosine triphosphate hydrolysis of RalA. Knockdown of RGC proteins leads to increased RalA activity and glucose uptake in adipocytes. Insulin inhibits the GAP complex through Akt2-catalyzed phosphorylation of RGC2 in vitro and in vivo, while activated Akt relieves the inhibitory effect of RGC proteins on RalA activity. The RGC complex thus connects PI 3-kinase/Akt activity to the transport machineries responsible for GLUT4 translocation.
INTRODUCTION
Insulin-stimulated glucose transport plays a key role in the maintenance of glucose homeostasis (Saltiel and Kahn, 2001; Watson et al., 2004). The hormone increases glucose uptake into insulin-responsive tissues including muscle and fat by recruitment of the facilitative glucose transporter 4 (GLUT4) from intracellular storage vesicles to the plasma membrane (Watson et al., 2004; Huang and Czech, 2007). While our understanding of the precise mechanisms underlying this process remains incomplete, it is clear that the translocation of GLUT4 vesicles requires concerted actions of distinct transport machineries that are governed by multiple signaling cascades (Watson and Pessin, 2006; Hou and Pessin, 2007). Activation of the protein kinase Akt downstream of PI 3-kinase plays a central role in the trafficking of GLUT4 to the plasma membrane (Whiteman et al., 2002; Hou and Pessin, 2007; Sakamoto and Holman, 2008), although the exact mechanisms by which Akt signals cellular machineries involved in the transport process remain to be elucidated.
One possible target of the PI 3-kinase/Akt pathway is the family of small GTPases (Watson and Pessin, 2006; Welsh et al., 2006; Ishikura et al., 2007), which act as “molecular switches” that operate at the crossroads of signaling and vesicle trafficking (Bourne et al., 1991; Takai et al., 2001; Grosshans et al., 2006). Once activated, these G proteins can mobilize transport machineries that facilitate specific steps in exocytosis, including vesicle budding, transport along cytoskeletal tracks, and tethering and docking at the plasma membrane (Ferro-Novick and Novick, 1993; Hall, 1998; Cai et al., 2007). A number of small GTPases, including Ras, Rho, and Rab family members, have been implicated in insulin-stimulated GLUT4 trafficking (Welsh et al., 2006; Hou and Pessin, 2007; Ishikura et al., 2008). We recently reported that activation of the small GTPase RalA by insulin plays an important role in plasma membrane targeting of GLUT4 vesicles through recognition of the exocyst (Chen et al., 2007), an evolutionarily conserved vesicle tethering complex (Novick and Guo, 2002; Schekman and Novick, 2004). RalA also interacts with the molecular motor Myo1c complex that facilitates recruitment of GLUT4 vesicles to the plasma membrane (Bose et al., 2002; Chen et al., 2007). However, the exact mechanism by which insulin activates RalA remains to be established.
The activation status of small GTPases is determined by specific guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) (McCormick, 1998; Bos et al., 2007). In many scenarios, rather than simply terminating the activity of their cognate GTPase substrates, GAPs act as the primary determinants of GTPase activity based on upstream inputs (Kwiatkowski and Manning, 2005; Watson and Pessin, 2006). One example is the TSC1/2 complex, a tumor suppressor complex that functions as a GAP for the Rheb GTPase and serves as the primary mechanism whereby Rheb is activated (Manning and Cantley, 2003). This GAP complex is directly phosphorylated by Akt, in the process reducing its activity and thus relieving its inhibitory effect on Rheb GTPases, leading to activation of its downstream target, the mTORC1 kinase (Garami et al., 2003; Inoki et al., 2003; Patel et al., 2003; Saucedo et al., 2003; Stocker et al., 2003; Tee et al., 2003; Zhang et al., 2003). Likewise, it is thought that phosphorylation of the GAP AS160/TBC1D4 by Akt inhibits its activity toward Rab target proteins involved in insulin-stimulated GLUT4 exocytosis (Sano et al., 2003, 2007; Miinea et al., 2005). AS160 is thought to inactivate Rabs 8, 10, and/or 14 (Ishikura et al., 2007; Sano et al., 2007, 2008); however, the downstream machineries mobilized by these proteins have not yet been determined. Although the exact mechanism by which Akt inhibits TSC2 and AS160 GAP activities remains unknown, these findings point to a regulatory paradigm in which GAPs integrate signals from Akt to activate small GTPases, which in turn engage with downstream effectors (Inoki and Guan, 2006; Huang and Manning, 2009). Here we report the identification and characterization of a Ral GAP complex (RGC) that connects the PI 3-kinase/Akt pathway to the activation of RalA, a small GTPase that mobilizes the exocyst complex to facilitate trafficking of GLUT4 vesicles.
RESULTS
Identification of the RGC in adipocytes
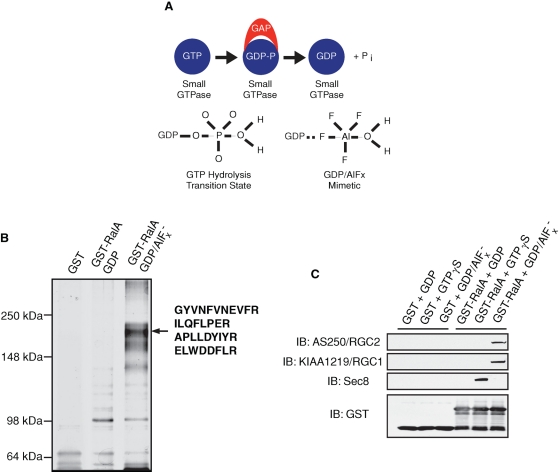
Previous studies have demonstrated that GAPs preferentially interact with their target GTPases during the transition state of guanosine triphosphate (GTP) hydrolysis (Bos et al., 2007; Scheffzek et al., 1997). This interaction is transient and occurs with lower affinity when wild-type GTPases are locked in either guanosine diphosphate (GDP)– or GTP-bound states. However, binding of GTPases to GDP in complex with aluminum fluoride (GDP/AlFx) mimics the GTP hydrolysis transition state, thus permitting stabilized GTPase–GAP interaction (Scheffzek et al., 1997; Wittinghofer, 1997; see Figure 1A). We took advantage of this property in a proteomic approach to search for potential GAPs that may interact with immobilized Ral loaded with GDP/AlFx in 3T3-L1 adipocytes since wild-type RalA displayed a much lower GTP loading profile than did its mutant form that fails to catalyze GTP hydrolysis in these cells (Supplemental Figure S1). Interestingly, several proteins were found to preferentially associate with glutathione S-transferase (GST)–RalA loaded with GDP/AlFx but not with GDP-loaded GST-RalA or GST alone, and the most prominent of these proteins migrated at ∼200 kDa (p200; Figure 1B). When subjected to tandem mass spectrometry, four unique peptides were recovered with the protein; each sequence matched that of a protein named AS250 (Figure 1B). AS250, or Akt substrate of 250 kDa, was originally identified as a phosphoprotein of 1984 amino acids that was purified from adipocytes by virtue of its interaction with a phospho-Akt substrate antibody (Gridley et al., 2006). The protein contains a putative GAP domain homologous to TSC2 and RapGAP and was found to associate with the protein KIAA1219 (Gridley et al., 2006). In agreement with the mass spectrometry data, Western blotting (WB) revealed that p200 GAP (AS250) and its associating protein KIAA1219 specifically interacted with RalA bound to GDP/AlFx, but not with RalA bound to GDP or GTPγS, or GST alone (Figure 1C). Meanwhile, subunits of the exocyst proteins preferentially interacted with RalA loaded with GTPγS (Figure 1C), further confirming the specificity of the transition state–dependent binding between KIAA1219/p200 GAP and RalA that is characteristic of a GAP–substrate GTPase interaction. We thus named these two proteins RGC1 and RGC2, to reflect their function and homology with TSC1/2, as will be further described. RGC1 represents the regulatory subunit of the complex, while RGC2 contains the GAP domain and represents the catalytic subunit of the complex.
FIGURE 1:
RGC2/AS250 preferentially interacts with RalA in the transition state. (A) Schematic view of the transition state interaction between GAPs and their cognate small GTPase targets. Top: GAPs interact preferentially with their cognate targets during the transition state of GTP hydrolysis. Bottom: Transition state of GTP hydrolysis mimicked by GDP in complex with aluminum fluoride (AlFx). (B) 3T3-L1 adipocyte lysates were incubated with immobilized GST alone, GST-RalA bound to GDP, or GST-RalA bound to GDP/AlFx. Associated proteins were separated using 6% SDS–PAGE and visualized by silver staining; protein bands were cut from the gel and subjected to tandem mass spectrometry analysis. (C) Cell lysates from a 10-cm plate of 3T3-L1 adipocytes were incubated with 5 μg of immobilized GST or GST-RalA bound to GDP, GTPγS, or GDP/AlFx. Associated proteins were resolved on SDS–PAGE and visualized by WB using the indicated antibodies.
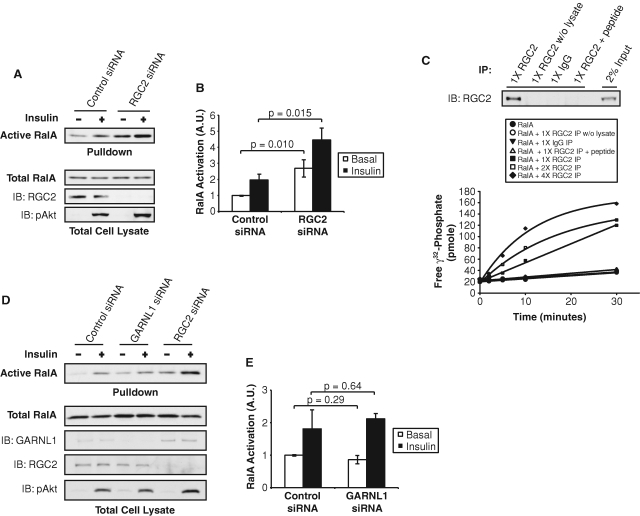
We sought to explore the importance of the RGC in the regulation of Ral activity by insulin. Consistent with the characteristic GAP–substrate GTPase interaction, siRNA-mediated depletion of the GAP RGC2 in 3T3-L1 adipocytes led to increased RalA activity in both the basal and insulin-stimulated states, as determined by a pulldown assay using the immobilized effector domain of Sec5 (Figure 2, A and B). This gain-of-function phenotype was specific to the loss of RGC2, as knockdown of other GAPs including p120 Ras GAP, RapGAP, AS160, or TSC2 had little effect on RalA activity (Supplemental Figure S2). When immunoprecipitated from cell lysates of 3T3-L1 adipocytes by an anti-RGC2 antibody, RGC proteins efficiently enhanced GTP hydrolysis of recombinant RalA in vitro, compared with RalA alone or RalA incubated with various control immunoprecipitates (Figure 2C). The immunoprecipitated RGC proteins also catalyzed GTP hydrolysis of the Ral family member RalB (Figure S3A). These results are in contrast to a previous report that failed to detect RGC GAP activity toward RalA or other G proteins (Gridley et al., 2006), perhaps due to differences in the assay used to detect activity. Immunoprecipitated RGC did not stimulate GTP hydrolysis of other GTPases of close homology to Ral, such as Rheb (Supplemental Figure S3B) or H-Ras (Supplemental Figure S3C), demonstrating the specificity of the GAP activity of the RGC proteins for Ral GTPases.
FIGURE 2:
RGC2 negatively regulates RalA activity in adipocytes. (A) 3T3-L1 adipocytes were transfected with control siRNA or siRNA oligos that deplete RGC2. Four days after transfection, cell lysates were subjected to a pulldown assay to detect RalA activation. Pulldowns represent the amount of active RalA from half of a 10-cm plate of adipocytes. Lysates represent 2% of total protein from a 10-cm plate of adipocytes. (B) Quantification of RalA activity upon RGC2 knockdown from three independent experiments. Significance was determined by Student's t test. (C) E. coli–expressed recombinant RalA loaded with γ32-GTP was incubated with increasing amounts of immune complexes from immunoprecipitations (IP) with the indicated antibodies (1X = immune complex from half of a 15-cm dish; 2X = immune complex from one 15-cm dish; 4X = immune complex from two 15-cm dishes). Free γ32-phosphate was measured as a reflection of GTP hydrolysis. Top: RGC2 proteins present in the IP were determined by WB analysis. (D) 3T3-L1 adipocytes were transfected with control siRNA, siRNA oligos that deplete GARNL1, or siRNA oligos that deplete RGC2. Four days after transfection, cell lysates were subjected to a pulldown assay to detect RalA activation. Pulldowns represent the amount of active RalA from half of a 10-cm plate of adipocytes. Lysates represent 2% of total protein from a 10-cm plate of adipocytes. (F) Quantification of RalA activity upon GARNL1 or RGC2 knockdown from two independent experiments.
A protein named GARNL1 was recently isolated in complex with RalAQ72L and RGC1 in brain lysates (Shirakawa et al., 2009). GARNL1 contains a GAP domain with 83% identity to the GAP domain of RGC2 (Supplemental Figure S4A) and has in vitro activity toward Ral GTPases (Shirakawa et al., 2009). SiRNA-mediated depletion of GARNL1 did not affect the activity of RalA in 3T3-L1 adipocytes (Figure 2, D and E), despite efficient knockdown of GARNL1 protein (∼82% knockdown), whereas knockdown of RGC2 (∼95% knockdown) increased both basal and insulin-stimulated RalA activity. Surprisingly, we observed a ∼245% increase in GARNL1 protein levels when cells were transfected with siRNA to deplete RGC2 (Figure 2D). Analysis of the expression of GARNL1 and RGC2 during adipocyte differentiation revealed the up-regulation of GARNL1 protein levels during the clonal expansion stage of 3T3-L1 adipogenesis, followed by a marked down-regulation during later stages of differentiation, with little GARNL1 protein present in mature 3T3-L1 adipocytes (Supplemental Figure S3B). In contrast, RGC2 expression was induced during insulin treatment of adipocytes and remained high in mature adipocytes (Supplemental Figure S4B). Furthermore, RGC2 was the predominant Ral GAP expressed in primary white adipocytes (Supplemental Figure S4C). Taken together, these data indicate that RGC2 is the primary Ral GAP expressed in adipocytes and is responsible for regulating RalA activity in these cells. The identification of a specific Ral GAP in adipocytes points to a novel regulatory process targeting this G protein in insulin action.
RGC1 and RGC2 form a physical and functional complex to inactivate RalA
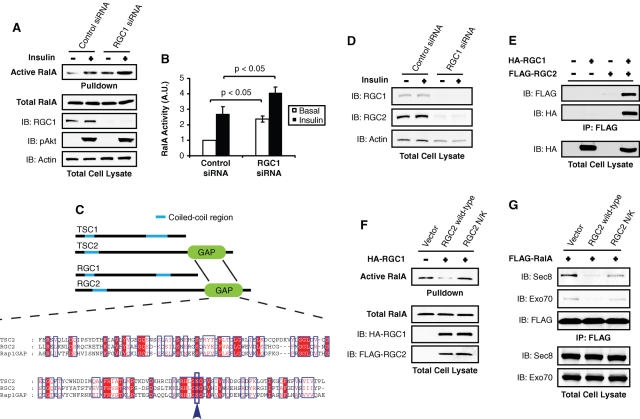
The precipitation of both RGC1 and 2 with the RalA-GDP/AlFx affinity matrix led us to test whether RGC1/2 forms a functional complex to inactivate RalA in vivo. We assayed RalA activity after depletion of RGC1 from 3T3-L1 adipocytes by siRNA-mediated knockdown. While proximal insulin signaling remained unaffected, as determined by p-Akt blotting, knockdown of RGC1 led to increased RalA activity in both the basal and insulin-stimulated states (Figure 3, A and B), indicating that both RGC proteins are required to form a functional GAP complex that inactivates RalA. The nature of this complex is reminiscent of the Rheb GAP TSC1/2 complex (Inoki et al., 2003; Li et al., 2004a), with which RGC1/2 shares a similar overall structure (Figure 3C). Furthermore, alignment of the GAP domains revealed that RGC2 shares a conserved catalytic asparagine residue with TSC2 and RapGAP (Figure 3C), indicating a similar catalytic mechanism among these GAPs (Li et al., 2004b). Interestingly, siRNA-mediated knockdown of RGC1 in adipocytes led to decreased levels of RGC2 (Figure 3D), suggesting that physical interaction is required for maximal stability of the subunits. Consistent with this notion, RGC2 protein was stabilized when coexpressed with RGC1, which interacts with the former protein (Figure 3E), further indicating that these proteins form a physical complex for stability.
FIGURE 3:
RGC1 and RGC2/AS250 form a physical and functional complex to inactivate RalA. (A) 3T3-L1 adipocytes were transfected with control siRNA or siRNA oligos that deplete RGC1 and were subjected to a pulldown assay to detect RalA activity. Pulldowns represent the amount of active RalA from half of a 10-cm plate of adipocytes. Lysates represent 2% of total protein from a 10-cm plate of adipocytes. (B) Quantification of RalA activity upon RGC1 knockdown from three independent experiments. Significance was determined by Student's t test. (C) ClustalW alignment of the RGC2 and TSC2 GAP domains. (D) 3T3-L1 adipocytes were transfected with control siRNA or siRNA oligos that deplete RGC1, and 2% of lysates from a 10-cm plate of adipocytes were resolved on SDS–PAGE and subjected to WB using the indicated antibodies. (E) COS-1 cells were transfected with the indicated DNA and subjected to immunoprecipitations (IP) with anti-FLAG antibody. Immune complexes were resolved on SDS–PAGE and analyzed by WB. (F) COS-1 cells were transfected with FLAG-RalA, HA-RGC1, and either wild-type or the N/K mutant FLAG-RGC2 construct as indicated. Cell lysates were subjected to pulldown assay to detect RalA activation. Pulldowns represent the amount of active RalA from half of a 10-cm plate of COS-1 cells. Lysates represent 2% of total protein from a 10-cm plate. (G) COS-1 cells were transfected with FLAG-RalA, HA-RGC1, and either wild-type or the N/K mutant FLAG-RGC2 constructs as indicated. Cell lysates were subjected to IP with FLAG antibody. Immune complexes were resolved on SDS–PAGE, and the interaction between FLAG-RalA and endogenous subunits of the exocyst was visualized by WB using the indicated antibodies.
We tested whether overexpression of RGC1/2 could inactivate RalA in vivo via its GAP activity. Consistent with the in vitro GAP assay, coexpression of RGC1 and 2 in cells significantly decreased the activity of the G protein, as determined by the pulldown assay (Figure 3F). This inhibitory effect was absent in cells expressing RGC1 and RGC2 N/K, in which the critical asparagine residue for GAP activity was substituted to lysine to abolish the catalytic activity (Figure 3F). Consistent with these data, overexpression of wild-type RGC1/2 decreased the interaction between wild-type RalA and the exocyst subunits Sec8 and Exo70, as assayed by coimmunoprecipitation of endogenous exocyst subunits (Figure 3G). The RGC2 N/K mutant had a minimal effect on the interaction between wild-type RalA and exocyst subunits, likely due to residual GAP activity of the mutant (Figure 3G). Taken together, these data demonstrate that RGC1 and RGC2 form a physical and functional GAP complex to inactivate RalA and prevent its interaction with the exocyst in a manner that resembles inactivation of Rheb by the TSC1/2 proteins.
RGC2 is an endogenous substrate for Akt2 downstream of PI 3-kinase
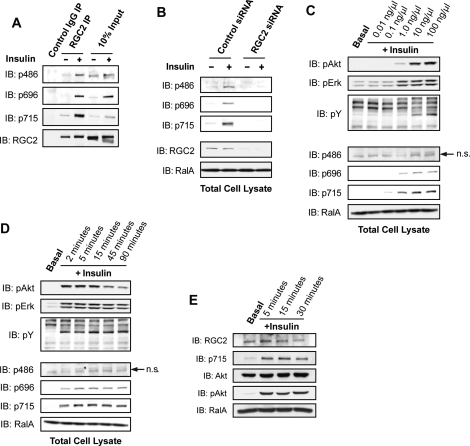
The similarity between the RGC1/2 complex and the Rheb GAP complex TSC1/2 (Figure 3), as well as the PI 3-kinase dependence of the activation of RalA by insulin, prompted us to investigate whether the GAP RGC2 may also be regulated by Akt-dependent phosphorylation downstream of PI 3-kinase activity. Previous mass spectrometry studies showed that insulin treatment increased the phosphorylation of Ser486, Ser696, and Thr715 on RGC2/AS250 in adipocytes (Gridley et al., 2006), although the protein kinase catalyzing these phosphorylation events was not defined. To elaborate these signaling events, we generated phosphospecific antibodies to each of the three sites. Insulin stimulated the phosphorylation of all three sites on RGC2 in 3T3-L1 adipocytes, as determined by blotting an anti-RGC2 immune complex or total cell lysates with the phosphospecific antibodies (Figure 4A). Phosphorylation of RGC2 in total cell lysates was depleted upon knockdown of the protein, further demonstrating the specificity of the phospho antibodies (Figure 4B). Insulin stimulated the phosphorylation of RGC2 in a dose-dependent manner that correlated well with the phosphorylation of Akt (Figure 4C). A time course of insulin treatment of adipocytes revealed that phosphorylation of the Ser696 and Thr715 sites was sustained for up to 90 min (Figure 4D). Phosphorylation at the Ser486 site peaked at 45 min and decreased by 90 min, perhaps indicating faster turnover at this site (Figure 4D). Consistent with the data from 3T3-L1 adipocytes, insulin also stimulated phosphorylation of RGC2 in white adipose tissue for up to 30 min (Figure 4E), providing in vivo evidence for this phosphorylation event.
FIGURE 4:
Insulin stimulates phosphorylation of RGC2 in adipocytes. (A) Serum-starved 3T3-L1 adipocytes were mock treated or stimulated with 100 nM insulin for 10 min, and lysates were immunoprecipitated using control rabbit IgG or anti-RGC2 antibody. The immune complexes from one 10-cm plate and 2% of total cell lysates were resolved on SDS–PAGE and analyzed by WB using the indicated antibodies. (B) 3T3-L1 adipocytes were transfected with control siRNA or siRNA oligos to deplete RGC2. Serum-starved cells were mock treated or stimulated with 100 ng/μl insulin for 10 min before lysis; 2% of cell lysates from a 10-cm plate of adipocytes were resolved by SDS–PAGE and subjected to WB analysis with the indicated antibodies. (C) Serum-starved 3T3-L1 adipocytes were mock treated or stimulated with insulin at the indicated doses for 10 min before lysis; 2% of cell lysates from a 10-cm plate of cells were resolved by SDS–PAGE and analyzed by WB with the indicated antibodies. (D) Serum-starved 3T3-L1 adipocytes were mock treated or stimulated with 100 ng/μl insulin for the indicated times; 2% of cell lysates from a 10-cm plate of cells were resolved by SDS–PAGE and analyzed by WB with the indicated antibodies. (E) Epididymal fat pads were excised from a mouse and starved ex vivo in serum-free DMEM for 30 min before stimulation with 100 nM insulin for the indicated times; 30 mg of protein per condition were resolved by SDS–PAGE, and RGC2 phosphorylation was determined by WB.
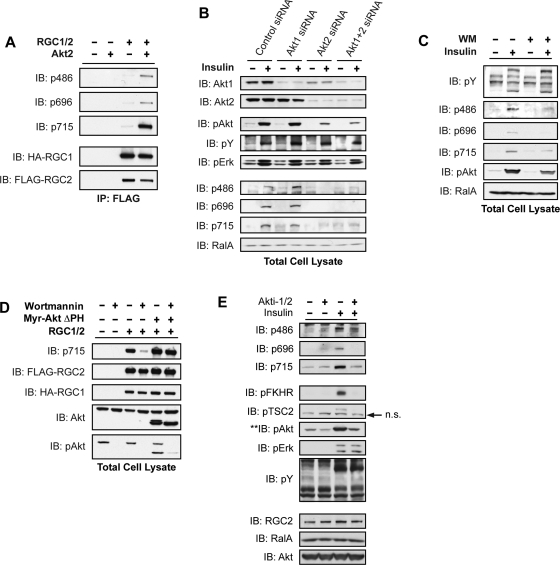
To test whether Akt catalyzed the phosphorylation of RGC2 on these sites, immunoprecipitated RGC1/2 complex was subjected to an in vitro phosphorylation assay in the presence of recombinant Akt2. Akt2 directly phosphorylated all three sites on RGC2 (Figure 5A). To ascertain the Akt isoform responsible for RGC2 phosphorylation in cells, we depleted Akt1, Akt2, or both isoforms in adipocytes by siRNA-mediated knockdown (Figure 5B, top). Knockdown of Akt1 produced minimal inhibition of Akt signaling, as determined by blotting with the phospho-Akt antibody, while knockdown of Akt2 alone or in combination with Akt1 dramatically attenuated Akt activation (Figure 5B, middle). SiRNA-mediated reduction in Akt2 activity led to inhibition of RGC2 phosphorylation on all three sites (Figure 5B, bottom), supporting the notion that the GAP RGC2 is an endogenous substrate of Akt2 in adipocytes. Interestingly, acute inhibition of Akt activity by incubation of cells with wortmannin blunted insulin-stimulated phosphorylation on Ser696 of RGC2, while phosphorylation of Ser486 and Thr715 was less sensitive, indicating that these sites may require lower Akt activity for phosphorylation (Figure 5C). Alternatively, it remains possible that Ser696 phosphorylation undergoes more efficient turnover. Collectively, the data indicate that activation of Akt2 via the PI 3-kinase pathway regulates RGC2 phosphorylation.
FIGURE 5:
RGC2 is an endogenous substrate of Akt2 in adipocytes. (A) COS-1 cells were transfected with HA-RGC1 and FLAG-RGC2 constructs as indicated. The RGC proteins were isolated by IP with FLAG antibody and subjected to an in vitro kinase assay with 100 ng recombinant Akt2 kinase and 100 μM ATP for 30 min at 32°C. The immune complexes were resolved on SDS–PAGE and analyzed by WB using the indicated antibodies. (B) 3T3-L1 adipocytes were transfected with control siRNA or siRNA oligos that deplete Akt1, Akt2, or both Akt isoforms. Four days after transfection, serum-starved cells were mock treated or stimulated with 100 nM insulin for 10 min; 2% of total lysates from a 10-cm plate of 3T3-L1 adipocytes were resolved on SDS–PAGE and subjected to WB analysis with the indicated antibodies. (C) Serum-starved 3T3-L1 adipocytes were pretreated with 100 nM wortmannin (WM) for 1 h before stimulation as in (B); 2% of total lysates from a 10-cm plate of 3T3-L1 adipocytes were resolved on SDS–PAGE and analyzed by WB using the indicated antibodies. (D) 293T cells were transfected with HA-RGC1, FLAG-RGC2, and Myr-Akt ΔPH constructs as indicated. Cells were treated with 100 nM WM for 1 h before lysis. RGC2 phosphorylation levels were determined by WB with the RGC2 phospho-715 antibody. (E) Serum-starved 3T3-L1 adipocytes were pretreated with 100 μM Akti-1/2 for 2.5 h before mock treatment or stimulation with 100 nM insulin for 10 min; 2% of total lysates from a 10-cm plate of 3T3-L1 adipocytes were resolved by SDS–PAGE and analyzed by WB using the indicated antibodies.
To further test this hypothesis, we used an active form of Akt2 in which the lipid-binding PH domain is replaced with a myristoylation sequence (Myr-Akt ΔPH). RGC2 phosphorylation was readily detectable when expressed in 293T cells that possess high endogenous Akt activity and was inhibited after blockade of PI 3-kinase/Akt activity by wortmannin, as confirmed by phospho-Akt blotting (Figure 5D). However, although wortmannin blocked signaling events upstream of Akt, inhibition of RGC phosphorylation with this compound could be bypassed by the expression of Myr-Akt ΔPH, demonstrating that this phosphorylation event is catalyzed directly by Akt (Figure 5D). To further confirm that phosphorylation of RGC2 is dependent on Akt activity, we also used Akti-1/2, an inhibitor with high specificity for Akt kinases by targeting the PH domain (Lindsley et al., 2005; Logie et al., 2007). Treating adipocytes with Akti-1/2 led to inhibition of Akt-dependent phosphorylation events, as detected by phospho-FKHR and phospho-TSC2 WB, whereas Akt-independent phosphorylation events remained intact, as visualized by phosphotyrosine and phospho-Erk blotting (Figure 5E). Interestingly, treatment of 3T3-L1 adipocytes with Akti-1/2 blunted insulin-stimulated phosphorylation of RGC2 at Ser486, Ser696, and Thr715 (Figure 5E). These data suggest that insulin, via activation of Akt2, differentially regulates RGC2 phosphorylation on different sites.
RalA is activated through inhibition of the Ral GAP complex
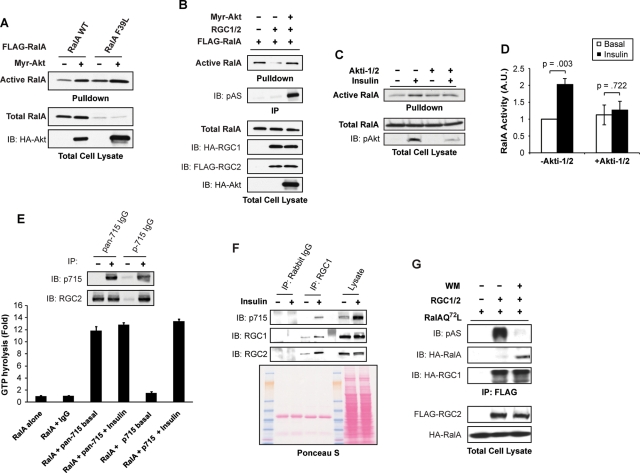
It has been suggested that the GAP activity of TSC2 and AS160 against their target G proteins is blocked by Akt-catalyzed phosphorylation (Inoki et al., 2002; Manning et al., 2002; Sano et al., 2003, 2007; Cai et al., 2006), although the molecular mechanism responsible remains unknown. We postulated that, like TSC2 and AS160, Akt might negatively regulate the inhibitory effect of RGC proteins on RalA. Consistent with this idea, expression of myristoylated, constitutively active Akt (Myr-Akt) produced an increase in wild-type RalA activity (Figure 6A). To determine whether the stimulation of RalA by Akt occurred through regulation of the GAP, we coexpressed Myr-Akt with RalAF39L, a fast-exchanging RalA that is activated independently of GEFs but is still responsive to GAPs (Figure 6A). Because RalAF39L displays high activity, low concentrations of the mutant were expressed with Myr-Akt so that the amount of RalA in the pulldown would remain within the linear range of the assay. Interestingly, expression of Myr-Akt activated RalAF39L, suggesting that Akt can activate RalA by inhibiting its GAPs (Figure 6A). Moreover, coexpression of activated Akt with RGC1/2 alleviated the inhibition of RalA by the GAP complex (Figure 6B). In addition, inhibition of Akt activity by treatment of adipocytes with Akti-1/2 blunted RalA activation by insulin (Figure 6, C and D). Taken together, these data suggest that the RGC1/2 proteins provide a direct link for the activation of the small GTPase RalA by Akt.
FIGURE 6:
Insulin activates RalA by inhibiting the Ral GAP complex. (A) COS-1 cells were transfected with FLAG-RalA, FLAG-RalAF39L, and Myr-Akt-HA as indicated. Cell lysates were subjected to a pulldown assay to detect RalA activation. Pulldowns represent the amount of active RalA from half of a 10-cm plate of COS-1 cells. Lysates represent 2% of total protein from a 10-cm plate. (B) COS-1 cells were transfected with FLAG-RalA, HA-RGC1, FLAG-RGC2, and Myr-Akt-HA constructs as indicated. Cell lysates were subjected to a pulldown assay to detect RalA activation. Pulldowns represent the amount of active RalA from half of a 10-cm plate of COS-1 cells. Lysates represent 2% of total protein from a 10-cm plate. (C) Serum-starved 3T3-L1 adipocytes were pretreated with dimethyl sulfoxide (DMSO) or 100 mM Akti-1/2 for 2.5 h. Cells were then mock treated or stimulated with 100 nM insulin as indicated. Cell lysates were subjected to a pulldown assay to detect RalA activation. Pulldowns represent the amount of active RalA from half of a 10-cm plate of 3T3-L1 adipocytes. Lysates represent 2% of total protein from a 10-cm plate. (D) Quantification of RalA activation in mock treated and Akti-1/2-treated cells in three independent experiments. Significance was determined by Student's t test. (E) Serum-starved 3T3-L1 adipocytes were mock treated or stimulated with 100 nM insulin for 10 min. Total cell lysates from a 10-cm plate of adipocytes were subjected to immunoprecipitation (IP) with the pan-715 antibody or the p715 antibody. The immune complexes were then incubated with 0.5 mg E. coli–expressed recombinant RalA loaded with γ32-GTP for 10 min. Free γ32-phosphate was measured as a reflection of GTP hydrolysis. Top: Western blot of immune complexes with the indicated antibodies. (F) Serum-starved 3T3-L1 adipocytes were mock treated or stimulated with 100 ng/μl of insulin for 10 min. Lysates from a 10-cm plate of cells were subjected to immunoprecipitation with control antibody or RGC1 antibody. The immune complexes were resolved on SDS–PAGE and analyzed by WB using the indicated antibodies. The nitrocellulose membrane was stained with Ponceau S dye to verify equal loading. (G) 293A cells were transfected with HA-RalAQ72L, HA-RGC1, and FLAG-RGC2 as indicated. Cells were treated with DMSO or 100 nM wortmannin for 1 h to decrease active Akt levels, and RGC proteins were immunoprecipitated with an anti-FLAG antibody. Immune complexes were resolved by SDS–PAGE and analyzed by WB with the indicated antibodies.
Previous investigations of GAPs downstream of Akt such as TSC2 or AS160 have suggested that phosphorylation may directly inhibit catalytic activity, though there is little data to definitively evaluate this model. Alternatively, phosphorylation could restrict substrate accessibility of the GAP, therefore leading to the activation of the downstream GTPases. To evaluate whether phosphorylation of RGC2 might directly attenuate its GAP activity, we phosphorylated the immunoprecipitated protein in vitro by incubating with Akt2 kinase and assayed its activity as a GAP for RalA. Akt-catalyzed phosphorylation produced little change in the GAP activity of the RGC immune complex toward recombinant RalA (Figure S5). Because it was possible that our failure to observe a change in GAP activity reflected a low degree of phosphorylation of the complex, we attempted to isolate stoichiometrically phosphorylated GAP complex by immunoprecipitation with the phosphospecific antibody toward the 715 site (p715) on RGC2, with the nonphospho antibody generated from the same epitope (pan-715) as a control (Figure 6E), followed by assay of GAP activity. RGC2 isolated from basal or insulin-stimulated adipocytes by the pan-715 antibody showed little difference in GAP activity, and phosphorylated RGC2 purified by the p715 antibody from insulin-stimulated adipocytes displayed essentially the same GAP activity, offering evidence that phosphorylation may not inhibit RGC2 catalytic capacity per se (Figure 6E).
Despite the fact that phosphorylation did not appear to modulate the catalytic activity of RGC2 in vitro, we noticed that the recognition of the native protein by the RGC1 antibody was increased in cells treated with insulin, as visualized by an increased amount of RGC1 in the immunoprecipitates (Figure 6F, top). This was not due to an increase in the amount of antibody or lysate that was included in the immunoprecipitation reaction (Figure 6F, Ponceau S staining). These data suggest that insulin augments exposure of the antigenic sequence to the RGC1 antibody, raising the possibility that phosphorylation produces a conformational change in the protein complex involving the regulatory subunit. These data prompted us to evaluate the interaction between RalA and the phosphorylated complex. When expressed in 293T cells with high endogenous Akt activity, the RGC had little interaction with RalAQ72L, a mutant capable of stabilizing GAP interactions (Figure 6G). Treating the cells with wortmannin lowered Akt activity and consequently RGC2 phosphorylation but increased the amount of RalA in complex with immunoprecipitated RGC1/2 (Figure 6G), suggesting that phosphorylation led to reduced access of RGC2 GAP to its substrate. Together, these data suggest that insulin activates the protein kinase Akt to directly phosphorylate the RGC, which produces a conformational change in the complex that decreases its access to RalA, resulting in increased activity of the G protein.
RGC proteins regulate GLUT4 trafficking in 3T3-L1 adipocytes
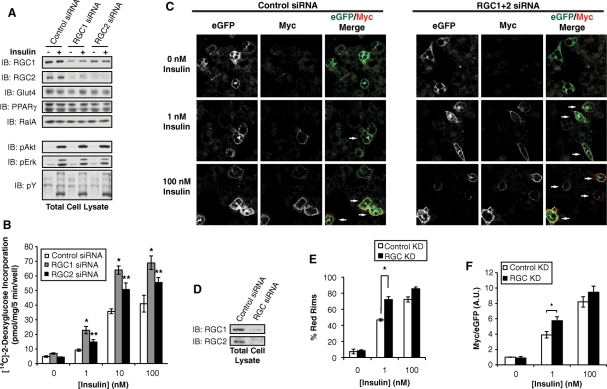
Activation of Akt kinases downstream of PI 3-kinase plays an essential role in insulin-stimulated GLUT4 exocytosis (Whiteman et al., 2002; Watson and Pessin, 2006), and expression of an activated version of the kinase can promote glucose uptake in adipocytes, although to an extent less than that seen with insulin (Kohn et al., 1996; Ng et al., 2008). We and others have reported that the exocyst complex is required for targeting of GLUT4 vesicles to the plasma membrane in 3T3-L1 adipocytes (Inoue et al., 2003; Ewart et al., 2005), a process that requires activation of RalA in a PI 3-kinase–dependent manner (Chen et al., 2007). This led us to speculate that the RGC proteins, which inhibit RalA activity, may function as negative regulators of GLUT4 exocytosis. Consistent with this idea, Lienhard and colleagues found that adipocytes differentiated from fibroblasts stably expressing AS250/RGC2 knockdown shRNA displayed increased GLUT4 exocytosis (Gridley et al., 2006). However, it remains possible that stable knockdown of RGC2 achieved in these experiments may have affected the differentiation states of adipocytes or led to adaptation of cells to the increased activity of Ral GTPases. To avoid these caveats, we acutely depleted RGC1 or RGC2 by siRNA-mediated knockdown in mature 3T3-L1 adipocytes. Loss of RGC2 in adipocytes led to a decrease of RGC1 (Figure 7A), a phenomenon not evident in cells in which RGC2 was stably knocked down (Gridley et al., 2006), further implying a potential compensatory process in these cells. Depletion of RGC1 or RGC2 in mature adipocytes did not affect proximal signaling events, including phosphorylation of insulin receptor, Akt, or Erk, nor were there changes in the expression of adipocyte-specific proteins such as GLUT4 or PPARγ, as determined by WB with the specific antibodies (Figure 7A). Knockdown of RGC1 or RGC2 had little effect on basal glucose uptake despite an increase in basal RalA activity (Figures 2D, 3A, and 7B), perhaps because RalA activation alone is not sufficient for glucose uptake or because the RalA activity observed in an RGC knockdown is not sufficient to increase basal glucose uptake. However, loss of RGC1 or RGC2 caused an increase in glucose uptake after stimulation with submaximal (1 nM) and maximal (100 nM) insulin concentrations when compared with cells transfected with control oligos (Figure 7B). Considering that GLUT1 transporters also contribute to glucose uptake in cultured adipocytes (Liao et al., 2006), and the fact that siRNA cannot completely abolish gene expression, these data suggest that the RGC regulates insulin-stimulated GLUT4 trafficking in adipocytes. Previous studies have shown that knockdown of the Rab GAP AS160 increases basal glucose uptake with little effect on insulin-stimulated glucose transport (Eguez et al., 2005), indicating that AS160 and the RGC proteins may engage different mechanisms to regulate GLUT4 trafficking.
FIGURE 7:
The Ral GAP complex regulates glucose uptake and GLUT4 trafficking in adipocytes. (A) 3T3-L1 adipocytes were transfected with control siRNA or siRNA to deplete RGC1 or RGC2. After 7 d, cells were serum starved for 6 h and then mock treated or stimulated with 100 nM insulin for 15 min; 2% of lysates from a 10-cm dish of cells were subjected to WB analysis with the indicated antibodies. (B) 3T3-L1 adipocytes were transfected with control siRNA or siRNA to deplete RGC1 or RGC2. Seven days after transfection, adipocytes were serum-starved for 6 h and then stimulated with 0, 1, 10, or 100 nM insulin for 30 min. [14C]2-Deoxyglucose uptake was measured as described in Materials and Methods. The data are a representative replicate of at least five independent experiments. Significance was determined by Student's t test. *p value < 0.02 between control and RGC1 knockdown; **p value < 0.02 between control and RGC2 knockdown. (C) Three days after transfection with siRNA to deplete RGC proteins, 3T3-L1 adipocytes were infected with lentivirus expressing Myc-GLUT4-eGFP. Four days after infection, cells were serum starved for 6 h and then stimulated with 0, 1, or 100 nM insulin. The cells were fixed without permeabilization, stained with anti-Myc antibody, and imaged by confocal microscopy. Arrows denote GFP+ cells with Myc rim staining. (D) 3T3-L1 adipocytes were transfected with control siRNA or siRNA to deplete both RGC1 and RGC2. Seven days after transfection, total cell lysates were subjected to WB analysis with the indicated antibodies. (E) Quantification of Myc rim staining. At least 100 cells per condition per experiment were counted for anti-Myc rim staining in at least two separate experiments. Error bars represent ± SEM. Significance was determined by Student's t test. *p value < 0.02. (F) Quantification of ratio of Myc fluorescence and GFP fluorescence. Total eGFP signal or the Myc signal in the outer 15% of the cell was quantified using the CellProfiler program (Broad Institute, Cambridge, MA). At least 100 cells for each condition tested were quantified. The experiment shown is representative of two independent experiments. Error bars represent ± SEM. Significance was determined by Student's t test. *p value < 0.02.
To examine the effect of RGC knockdown on GLUT4 trafficking, 3T3-L1 adipocytes were infected with a lentivirus expressing a Myc-GLUT4-eGFP construct that allows for quantification of fused GLUT4 in nonpermeabilized cells. Cells that were transfected with siRNA to deplete both RGC1 and RGC2 demonstrated a −70–80% knockdown of each protein (Figure 7D). Consistent with the glucose uptake data, cells that were depleted of RGC1/2 by siRNA exhibited a 25% increase of cells with exofacial Myc staining when stimulated with 1 nM insulin and a trend of increased GLUT4 insertion when stimulated with 100 nM insulin (Figure 7, C and E). To quantitatively assess the effect of RGC1/2 knockdown on GLUT4 insertion into the plasma membrane, the amount of Myc rim staining versus total eGFP fluorescence was determined. Cells depleted of RGC1/2 displayed an increase in Myc rim staining versus total eGFP fluorescence when stimulated with 1 nM insulin (Figure 7F). Together these data indicate that loss of the RGC results in increased plasma membrane GLUT4 levels, either by increased exocytosis, decreased endocytosis, or perhaps regulating both endo- and exocytosis of the glucose transporter. Thus, the RGC is a novel component of the insulin signaling pathway that regulates GLUT4 trafficking and glucose uptake in adipocytes.
DISCUSSION
Insulin-stimulated glucose transport is a pivotal physiological process that maintains glucose homeostasis (Saltiel and Kahn, 2001; Watson et al., 2004) and serves as a model for signaling-regulated exocytosis. While it is clear that exocytic trafficking of GLUT4 vesicles is under the control of several insulin signaling pathways (Huang and Czech, 2007), how these signals govern the transport machineries responsible for GLUT4 vesicle trafficking remains uncertain (Hou and Pessin, 2007). Investigations of several small G proteins, and their upstream GAP and GEFs, indicate that these proteins may serve as integrators of signaling and trafficking events (Watson and Pessin, 2006; Welsh et al., 2006; Ishikura et al., 2008). One such pathway is the Akt substrate and Rab GAP AS160/TBC1D4 (Sakamoto and Holman, 2008). Consistent with its proposed action as a GAP that regulates an important G protein (Sano et al., 2003; Miinea et al., 2005), knockdown of AS160 results in a small increase in basal glucose uptake and GLUT4 plasma membrane localization, in a process dependent on its GAP activity (Sano et al., 2007). Although several reports have shown that AS160's GAP activity regulates Rabs 8, 10, and 14 (Ishikura et al., 2007; Sano et al., 2007, 2008), further investigation is required to define the mechanism by which it controls transport machineries.
In addition to the target(s) of AS160, data also support the involvement of the small GTPase RalA in insulin-stimulated glucose transport (Chen et al., 2007). A pool of RalA resides on GLUT4 vesicles in adipocytes and upon its activation interacts with the exocyst complex that directs the exocytic trafficking of GLUT4 vesicles. While RalA was activated by insulin in a PI 3-kinase–dependent manner (Chen et al., 2007), the upstream components involved in this process were not known. Here we identify and characterize RGC1/2, a GAP complex that connects Akt to RalA. RGC1 and RGC2 form a physical and functional complex to inactivate RalA in a manner dependent on the GAP activity of RGC2. In response to insulin, Akt2 directly phosphorylates RGC proteins; this process may lead to a conformational change of the complex that decreases the interaction between RGC and RalA. Loss of RGC proteins causes increased RalA activity and glucose uptake and increased plasma membrane GLUT4 levels in adipocytes. These data support a regulatory role of the RalA/exocyst complex in insulin action and more broadly highlight a novel pathway that integrates insulin signaling with the mobilization of transport machineries for GLUT4 trafficking. During the preparation of this article, Shirakawa et al. (2009) reported the purification of a Ral GAP complex from brain and lung that appears identical to the RGC complex described here. Whether Ral GAP is subject to similar hormonal regulation by phosphorylation in other tissues, or by other protein kinases, will be of interest.
The RGC1/2 complex shares notable similarities with the tumor suppressor complex TSC1/2 that also lies downstream of Akt but functions as a GAP for the Rheb GTPase (Manning and Cantley, 2003; Li et al., 2004b). Both complexes are comprised of a catalytic subunit containing a C-terminal GAP domain and a regulatory/targeting subunit that can stabilize its binding partner. While the Rheb GTPase primarily activates the mTORC1 pathway responsible for regulation of protein synthesis and cell growth (Inoki and Guan, 2006), Ral GTPases mobilize the exocyst complex to direct the transport of cargo proteins and membranes (Novick and Guo, 2002). Efficient relocation of GLUT4 or perhaps other nutrient transporters in various cell types could supply energy for protein synthesis and cell growth. Thus, the activation of these two G proteins by Akt via structurally related GAP complexes suggests a coordinated yet specialized regulation of cellular metabolism that merits further investigation.
How does Akt precisely regulate the GAP activity of RGC proteins? Our data clearly demonstrate that RGC2 is an endogenous substrate of Akt and further that activation of the kinase leads to increased activity of RalA. This resembles the scenario in which Akt negatively regulates the GAP activity of TSC2 toward the small GTPase Rheb, although the exact mechanism underlying this intensively investigated process remains unknown (Huang and Manning, 2009). The similarities between the RGC1/2 and TSC1/2 proteins lead us to speculate that phosphorylation-mediated inhibition of GAP activity could be a universal mechanism to increase the activity of small GTPases. While the composition of the GAP complex is not altered by insulin, our data support a model in which the hormone may induce conformational changes in these proteins that prevent substrate access. Phosphorylation does not render the complex incapable of binding to RalA in vitro, as the complex still exhibits catalytic activity in this setting. Perhaps phosphorylation instead induces a change in localization of the complex that prevents it from binding to RalA. Additional proteins that are recruited to the RGC complex upon its phosphorylation may facilitate the process. Further investigations into the precise mechanisms involved in RGC regulation by phosphorylation are ongoing.
MATERIALS AND METHODS
DNA constructs
The human RGC1 cDNA and RGC2 cDNA were amplified by polymerase chain reaction from total cDNA generated from MCF7 cells. RGC1 cDNA was cloned into a pKH3 vector (Chen et al., 2007). RGC2 cDNA was cloned into a pEGFP vector (Clontech, Mountain View, CA) with the eGFP sequence replaced with a FLAG tag. The RGC2 N/K point mutant was introduced by site-directed mutagenesis. Automatic DNA sequencing was performed at the University of Michigan DNA Sequencing Core (Ann Arbor, MI). The Myr-Akt-HA construct was kindly provided by A. Vojteck (University of Michigan). All other constructs were described previously (Chen et al., 2007).
Antibodies
Rabbit polyclonal antibodies were generated by Pacific Immunology (Ramona, CA). The peptide sequences used for immunization of rabbits are as follows: RGC1 (aa 1433–1460, RRKRLESDSYSPPHVRRK), RGC2 (aa 451–467, AQEDADKLGLSETDSKE), RGC2 pS486 (aa 476–495 KRSSSWGRTYpSFTSAMSRGC), RGC2 pS696 (aa 687–704, KGTAVGRSFpSLSWRSHPD), and RGC2 pT715 (aa 707–724, EPMRFRSApTTSGAPGVEK). Total immunoglobulin G (IgG) fractions were purified from crude serum using a Protein A IgG purification kit (Pierce, Rockford, IL). For pan-antibodies, the IgG fractions were purified using affinity columns conjugated to protein-specific peptides as previously described. For phosphospecific antibodies, the total IgG fraction was first depleted using an affinity column conjugated to a nonphospho form of the antigenic peptide. The flow through was subjected to affinity columns conjugated to the phosphopeptides of specific proteins. Purified antibodies were once more passed through the nonphospho peptide columns to deplete the residual nonphospho antibodies and ensure the recognition with phosphorylated epitope only. Commercially available antibodies are described in the Supplemental Material (see the Experimental Procedures).
Cell culture, transfections, and inhibitors
COS-1 and 293T cells were maintained as previously described (Liu et al., 2005). 3T3-L1 fibroblasts were maintained and differentiated into adipocytes as previously described (Liu et al., 2005). COS-1 cells were transfected with DNA using FuGENE 6 transfection reagent (Roche, Indianapolis, IN), 293T cells were transfected with DNA using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA), and 3T3-L1 adipocytes were transfected with Stealth siRNA (Invitrogen) by electroporation as previously described (Inoue et al., 2006). SiRNA sequences are listed in the Supplemental Material. Cells were treated with 100 nM Wortmannin (Sigma-Aldrich, St. Louis, MO) for 2 h or 1 mM Akti-1/2 (Calbiochem, San Diego, CA) for 2.5 h before lysis.
Tissue lysis
Epididymal fat pads were isolated from 3-mo-old male C57BL/6 mice. Fat pads were cut into four equal-weight pieces and incubated in serum-free DMEM for 30 min at 37°C before stimulation with 100 nM insulin for the indicated times. The fat pads were then lysed by sonication and solubilized in 2X SDS sample buffer.
Immunoprecipitations, pulldowns, and Western blotting
Cells were washed two times with ice-cold phosphate-buffered saline and then lysed on ice for 5 min with 1 ml lysis buffer (100 mM Tris, pH 7.5, 1% NP-40, 130 mM NaCl, 1 mM Na3VO4, 5 mM MgCl2, 1 mM EDTA, and 10 mM NaF) supplemented with Complete, EDTA-free protease inhibitor tablets (Roche). Whole cell lysates were cleared by centrifugation for 10 min at 13,000 × g. Lysates were incubated with 4 μg of the indicated antibody for 2 h at 4°C, and then protein A/G beads (Santa Cruz Biotechnology, Santa Cruz, CA) were added for 2 h at 4°C to precipitate the antibody. Beads were washed three times in lysis buffer and then resuspended in 2X SDS sample buffer.
Expression of GST fusion proteins was induced in Rosetta(DE3)pLysS E. coli (Novagen, Gibbstown, NJ) overnight at 20°C with 0.1 mM IPTG. Proteins were purified as previously described. For GAP assays, the GST fusion proteins were further purified on a Superdex 200 size exclusion column (GE, Piscataway, NJ). For pulldown experiments, GST-RalA bound to glutathione beads was incubated for 30 min at 25°C in loading buffer (20 mM Tris, pH 7.5, 1 mM dithiothreitol (DTT), 50 mM NaCl) supplemented with 2 mM EDTA and Complete, EDTA-free protease inhibitor tablets. GST-RalA was then loaded with nucleotide by incubating in loading buffer supplemented with 2 mM GDP or 200 mM GTPγS for 1 h at 25°C. To stop loading, 10 mM MgCl2 was added for 5 min at 25°C. When indicated, 30 mM AlCl3 and 10 mM NaF were included in the GDP loading buffer to induce GDP/AlFx-bound RalA. Loaded GST-RalA beads were then added to 3T3-L1 adipocytes that were lysed in buffer (25 mM Tris, pH 7.5, 137 mM NaCl, 10% glycerol, 1% NP-40, 5 mM MgCl2, and 1 mM DTT) supplemented with 10 μM GDP; 10 μM GTPγS; or 10 μM GDP, 30 μM AlCl3, and 10 mM NaF, and Complete, EDTA-free protease inhibitor tablets. The reaction was incubated for 2 h at 4°C, and then beads were washed three times in wash buffer (25 mM Tris, pH 7.5, 40 mM NaCl, 30 mM MgCl2, 1% NP-40, and 1 mM DTT) and once in rinse buffer (25 mM Tris, pH 7.5, 40 mM NaCl, 30 mM MgCl2, and 1 mM DTT) supplemented with 10 μM GDP; 10 μM GTPγS; or 10 μM GDP, 30 mM AlCl3, and 10 mM NaF, and Complete EDTA-free protease inhibitor tablets. The pulldowns were solubilized by addition of 2X SDS sample buffer and resolved by SDS–PAGE. The SDS–PAGE gel was either transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA) for WB or silver stained using the SilverQuest Silver Staining Kit (Invitrogen). Silver-stained protein bands were excised from the gel and subjected to tandem mass spectrometry at the Michigan Proteome Consortium (Ann Arbor, MI). RalA activation assays were performed as described previously (Chen et al., 2007). Samples were resolved by SDS–PAGE and then transferred to nitrocellulose membranes. Membranes were incubated with the indicated primary antibody and horseradish peroxidase secondary antibody (Pierce, Rockford, IL). Blots were reacted with ECL Western Blotting Substrate (Pierce).
Kinase assay and GAP assay
In vitro kinase assays were performed in 50 μl of kinase buffer (50 mM MOPS pH 7.2, 50 mM NaCl, 25 mM β-glycerolphosphate, 10 mM MgCl2, 1 mM EGTA, 1 mM NaF, and 100 μM ATP). The reaction was incubated with 100 ng of His-Akt2 S474D (Millipore, Billerica, MA) for 30 min at 32°C. The reaction was stopped by the addition of 2X SDS sample buffer, and proteins were resolved by SDS–PAGE. Phosphorylated proteins were then visualized by WB using the indicated antibodies.
For GAP assays, GST-RalA (final concentration 3 mM) was loaded in 30 μl of load buffer (20 mM Tris, pH 8.0, 5 mM EDTA, 1 mM MgCl2, 50 μM cold GTP, 0.1 mg/ml bovine serum albumin [BSA], and 5 μCi [γ-32P]GTP) for 20 min at 20°C. Loaded GST-RalA was purified from unbound nucleotide using a 0.5-ml Zeba desalt column (Pierce). Loaded GST-RalA (30 μl) was then added to 270 μl of reaction buffer (20 mM Tris, pH 8.0, 2 mM MgCl2, 100 mM NaCl, 0.01 mg/ml BSA, 4 mM DTT, and 1 mM cold GTP) with or without immunoprecipitated RGC2 to start the reaction. Reactions were incubated at 20°C. At the indicated time points, the reaction was terminated by adding 50 μl of the reaction to 750 μl of quench buffer (50 mM NaH2PO4, 5% activated charcoal, pH 2.0) on ice. To monitor hydrolysis, 300 μl of the quench buffer supernatant was quantified by liquid scintillation counting.
[14C]2-deoxyglucose uptake assay and Myc-GLUT4-eGFP assay
3T3-L1 adipocytes were transfected with 1.5 nmol of stealth siRNA per 150-mm plate of cells. Electroporated cells from a 150-mm plate were reseeded into a 12-well dish. Six days after transfection, cells were starved in low-glucose DMEM containing 0.5% fetal bovine serum (FBS) for 6 h and then subjected to a [14C]2-deoxyglucose uptake assay as previously described (Inoue et al., 2006).
3T3-L1 adipocytes were transfected with 1.5 nmol of stealth siRNA per 150-mm plate of cells and then reseeded onto cover slips in a 6-well dish. Three days after electroporation, cells were infected with lentiviral particles containing Myc-GLUT4-eGFP (Chen et al., 2007). Four days after infection, cells were starved for 6 h in low-glucose DMEM containing 0.5% FBS and then stimulated with 0, 1, or 100 nM insulin for 20 min. Cells were fixed in 10% Formalin for 10 min at room temperature, and then fixation was stopped with 100 mM glycine in phosphate-buffered saline (PBS). Cells were incubated in blocking buffer (1% chicken egg serum, 1% BSA, 2% sodium azide in PBS) for 1 h at room temperature before immunostaining with an anti-Myc antibody (1:500 dilution) overnight at 4°C. Cells were incubated with Alexa-Fluor conjugated goat anti-mouse secondary antibody (Molecular Probes, Carlsbad, CA) for 1 h at room temperature and then mounted on glass slides with VectaShield (Vector Laboratories, Burlingame, CA). Confocal images were captured on an inverted Olympus confocal microscope and processed using FluoView version 5.0 (Olympus, Melville, NY). Myc-positive cells were manually counted in a blind experiment.
Supplementary Material
Acknowledgments
We are grateful to Francis Barr and Ben Margolis for helpful discussions and Yong Li and John Tesmer for their advice on the in vitro GAP assay. The work is supported by National Institutes of Health Grants DK-076956 and DK-061618 to A.R.S.
Abbreviations used:
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- GLUT4
glucose transporter-4
- GDP
guanosine diphosphate
- GST
glutathione S-transferase
- GTP
guanosine triphosphate
- IgG
immunoglo-bulin G
- RGC
Ral GAP complex.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-08-0665) on December 9, 2010.
REFERENCES
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Bose A, Guilherme A, Robida SI, Nicoloro SM, Zhou QL, Jiang ZY, Pomerleau DP, Czech MP. Glucose transporter recycling in response to insulin is facilitated by myosin Myo1c. Nature. 2002;420:821–824. doi: 10.1038/nature01246. [DOI] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Cai SL, Tee AR, Short JD, Bergeron JM, Kim J, Shen J, Guo R, Johnson CL, Kiguchi K, Walker CL. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol. 2006;173:279–289. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XW, Leto D, Chiang SH, Wang Q, Saltiel AR. Activation of RalA is required for insulin-stimulated Glut4 trafficking to the plasma membrane via the exocyst and the motor protein Myo1c. Dev Cell. 2007;13:391–404. doi: 10.1016/j.devcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Eguez L, Lee A, Chavez JA, Miinea CP, Kane S, Lienhard GE, McGraw TE. Full intracellular retention of GLUT4 requires AS160 Rab GTPase activating protein. Cell Metab. 2005;2:263–272. doi: 10.1016/j.cmet.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Ewart MA, Clarke M, Kane S, Chamberlain LH, Gould GW. Evidence for a role of the exocyst in insulin-stimulated Glut4 trafficking in 3T3-L1 adipocytes. J Biol Chem. 2005;280:3812–3816. doi: 10.1074/jbc.M409928200. [DOI] [PubMed] [Google Scholar]
- Ferro-Novick S, Novick P. The role of GTP-binding proteins in transport along the exocytic pathway. Annu Rev Cell Biol. 1993;9:575–599. doi: 10.1146/annurev.cb.09.110193.003043. [DOI] [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- Gridley S, Chavez JA, Lane WS, Lienhard GE. Adipocytes contain a novel complex similar to the tuberous sclerosis complex. Cell Signal. 2006;18:1626–1632. doi: 10.1016/j.cellsig.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Hou JC, Pessin JE. Ins (endocytosis) and outs (exocytosis) of GLUT4 trafficking. Curr Opin Cell Biol. 2007;19:466–473. doi: 10.1016/j.ceb.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans. 2009;37:217–222. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Czech MP. The GLUT4 glucose transporter. Cell Metab. 2007;5:237–252. doi: 10.1016/j.cmet.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Inoki K, Guan KL. Complexity of the TOR signaling network. Trends Cell Biol. 2006;16:206–212. doi: 10.1016/j.tcb.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Inoue M, Chang L, Hwang J, Chiang SH, Saltiel AR. The exocyst complex is required for targeting of Glut4 to the plasma membrane by insulin. Nature. 2003;422:629–633. doi: 10.1038/nature01533. [DOI] [PubMed] [Google Scholar]
- Inoue M, Chiang SH, Chang L, Chen XW, Saltiel AR. Compartmentalization of the exocyst complex in lipid rafts controls Glut4 vesicle tethering. Mol Biol Cell. 2006;17:2303–2311. doi: 10.1091/mbc.E06-01-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikura S, Bilan PJ, Klip A. Rabs 8A and 14 are targets of the insulin-regulated Rab-GAP AS160 regulating GLUT4 traffic in muscle cells. Biochem Biophys Res Commun. 2007;353:1074–1079. doi: 10.1016/j.bbrc.2006.12.140. [DOI] [PubMed] [Google Scholar]
- Ishikura S, Koshkina A, Klip A. Small G proteins in insulin action: Rab and Rho families at the crossroads of signal transduction and GLUT4 vesicle traffic. Acta Physiol. 2008;192:61–74. doi: 10.1111/j.1748-1716.2007.01778.x. [DOI] [PubMed] [Google Scholar]
- Kohn AD, Summers SA, Birnbaum MJ, Roth RA. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J Biol Chem. 1996;271:31372–31378. doi: 10.1074/jbc.271.49.31372. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Manning BD. Tuberous sclerosis: a GAP at the crossroads of multiple signaling pathways. Hum Mol Genet. 2005;14(suppl 2):R251–258. doi: 10.1093/hmg/ddi260. [DOI] [PubMed] [Google Scholar]
- Li Y, Corradetti MN, Inoki K, Guan KL. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem Sci. 2004a;29:32–38. doi: 10.1016/j.tibs.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Li Y, Inoki K, Guan KL. Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol Cell Biol. 2004b;24:7965–7975. doi: 10.1128/MCB.24.18.7965-7975.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Nguyen MT, Imamura T, Singer O, Verma IM, Olefsky JM. Lentiviral short hairpin ribonucleic acid-mediated knockdown of GLUT4 in 3T3-L1 adipocytes. Endocrinology. 2006;147:2245–2252. doi: 10.1210/en.2005-1638. [DOI] [PubMed] [Google Scholar]
- Liu J, DeYoung SM, Zhang M, Cheng A, Saltiel AR. Changes in integrin expression during adipocyte differentiation. Cell Metab. 2005;2:165–177. doi: 10.1016/j.cmet.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. Rheb fills a GAP between TSC and TOR. Trends Biochem Sci. 2003;28:573–576. doi: 10.1016/j.tibs.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- McCormick F. Going for the GAP. Curr Biol. 1998;8:R673–R674. doi: 10.1016/s0960-9822(98)70431-2. [DOI] [PubMed] [Google Scholar]
- Miinea CP, Sano H, Kane S, Sano E, Fukuda M, Peranen J, Lane WS, Lienhard GE. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem J. 2005;391:87–93. doi: 10.1042/BJ20050887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng Y, Ramm G, Lopez JA, James DE. Rapid activation of Akt2 is sufficient to stimulate GLUT4 translocation in 3T3-L1 adipocytes. Cell Metab. 2008;7:348–356. doi: 10.1016/j.cmet.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Novick P, Guo W. Ras family therapy: Rab, Rho and Ral talk to the exocyst. Trends Cell Biol. 2002;12:247–249. doi: 10.1016/s0962-8924(02)02293-6. [DOI] [PubMed] [Google Scholar]
- Patel PH, Thapar N, Guo L, Martinez M, Maris J, Gau CL, Lengyel JA, Tamanoi F. Drosophila Rheb GTPase is required for cell cycle progression and cell growth. J Cell Sci. 2003;116:3601–3610. doi: 10.1242/jcs.00661. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Holman GD. Emerging role for AS160/TBC1D4 and TBC1D1 in the regulation of GLUT4 traffic. Am J Physiol Endocrinol Metab. 2008;295:E29–E37. doi: 10.1152/ajpendo.90331.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- Sano H, Eguez L, Teruel MN, Fukuda M, Chuang TD, Chavez JA, Lienhard GE, McGraw TE. Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab. 2007;5:293–303. doi: 10.1016/j.cmet.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Sano H, Kane S, Sano E, Miinea CP, Asara JM, Lane WS, Garner CW, Lienhard GE. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J Biol Chem. 2003;278:14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- Sano H, Roach WG, Peck GR, Fukuda M, Lienhard GE. Rab10 in insulin-stimulated GLUT4 translocation. Biochem J. 2008;411:89–95. doi: 10.1042/BJ20071318. [DOI] [PubMed] [Google Scholar]
- Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Schekman R, Novick P. 23 genes, 23 years later. Cell. 2004;116:S13–S15. doi: 10.1016/s0092-8674(03)00972-3. [DOI] [PubMed] [Google Scholar]
- Shirakawa R, et al. Tuberous sclerosis tumor suppressor complex-like complexes act as GTPase-activating proteins for Ral GTPases. J Biol Chem. 2009;284:21580–21588. doi: 10.1074/jbc.M109.012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- Watson RT, Pessin JE. Bridging the GAP between insulin signaling and GLUT4 translocation. Trends Biochem Sci. 2006;31:215–222. doi: 10.1016/j.tibs.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Watson RT, Kanzaki M, Pessin JE. Regulated membrane trafficking of the insulin-responsive glucose transporter 4 in adipocytes. Endocr Rev. 2004;25:177–204. doi: 10.1210/er.2003-0011. [DOI] [PubMed] [Google Scholar]
- Welsh GI, Hers I, Wherlock M, Tavare JM. Regulation of small GTP-binding proteins by insulin. Biochem Soc Trans. 2006;34:209–212. doi: 10.1042/BST20060209. [DOI] [PubMed] [Google Scholar]
- Whiteman EL, Cho H, Birnbaum MJ. Role of Akt/protein kinase B in metabolism. Trends Endrocrinol Metab. 2002;13:444–451. doi: 10.1016/s1043-2760(02)00662-8. [DOI] [PubMed] [Google Scholar]
- Wittinghofer A. Signaling mechanistics: aluminum fluoride for molecule of the year. Curr Biol. 1997;7:R682–R685. doi: 10.1016/s0960-9822(06)00355-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.