Cand1 can be separated into two functional polypeptides. C-terminal Cand1 binds first at the cullin adaptor site. N-terminal Cand1 blocks the neddylation site subsequently. Defects in the split fungal Cand1 impair development more than defects in CSN.
Abstract
Cand1 inhibits cullin RING ubiquitin ligases by binding unneddylated cullins. The Cand1 N-terminus blocks the cullin neddylation site, whereas the C-terminus inhibits cullin adaptor interaction. These Cand1 binding sites can be separated into two functional polypeptides which bind sequentially. C-terminal Cand1 can directly bind to unneddylated cullins in the nucleus without blocking the neddylation site. The smaller N-terminal Cand1 cannot bind to the cullin neddylation region without C-terminal Cand1. The separation of a single cand1 into two independent genes represents the in vivo situation of the fungus Aspergillus nidulans, where C-terminal Cand1 recruits smaller N-terminal Cand1 in the cytoplasm. Either deletion results in an identical developmental and secondary metabolism phenotype in fungi, which resembles csn mutants deficient in the COP9 signalosome (CSN) deneddylase. We propose a two-step Cand1 binding to unneddylated cullins which initiates at the adaptor binding site and subsequently blocks the neddylation site after CSN has left.
INTRODUCTION
Cullins represent stalk-like eukaryotic scaffold proteins with three repeats of a five-helix bundle. They serve as a platform for the formation of various multisubunit complexes (Zheng et al., 2002b; Wu et al., 2003). The minimal set of a eukaryotic cell comprises the three cullins Cul1, Cul3, and Cul4; humans express seven cullins and additional proteins with a cullin homology domain (Pintard et al., 2004; Petroski and Deshaies, 2005). The cullin C-terminus forms a globular domain and stably interacts with the RING H2 finger protein Roc1/Rbx1/Hrt1 (Kamura et al., 1999; Seol et al., 1999; Tan et al., 1999), which attracts E2 ubiquitin-conjugating enzymes. The cullin RING ligases (CRLs) make up the largest group of E3 ubiquitin ligases. They contain a variable substrate recognition subunit (SRS) and in most cases an adaptor that links the SRS to the complex. The CRLs are the specificity factors for the covalent binding of ubiquitin to substrates. They regulate a wide range of dynamic cellular and developmental responses by triggering 26S proteasome-mediated protein degradation. The largest group of CRLs are Cul1-based Skp1-Cul1-F-box (SCF) protein complexes. The F-box proteins represent the large class of substrate recognition proteins, which are linked to Cul1 by the adaptor protein Skp1 (Feldman et al., 1997; Skowyra et al., 1997).
Activation of CRLs requires neddylation, which is the posttranslational covalent linkage of the ubiquitin-like protein Nedd8 to a conserved C-terminal lysine residue of the cullin (Pan et al., 2004). Neddylation opens the closed conformation of the compact globular domain formed by cullin’s C-terminus and the RING protein. This frees the RING domain from interactions with cullin and enables flexible positioning of the attached E2 for substrate ubiquitination (Duda et al., 2008). Neddylation-induced activation therefore increases the interaction surface between CRLs and charged E2 ubiquitin–conjugating enzyme (Kawakami et al., 2001; Sakata et al., 2007). CRL activity is inhibited in vitro by the COP9 signalosome (CSN), which is a deneddylase removing Nedd8 from CRLs (Cope et al., 2002; Cope and Deshaies, 2003).
Cullin-associated Nedd8-dissociated protein 1 (Cand1) is another in vitro inhibitor of CRLs, which stably binds to unneddylated cullin-RING complexes and colocalizes with cullin mainly in the nucleus (Yogosawa et al., 1996; Zheng et al., 2002a; Oshikawa et al., 2003). Cand1 inhibits the cullin interaction of proteins which, as the DDB1 homologue SAP130, interact to neddylated cullins in vivo (Menon et al., 2008). Cand1 binds to the closed conformation of the cullin-RING complex and the numerous contacts between Cand1’s C-terminus and cullin induce a slightly less curved conformation of cullin’s N-terminal domain. In the crystal structure, Cand1 blocks the Lys720 neddylation site in the catalytic C-terminal part of Cul1 of the Cand1-cullin-RING complex. In addition, the Skp1 adaptor binding site in the Cul1 N-terminal region is blocked by the β-hairpin protrusion of the C-terminal Cand1 (Goldenberg et al., 2004).
Impairment of Cand1 or CSN results in decreased CRL activities in vivo, which is called a CSN paradox because it is in contrast to the observed biochemical CRL inhibition in vitro (Liu et al., 2002; Busch et al., 2003; Feng et al., 2004; Bosu and Kipreos, 2008). This is presumably due to an increased autoubiquitination activity of CRLs that lack substrates. This results in the destabilization of F-box substrate binding proteins. Deneddylation and inhibition by CSN seem necessary for stabilization of CRL subunits, thus promoting CRL activity in vivo (Zheng et al., 2002a; Wee et al., 2005; Cope and Deshaies, 2006; Chew et al., 2007; Dubiel, 2009; Schmidt et al., 2009). The composition of CRLs has been proposed to be regulated by cycles of assembly and disassembly resulting in active neddylated CRLs and inactive unneddylated cullin-RING subcomplexes, respectively. A small fraction of unneddylated subcomplexes is sequestered by Cand1 for stability-independent recycling of CRL substrate recognition proteins (Lo and Hannink, 2006). When a new substrate becomes available, Cand1 can be replaced by Skp1 and another F-box protein for a new round of CRL assembly (Bornstein et al., 2006; Siergiejuk et al., 2009). Replacement of Cand1 by the substrate adaptors might be facilitated by additional factors or the neddylation of Cand1 itself. This was recently observed for the Cand1 homologue Lag2 in baker’s yeast (Siergiejuk et al., 2009). Thus, availability of substrates transfers CRLs from a Cand1 cycle to a CSN cycle, which starts with substrate binding followed by cullin neddylation. The Cand1 cycle thus allows the incorporation of rare adapters into a subset of CRL complexes (Bosu and Kipreos, 2008; Schmidt et al., 2009).
The exchange of different substrate binding proteins owing to changes in substrate levels for CRLs is important during the development of multicellular organisms. CSN dysfunction results in early lethality or developmental defects in animals, plants, and filamentous fungi (Castle and Meinke, 1994; Freilich et al., 1999; Lykke-Andersen et al., 2003; Busch et al., 2007). Cand1 is present during all analyzed stages of development of plants or mice, and plant cand1 mutants show severe defects in fertility, photomorphogenesis, and flowering (Aoki et al., 1999; Yogosawa et al., 1999; Cheng et al., 2004; Chuang et al., 2004).
We show that Cand1 can be split into two functional proteins binding to each other and to cullins. Binding of the C-terminal Cand1 peptide to cullin’s N-terminal adaptor interaction site mediates the binding of the N-terminal Cand1 entity to the neddylation site on cullin’s C-terminus. C-terminal Cand1 can only associate to unneddylatable cullin in vivo. N-terminal Cand1 is unable to interact with any form of cullin without prior binding of the C-terminal part. The separation of a single Cand1 encoding gene corresponds to the in vivo situation of the filamentous fungus Aspergillus nidulans. Deletion of either of the two Cand1 encoding genes in the fungus results in identical phenotypes and an even stronger impact on development than deletion of genes for CSN subunits.
RESULTS
Aspergillus nidulans Cand1 is encoded by two separated genes, candA-N and candA-C
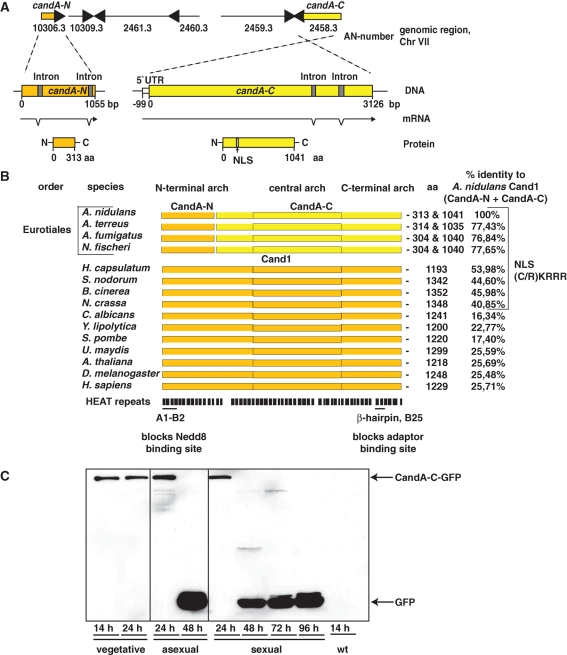
Whereas the crystal structure of the Cand1-cullin complex has been resolved, the molecular process of how the Cand1 protein binds to unneddylated cullins is unknown. The genome of the mold A. nidulans (Galagan et al., 2005) revealed a cand1 homologue divided into candA-N (AN10306.3) encoding the smaller Cand1 N-terminus and candA-C (AN2458.3) for the larger C-terminal part (Figure 1A). Both genes are located on chromosome VII in relative proximity to each other, separated by four open reading frames of conserved hypothetical proteins. The distribution of a split Cand1 protein system in the fungal kingdom is restricted to the Eurotiales including various Aspergilli. Other fungi possess a single cand1 encoding an approximately 1300-amino-acid protein as in plants or animals (Figure 1B). The percentage of identities of A. nidulans CandA to other Cand1 sequences ranges from 77% for Cand1 of other Aspergilli to 26% for human Cand1. The identity of A. nidulans CandA is only 10% to the recently discovered Cand1/Lag2 of Saccharomyces cerevisiae, which differs from the actual Cand1 in sequence and in a much smaller size of only 660 amino acids (Siergiejuk et al., 2009). There is high conservation of both HEAT repeats A1/B2 and the β-hairpin protrusion of the mammalian Cand1 (Goldenberg et al., 2004). A1/B2 is located in the smaller CandA-N of A. nidulans, representing the N-terminal part of the Cand1 protein, and blocks the binding site for the ubiquitin-like modifier Nedd8 on Cul1 in mammalian cells. In CandA-N, residues Asp22 and Asp24 are conserved, corresponding to the two important residues of Cand1, which block the cullin Lys720 neddylation site. The β-hairpin protrusion domain, which partially occupies the adaptor binding site on Cul1, is located in the larger CandA-C corresponding to the C-terminal part of Cand1.
FIGURE 1:
Split Cand1 in A. nidulans and relatives. (A) candA-N (orange) and candA-C (yellow) genomic locus, mRNA, and protein. CandA-C carries NLS (RKRRR) at position 197–201. (B) Cand1/A alignments determined by Clustal W. Amino acid (aa) numbers represent protein lengths. Fungi with conserved Cand1/A NLS are indicated. A1-B2 HEAT repeats block Cul1 neddylation; β-hairpin and B25 impair Skp1 adaptor binding. (C) Expression of CandA-C (AGB266) during vegetative growth and early development. Western analysis of 60 μg of A. nidulans crude extract with functional candA-C::gfp (AGB266) or as control wild-type A4 (wt). Indicated sizes: CandA-C-GFP, 140 kDa; GFP, 27 kDa.
Sequencing of candA-C genomic and cDNA confirmed two introns at the 3′-end of candA-C. The 5′-untranslated region of the transcript was determined and is located 99 base pairs upstream of the start codon corresponding to the first AUG of the mRNA. The deduced sequence of 1041 amino acids results in a 113.5-kDa protein. Genomic and cDNA of candA-N also revealed two introns and a deduced 313-amino-acid protein of 33.6 kDa making up one quarter in length of the combined CandA protein. Therefore, both candA genes of A. nidulans are transcribed and can result in two separate proteins with a combined size of the Cand1 of higher eukaryotes (Figure 1A). The restriction of the split cand1 genes to a small group of organisms suggests that the common ancestor of these filamentous fungi had a single cand1 that has been separated by a recombination event.
Both candA genes are required for fungal development
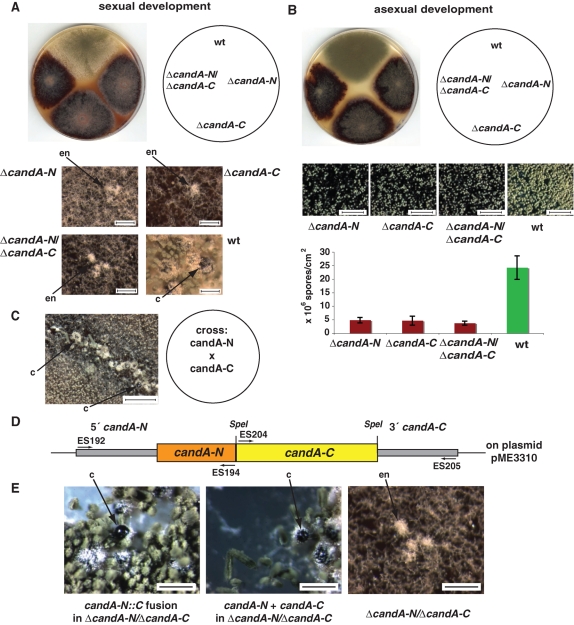
Single-deletion strains of candA-N and candA-C as well as a double-deletion strain candA-N/candA-C were constructed. All phenotypes could be complemented by ectopic integration of the corresponding wild-type genes. The three ΔcandA strains were similar and appeared dark red when grown on an air-medium interface that induces development (Figure 2). The red color was not present in wild type but was reminiscent to various A. nidulans mutants impaired in the CSN (Busch et al., 2003, 2007). A. nidulans induces the sexual cycle in the dark and under oxygen limitation. First, filaments aggregate and form so-called nests. Then primordia are formed, which finally differentiate to closed fruit bodies termed cleistothecia. All ΔcandA strains were able to initiate the cycle but were blocked in the initial stage of early nest formation. Hyphae aggregated to the small white or yellow nest structure including the nursing Hülle cells but did not develop primordia as a prerequisite for sexual fruit bodies (Figure 2A). A. nidulans csn mutants defective in the COP9 signalosome are also blocked in sexual development but are still able to proceed one step further and form primordia (Busch et al., 2003, 2007). The similar but less pronounced phenotype of A. nidulans csn mutants in fungal development suggests a related developmental function (Busch et al., 2003, 2007).
FIGURE 2:
ΔcandA-N and ΔcandA-C mutants impaired in development and secondary metabolism. (A) Sexual development of same strains grown for 6 d on a sealed agar plate in the dark. Close-ups for stop of development of ΔcandA strains at early nests (en) when wild type produces mature fruit bodies (c: cleistothecia). Bar, 200 μm. (B) Asexual development of A. nidulans ΔcandA-N (AGB264) and ΔcandA-C (AGB262), double mutant (AGB268) and wild-type AGB160 (wt) at 37°C for 6 d in light. Conidia shown in the close-up (bar, 600 μm) are quantified with indicated standard deviation (n = 3). (C) Crossing of hyphae of both candA deletion strains results in heterokaryon, which develops mature cleistothecia. Bar, 600 μm. (D) candA-N::C fusion construct (plasmid pME3310) with primer and restriction sites. The candA-N stop codon was replaced by SpeI restriction site. (E) Development of integrated candA-N::C fusion (AGB332) or integrated single genes candA-N and candA-C (AGB331) into the ΔcandA-N/ΔcandA-C deletion strain (AGB268) complementing the deletion phenotype. Arrows indicate mature cleistothecia (left and middle) and early nest structures (right). Bar, 200 μm.
Fungal asexual development is the alternative life cycle and is promoted by light. Asexual differentiation of wild type and the different ΔcandA mutants was induced by illumination in the presence of sufficient oxygen. Asexual spore quantification of all three ΔcandA strains showed a significantly decreased number of asexual conidia compared to the wild-type strain (Figure 2B). In contrast, the A. nidulans csnE (CSN5) mutant impaired in the CSN denedddylase formed similar numbers of asexual spores as the wild type supporting a similar but more prominent role of Cand1 in fungal development (Busch et al., 2003).
We investigated the time points when CandA-N and CandA-C were present. Replacement by gfp::candA-N and candA-C::gfp including the native promoter complemented the phenotypes of the corresponding deletion strains. A single band representing the CandA-C-GFP protein was present during vegetative growth and during early asexual or sexual development (Figure 1C). During later development, CandA-C was unstable, resulting only in the smaller band representing the stable GFP-tag. The GFP-CandA-N fusion only resulted in a stable GFP signal, suggesting that the functional fusion is not very stable. This suggests that at least CandA-C fulfills its function primarily during vegetative growth and at the beginning of asexual or sexual development.
All three candA deletion strains exhibited identical reduction in asexual and block in sexual development. A genetic crossing experiment was performed to examine whether the ΔcandA-N and ΔcandA-C deletions present in different nuclei can complement each other. Hyphae of the ΔcandA-N strain were able to fuse to hyphae of ΔcandA-C strains and to develop primordia within the nests and ultimately mature cleistothecia (Figure 2C). This supports a common function of CandA-N and CandA-C within the cell and suggests a cooperation of both proteins on the molecular level.
candA mutants produce the same pattern of secondary metabolites and separation of CandA-N and CandA-C can be reversed by gene fusion
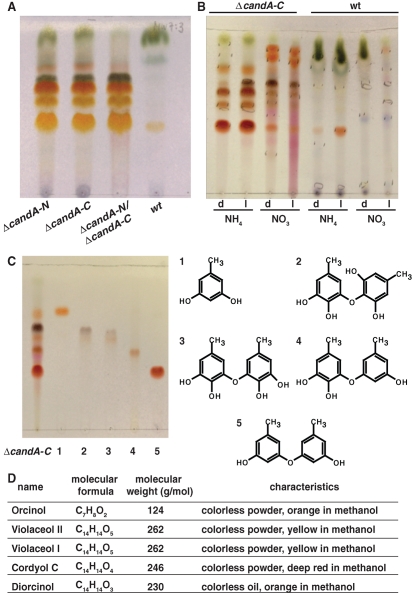
The red color of the ΔcandA-N, ΔcandA-C, and ΔcandA-N/ΔcandA-C mutant strains suggested a similar defect in secondary metabolism. The culture filtrate contained several substances especially with ammonium as a nitrogen source that are only produced in deletion strains but not in wild type (Figures 3A and 3B). The culture filtrate was extracted with ethyl acetate, and the organic phase was concentrated in vacuum and analyzed by thin-layer chromatography and high-performance liquid chromatography/mass spectrometry (HPLC/MS/UV) detection (Figures S1 and S2). Metabolites were isolated by column chromatography, gel chromatography, and HPLC to yield the pure compounds in microscale. The structural elucidation was performed using spectroscopic methods and databases and revealed orcinol, violaceol II, violaceol I, cordyol C, and diorcinol that were isolated from all ΔcandA strains, but not from wild type (Figures 3C and 3D). These data indicate that the lack of CandA-N, CandA-C, or both proteins resulted in an identical change in secondary metabolite production and further supports a common function of both fungal CandA proteins.
FIGURE 3:
candA mutant secondary metabolites. (A) Secondary metabolite production of ΔcandA-N (AGB264), ΔcandA-C (AGB262), ΔcandA-N/ΔcandA-C (AGB268), and wild type (wt, AGB160) grown for 10 d on an NH4 nitrogen source in the dark. (B) Metabolite pattern of ΔcandA-C and wild type (wt) grown in medium containing NH4 or NO3 in the dark (d) or light (l). (C) Crude extract and isolated and identified substances (lane 1–5) (left) and chemical structure (right) 1, Orcinol; 2, Violaceol II; 3, Violaceol I; 4, Cordyol C; and 5, Diorcinol. (A/B) Thin-layer chromatography MeOH:H2O (7:3) developed with anisaldehyde. (D) Summary of substances.
We examined whether CandA of A. nidulans has to be split into CandA-N and CandA-C to fulfill its function. This could reflect a change in molecular mechanism for the split fungal CandA during evolution in comparison to the single Cand1 proteins present in most eukaryotic organisms. A candA‑N::C fusion under the control of the native candA-N promoter and candA-C terminator (Figure 2D) was integrated ectopically into the ΔcandA-N/ΔcandA-C strain. As a control, both candA-N and candA-C genes were reintegrated separately into the double-deletion strain. The candA-N::C fusion strain lost the mutant colony color and grew like the wild type as the strain where the double mutation was complemented by the two separate candA genes. The development of asexual spores and the formation of sexual fruit bodies including mature sexual spores could also be restored (Figure 2E). This shows that both the CandA-N-C fusion protein and the combination of the two CandA proteins fulfill their molecular function in the fungus and support a molecular mechanism for the split fungal CandA similar to that for the single peptide Cand1 of higher eukaryotes.
CandA-N requires CandA-C for nuclear localization and only CandA-C but not CandA-N interacts with cullins in the yeast two-hybrid system
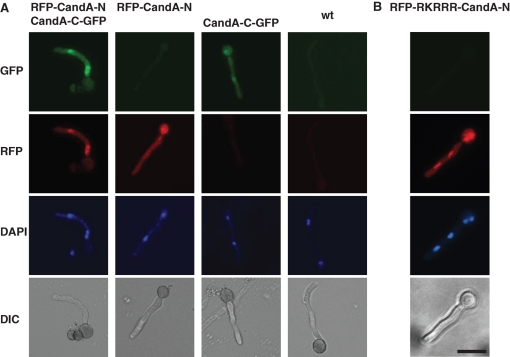
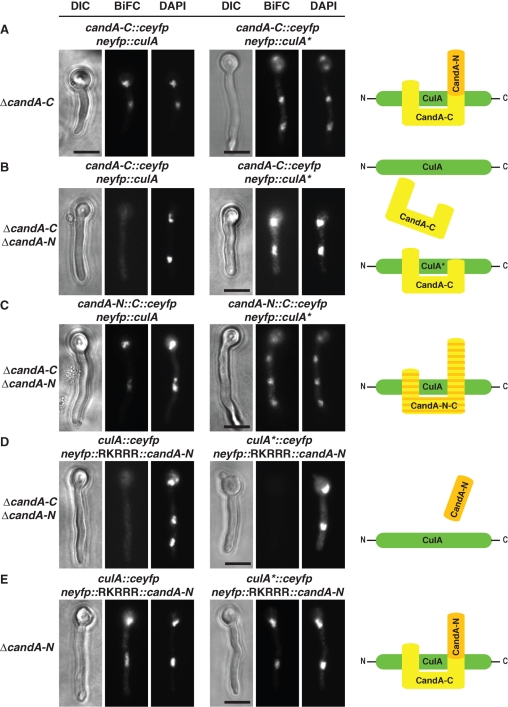
Functional mRFP-CandA-N and CandA-C-GFP fusions under the control of the inducible alcA promoter were expressed in the ΔcandA-N/ΔcandA-C strain (Figure S3A) to examine where both CandA proteins are localized. The strain with both fusions was phenotypically indistinguishable from wild type and both fusion proteins were exclusively detected in nuclei (63% of n = 30; Figure 4A). This suggests functions for both CandA proteins in the nucleus. We further analyzed whether CandA-N depends on CandA-C to be transported into the nucleus, because only CandA-C harbors a predicted nuclear localization sequence (NLS) (RKRRR). Therefore, we expressed mrfp::candA-N or candA-C::gfp2–5 separately in the ΔcandA double-deletion strain. CandA-C-GFP accumulated in the nuclei (64% of n = 62), whereas we never detected RFP-CandA-N in the nucleus (n = 60) (Figure 4A). This suggested that CandA-C is essential for the transport of CandA-N into the nucleus. We fused the RKRRR peptide N-terminally to CandA-N to investigate the importance of the putative NLS of CandA-C for CandA-N. The functional mRFP-RKRRR-CandA-N fusion localized in the nuclei (84% of n = 64; Figure S3B) in the candA-N single-deletion strain. The fusion also localized in the nuclei of the double-deletion strain (83% of n = 66; Figure 4B) but was unable to complement the candA deletion phenotype. This demonstrated that CandA-N cannot fulfill its function in the nucleus without CandA-C.
FIGURE 4:
CandA-N requires CandA-C for nuclear localization. (A) Fluorescence microscopy of RFP-CandA-N and/or CandA-C-GFP fusions expressed from the inducible alcA promoter in a ΔcandA-N/ΔcandA-C strain (AGB268) compared to wild type (AGB152). DAPI signal for nucleus. CandA-C-GFP in the strain lacking CandA-N (AGB386); RFP-CandA-N in a strain lacking CandA-C (AGB385). (B) mRFP-RKRRR-CandA-N (AGB571) from the alcA promoter in a ΔcandA-N/ΔcandA-C strain (AGB268 as control). Bar, 10 μm.
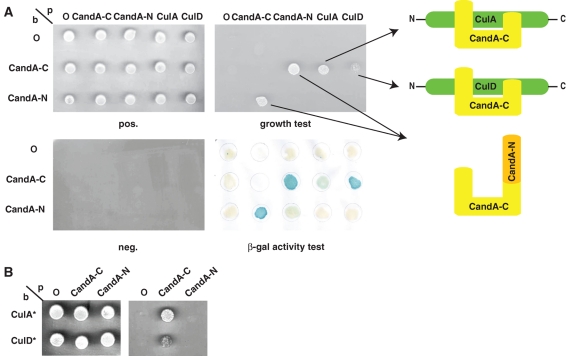
The ability of CandA-N and CandA-C to interact with A. nidulans cullins Cul1/CulA and Cul4/CulD (Busch et al., 2007) was analyzed in the yeast two-hybrid system (Figure 5A). An interaction of CandA-C which contains the protrusion domain that partially occupies the adaptor-binding site on Cul1 was observed with both fungal cullins. In analogy to mammals, CandA-C interacted with neither CsnB/CSN2 nor any other CSN subunit (data not shown). In contrast, CandA-N containing the HEAT repeats that blocks the binding site for Nedd8 did not interact with any cullin. We replaced both putative neddylation sites, Lys710 of CulA and Lys826 of CulD, respectively, by arginine to exclude the possibility that neddylation/rubbylation of A. nidulans cullins impedes the binding of CandA-N to the cullins. These proteins, CulA* and CulD*, did not bind to CandA-N in the yeast two-hybrid assay either (Figure 5B). Thus, our results suggest that the 1041 amino acid–long CandA-C has to mediate the binding of the 313 amino acid–long protein CandA-N into a putative complex consisting of CandA-N, CandA-C, and a cullin.
FIGURE 5:
Yeast two-hybrid interaction of A. nidulans CandAs, CulA, and CulD. (A) Combinations of candA-C, candA-N, culA, and culD cDNAs, cloned into the bait and prey vector, and/or empty bait (b) and prey (p) control vectors (O) were viable (pos) but unable to grow under restrictive conditions (neg). Interaction resulted in leucine prototrophy or specific β-galactosidase activity (β-Gal). 10 μl of liquid cultures (OD546 = 0.01). (B) Interaction of deneddylated CulAK710R (CulA*) and CulDK826R (CulD*) with CandA-N or CandA-C.
CandA-N interacts only in combination with CandA-C with deneddylated cullin in vivo, whereas CandA-C can bind on its own
We investigated CandA-N–CandA-C–cullin complexes in vivo. The C-terminal half of eyfp (ceyfp) was fused to candA-C and the N-terminal half of eyfp (neyfp) to CUL1/culA. These fusions were integrated at the wA locus and controlled by the bidirectional nitrate promoter in a ΔcandA-C as well as a ΔcandA-C/ΔcandA-N background still carrying the wild-type culA because deletion is essential. The candA-C::ceyfp fusion is functional. A bimolecular fluorescence complementation resulted in nuclear signals (79% in n = 63) in the ΔcandA-C strain (Figure 6A). When CandA-N was missing, however, fluorescent nuclei significantly decreased (9% of n = 63), indicating that CandA-C is dependent on CandA-N to bind wild-type CulA in vivo (Figure 6B). The interaction of CandA-C with the CulA* variant,which cannot be neddylated due to the K710R substitution, revealed a different pattern. Again, when CandA-N was present, CandA-C was bound to CulA* (88% of n = 51; Figure 6A), but, even in the absence of CandA-N, CandA-C interacted with CulA* (81% of n = 57; Figure 6B). This result supports that CandA-C can interact with deneddylated cullin but is unable to sequester the deneddylated cullin subpopulation from the cullin pool without CandA-N. As expected, the regenerated CandA-N-C fusion, which simulates the situation in higher eukaryotes, interacts as well to CulA in nuclei (90% of n = 52) as to the CulA* variant (81% of n = 52; Figure 6C). These results further confirm that the molecular mechanism of Cand1 function can tolerate splitting Cand1 into two polypeptide entities, as happened in an ancestor of the Aspergilli. Both proteins act in concert to bind cullins similarly as the single polypeptide chain Cand1.
FIGURE 6:
CandA-C, but not CandA-N, interacts with deneddylated cullin in vivo. (A) Bimolecular fluorescence complementation (BiFC) of CandA-C with wild-type (CulA; AGB556) or deneddylated (CulA*; AGB557) CulA in the presence of intact untagged CandA-N. (B) Same as (A), but in strains that lack CandA-N (CulA: AGB559 and CulA*: AGB560). (C) BiFC with CandA-N-C fusion and both CulA variants in candA double-deletion background (AGB561, AGB562). (D) BiFC using both CulA variants in combination with nuclear CandA-N (AGB570) and in the absence CandA-C (AGB568). (E) Same as (D), but in strains expressing candA-C (CulA: AGB569 and CulA*: AGB567). Bar, 10 μm. Right-hand schemes summarize interactions.
We then examined CandA-N’s ability to bind the two variants of CulA in the presence or absence of CandA-C. The yeast two-hybrid data did not support a CandA-N interaction to cullins, but CandA-N harbors the conserved binding site covering cullin’s neddylation site, which might be important for Cand1 in vivo to discriminate between the unneddylated and neddylated form. We N-terminally fused candA-N to neyfp and furthermore included the sequence encoding the NLS of CandA-C (RKRRR) to overcome the need for CandA-C to enter the nucleus. The functional neyfp::RKRRR::candA-N fusion resulted in signals like the controls (Figure S3C) and could not interact with either CulA or CulA* when CandA-C was missing (n = 60 and n = 60; Figure 6D). CandA-N’s affinity to unneddylated cullin was obviously too low to cause substantial, visible interaction. Complex formation of CandA-N with both variants of CulA was observed in vivo, however, in a combined action of the two CandA proteins when CandA-C was present in the hyphae (Figure 6E; 83% of n = 70 for CulA, 87% of n = 94 for CulA*).
These experiments demonstrate that CandA-N needs CandA-C in vivo not only for transport into the nucleus but also to support its binding to unneddylated cullins. Thus, in vivo binding of the larger CandA-C is a prerequisite for subsequent binding of the smaller CandA-N, which blocks the neddylation site and completes the interaction of CandA to deneddylated cullin.
DISCUSSION
A single Cand1 protein is typical for most eukaryotes, whereas the filamentous fungus A. nidulans and its relatives are an exception. A. nidulans possesses two genes for Cand1, but only the combination of the two genes candA-N and candA-C corresponds to mammalian Cand1. The two open reading frames oppose each other and are separated by four open reading frames. This suggests that an originally fused Cand1 gene was torn apart by DNA rearrangement. Both gene parts had to end up with all signal sequences, and the molecular mechanism of Cand1 function had to operate as a single peptide or as two peptides. The appearance of a split gene in only one taxonomic order suggests a single occasion for the rearrangement during evolution. The ancient recombination event is reversible, because a fused gene derived from the split candA genes is functional. This makes the split fungal CandA an interesting model to explore the molecular Cand1 function.
Deletions of both fungal candA genes resulted in identical developmental phenotypes and include an intense red color. The candA mutants synthesize orsellinic acid derivatives, which are produced when A. nidulans is under stress conditions (Schroeckh et al., 2009). Similar phenotypes including the red color are the result of deletions for genes encoding COP9 subunits (Busch et al., 2003, 2007). This corroborates the cross-talk between CandA and CSN at a developmental level in fungi. Both types of mutants are unable to form sexual fruit bodies, although sexual development stops even earlier in ΔcandA than in Δcsn strains. This is different in Cand1 defective plants, which flower later than wild type, whereas csn mutants stop in early development (Castle and Meinke, 1994; Freilich et al., 1999; Lykke-Andersen et al., 2003; Cheng et al., 2004; Chuang et al., 2004; Feng et al., 2004; Busch et al., 2007). However, none of the described plant mutants corresponded to full deletions (Cheng et al., 2004; Chuang et al., 2004; Feng et al., 2004; Alonso-Peral et al., 2006). A plant deletion strain should elucidate the significance of the csn and cand1 mutant phenotypes between plants and fungi. The stronger developmental impact of fungal candA in comparison to csn deletions might reflect yet unexplored additional deneddylase activities in A. nidulans.
Fungal CandA-C is only present during vegetative growth and at the beginning of development. This might reflect a function for CandA at a specific developmental transition point. Similarly, plant mutants defective in Cand1 are impaired in the vegetative to reproductive growth transition of the primary shoot apical meristem (Cheng et al., 2004; Chuang et al., 2004; Feng et al., 2004; Alonso-Peral et al., 2006). However, Cand1 is expressed in all developmental stages in plants and in mouse embryos (Aoki et al., 1999; Yogosawa et al., 1999). A developmental transition point might have a special need for reorganization of CRLs by Cand1 and CSN assisted cycles of disassembly and assembly (Lyapina et al., 2001; Cope and Deshaies, 2003; Wu et al., 2006; Schmidt et al., 2009; Figure 7). Special developmental stages might require specific regulatory proteins, which have to be substituted by others during subsequent phases of development. Accordingly, in mammalian cells, silencing of Cand1 leads to a stabilization of the cell-cycle-dependent kinase inhibitor p27, whose ubiquitination is SCF dependent (Zheng et al., 2002a). In plant cand1 mutants, the developmental regulator protein Hy5 and the gibberellin hormone pathway repressor RGA are stabilized (Feng et al., 2004). In A. nidulans, the SCF F-box protein GrrA is expressed in late sexual development and is required to form fertile sexual spores (Krappmann et al., 2006). This suggests that a SCFGrrA substrate has to be degraded for complete development and CSN and Cand1 might be necessary for accurate CRL formation.
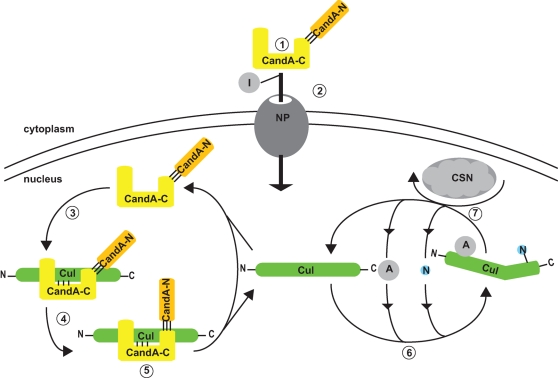
FIGURE 7:
Molecular function of split fungal CandA. 1) CandA-C/N forms heterodimer in the cytoplasm. 2) CandA-C nuclear localization signal interacts with importins (I) for transport through the nuclear pore (NP). 3) CandA-C sites inititate cullin (Cul) interaction, which 4) mediates binding between CandA-N and cullin’s C-terminal domain. 5) This leads to full inhibition of cullin-E3-ligase activity. 6) Release of CandA from cullin allows the formation of new cullin-E3-ligase complexes through recruitment of an adapter protein (A) and subsequent neddylation (N). 7) Removal of Nedd8 through CSN activity destabilizes the cullin complexes and starts the CandA cycle.
Wrapping of Cand1 around the cullin-RING complex covers the adaptor binding and the neddylation site and sequesters them from interacting with other proteins (Goldenberg et al., 2004). The two fungal CandA proteins bind to different sites on cullin and therefore allow a comparison of their cellular function. Alignment with the determined crystal structures suggests that fungal CandA-C can contact the stalk-like N-terminal domain (NTD) of CulA via three major anchoring sites (Goldenberg et al., 2004; Duda et al., 2008). The junction between both CandA proteins colocalizes with the region around Asp267 of Cand1. This is located on the opposite side of cullin’s C-terminal domain (CTD) in comparison to its winged helix binding (WHB) domain, Roc1 and the neddylation site. Thus, CandA-C’s N-terminus presumably forms only a small interaction surface for cullin’s four-helix bundle and α/β domain. Therefore, neddylation should primarily influence binding of CandA-N with its first two HEAT repeats forming the most prominent interactions with and around cullin’s neddylation site. This Cand1 binding site is lost upon neddylation due to opening the conformation of cullin’s CTD and Rbx1. The remainder of CandA-N interacts loosely with cullin’s CTD.
The broad rearrangement of cullin’s CTD and Rbx1 seems to influence the binding of CandA-C only to a minor extent through slightly changed bending of cullin’s NTD. Accordingly, CandA-C binds to cullin in a yeast two-hybrid assay, in contrast to the smaller CandA-N, and has the potential to interact even in the absence of CandA-N. CandA-N can form a heterodimer with CandA-C to be transported into the nucleus. However, the number of binding sites of the six HEAT repeats of CandA-N is not sufficient to mediate a stable interaction to cullins in the absence of the CandA-C partner protein. CandA-N is not even able to interact alone with unneddylatable cullin variants where the described Cand1 binding site of cullin WHB and RING domains is permanent.
The CandA-C/CandA-N heterodimer binds to cullin in the nucleus as stably as a single polypeptide chain in vivo. This requires a sequence of molecular interactions (Figure 7): 1) The two CandA proteins form a heterodimer in the cytoplasm. CandA-N needs CandA-C to enter the nucleus, because only CandA-C harbors a functional NLS. The A. nidulans CandA-C NLS is ancient, as it is conserved not only in split but also in fused Cand1/CandA homologues of distant fungal relatives. 2) CandA-C NLS interaction with importins results in nuclear transport. 3) CandA-C sites inititate cullin interaction, which 4) mediates binding between CandA-N and cullin’s CTD leading to 5) full inhibition of CRL activity.
CandA-C alone can only stably interact with unneddylatable cullin in vivo. Cullin interacts with numerous proteins, and in vivo binding of CandA-C might require that no other proteins with higher affinity compete for cullin. The affinity of CandA-N to CulA is too low to bind CulA alone in either form. The affinity of Cand1 to CUL1 is presumably higher than that of the deneddylating CSN. Therefore, Cand1 enhances the deneddylation reaction by sequestering unneddylated cullin (Min et al., 2005). The CSN binds to cullin’s 4HB and α/β domain via subunits 1, 2, 4, and 5, blocking access of Cand1. The region of amino acids 414 to 600 of Cul1 is important for CSN binding and is exclusively covered by CandA-N. Therefore, CandA-C binding might represent a still unstable intermediate after deneddylation when CSN still occupies CulA. Additional proteins might be involved in the cross-talk between Cand1 and CSN (Bosu and Kipreos, 2008). SAP130/SF3b-3 was identified as a protein that binds to the NTD and CTD of neddylated cullins in vivo and has to be replaced by Cand1 (Menon et al., 2008). A homologue to SAP130 exists in A. nidulans and might prevent CandA-C from binding to neddylatable cullin in vivo.
The stable binding of Cand1 to cullin has to be released to allow a new round of CRL assembly. There could be an asymmetric release to be initiated at either cullin end. Alternatively, CandA-N and CandA-C could be replaced simultaneously. A first release at the adaptor binding site is supported by the finding that adaptors competing with Cand1’s C-terminal part are required for dissociation of Cand1/Lag2 from cullins (Bornstein et al., 2006; Siergiejuk et al., 2009). Alternatively, displacement could start at the neddylation site, resulting in a transient intermediate of Cullin/Rbx1 and CandA-C. A CandA-C-cullin could even be a substrate to neddylation, because cullin can be neddylated while bound to Cand1 (Liu et al., 2002). Removal of the N-terminal part and exposure of the neddylation site could facilitate neddylation but would circumvent the process of adapter acquisition as a prerequisite for CRL function.
Cand1 release could also start with the modification of CandA-N lowering the affinity of the entire CandA to cullin’s CTD so that it can be more easily replaced by an adapter protein. The Cand1-related yeast protein Lag2 is neddylated in vivo when bound to cullin (Siergiejuk et al., 2009). Cand1 could be neddylated or ubiquitinated to trigger the release of the inhibitor. Lag2’s neddylated Lys16 is positioned in close proximity to cullin Lys720 so that neddylation of Lag2/Cand1 instead of cullin might be possible. This lysine is not strictly conserved; CandA-N bears a lysine closer to the N-terminus and it remains to be seen whether the mechanism of Cand1 release requires this residue.
MATERIALS AND METHODS
Cultivation of organisms
E. coli strains were propagated in Luria Bertani medium (1% bacto-tryptone, 0.5% yeast extract, 1% NaCl, pH 7.5) at 37°C. For selection, 100 μg/ml ampicillin, 25 μg/ml chloramphenicol, 20 μg/ml kanamycin, or 25 μg/ml Zeocin (Cayla, Toulouse, France) was used.
S. cerevisiae strains were grown at 30°C under nonselective conditions in yeast extract peptone dextrose (2% pepton, 1% yeast extract, 2% glucose) or under selective conditions in SC medium (0.15% yeast nitrogen base without amino acids, 0.5% (NH4)2SO4, 0.2 mM myo-Inositol, 0.2% amino acid mix containing either 2% glucose or 2% galactose/1% raffinose) supplemented as described (Guthrie and Fink, 1991).
A. nidulans strains were grown at 37°C in or on minimal medium (7 mM KCl, 11.2 mM KH2PO4 [pH 5.5], 2 mM MgSO4, trace elements; Käfer, 1965). As a carbon source, 1% glucose was used. As nitrogen sources, 70 mM NaNO3, 10 mM NaNO3, or 10 mM NH4Cl were added. The medium was supplemented with 4.8 μM pyridoxine HCl and/or 5 mM uridine when required. For plates, 2% agar was added. Selection for the ble marker of Streptoalloteichus hindustanus was performed using 10 μg/ml phleomycine (Cayla). For ptrA (TAKARA BIOKOM, Junki, Poland), 100 ng/ml pyrithiamine (Sigma-Aldrich Chemie GmbH, Munich, Germany) and for transformants containing the nourseothricin (nat) resistance (Goldstein and McCusker, 1999), 100 μg/ml nourseothricin dihydrogen sulfate (clonNAT, Werner BioAgents, Jena, Germany) was added to the medium.
Vegetative mycelia were obtained from submerged liquid cultures, inoculated with 106 spores/ml and grown on a rotary shaker for 14–30 h. For induction of development, 106 spores were spread on agar plates. Asexual development was obtained by incubating the plates in constant white light, whereas sexual development was induced under oxygen-limiting conditions on tape-sealed plates in the dark (Clutterbuck, 1974). Strains were grown on plates covered with cellophane film (Merck Chemicals, Nottingham, UK) when grown for harvesting. Colony growth was recorded as colony diameter with time. Conidiospore quantification (Bussink and Osmani, 1998) was performed as described (Busch et al., 2003).
Transformation procedures
For general cloning procedures, Escherichia coli DH5α [F−, Φ80dΔ(lacZ)M15−1, Δ(lacZYA-argF) U169, recA1, endA1, hsdR17 (rK−, mK+), supE44, λ−, thi1, gyrA96, relA1] (Woodcock et al., 1989) was used. Homologous and ectopic integration of constructs, respectively, was confirmed using PCR and/or Southern hybridization analyses (Southern, 1975). Recombinant DNA technologies were performed according to standard methods (Sambrook et al., 1989). For PCR reactions, Taq (Fermentas GmbH, St. Leon-Rot, Germany), Pfu (Fermentas), Kod (Novagen, Nottingham, UK), or Phusion (Finnzymes OY, Espoo, Finland) polymerases were used. Custom oligonucleotides were ordered from Operon Europe (Cologne, Germany) or Invitrogen GmbH (Karlsruhe, Germany). Restriction enzymes were ordered from Fermentas. 5′RACE was performed using the GeneRacer Kit (Invitrogen) according to the manual.
Transformations of E. coli, S. cerevisiae, and A. nidulans were performed as described (Inoue et al., 1990; Elble, 1992; Eckert et al., 2000). Strains are listed in Table 1; primer sequences and plasmids in Supplemental Tables S1 and S2.
Table 1.:
Aspergillus nidulans strains used and constructed in this study.
| Strain | Genotype | Reference/Construction |
|---|---|---|
| A4 | A. nidulans Glasgow wild type | FGSCa |
| TNO2A3 | pyrG89; pyroA4; ΔnkuA::argB | (Nayak et al., 2006) |
| AGB152 | pyrG89; pyroA4 | (Busch et al., 2003) |
| AGB160 | pyrG89/pyr-4; pyroA4 | (Busch et al., 2003) |
| AGB262 | ΔcandA-C::pyr-4; pyrG89; pyroA4 | pME3115 in AGB152 |
| AGB263 | ΔcandA-C::pyr-4; candA-C::ble; pyrG89; pyroA4 | pME3116 in AGB262 |
| AGB264 | ΔcandA-N::pyr-4; pyrG89; pyroA4 | pME3306 in AGB152 |
| AGB265 | ΔcandA-N::pyr-4; candA-N::ble; pyrG89; pyroA4 | pME3308 in AGB264 |
| AGB266 | candA-C::gfp2–5; pyrG89; pyroA4 | pME3120 in AGB262 |
| AGB268 | ΔcandA-N::pyr-4; ΔcandA-C::ble; pyrG89; pyroA4 | pME3127 in AGB264 |
| AGB331 | ΔcandA-N::pyr-4; candA-N::ptrA; pyrG89; pyroA4 | pME3311 and pME3114 in AGB268 |
| AGB332 | candA-N(p)::candA-N::candA-C::candA-C(t)::ptrA; ΔcandA-N::pyr-4; ΔcandA-C::ble; pyrG89; pyroA4 | pME3310 in AGB268 |
| AGB333 | gfp2–5::candA-N; pyrG89; pyroA4 | pME3490 in AGB264 |
| AGB384 | alcA(p)::mrfp::candA-N::natR; ΔcandA-N::pyr-4; pyrG89; alcA(p)::candA-C::gfp2–5::ptrA; ΔcandA-C::ble; pyroA4 | pME3394 and pME3395 in AGB268 |
| AGB385 | alcA(p)::mrfp::candA-N::natR; ΔcandA-N::pyr-4; ΔcandA-C::ble;pyrG89; pyroA4 | pME3395 in AGB268 |
| AGB386 | alcA(p)::candA-C::gfp2–5::ptrA; ΔcandA-N::pyr-4; ΔcandA-C::ble; pyrG89; pyroA4; | pME3394 in AGB268 |
| AGB553 | ΔcandA-C::ble; pyrG89; pyroA4; ΔnkuA::argB | pME3127 in TNO2A3 |
| AGB554 | ΔcandA-N::natR; ΔcandA-C::ble; pyrG89; pyroA4; ΔnkuA::argB | pME3601 in AGB553 |
| AGB555 | niiA(p)::neyfp::niiA(t); niaD(p)::candA-C::ceyfp::niaD(t); ΔwA; ΔcandA-C::ble; pyrG89/AfpyrG; pyroA4; ΔnkuA::argB | pME3740 in AGB553 |
| AGB556 | niiA(p)::neyfp::culA::niiA(t); pyrG89/AfpyrG; pyroA4; ΔwA; niaD(p)::candA-C::ceyfp::niaD(t); ΔcandA-C::ble; ΔnkuA::argB | pME3741 in AGB553 |
| AGB557 | niiA(p)::neyfp::culAK710R::niiA(t); pyrG89/AfpyrG; pyroA4; niaD(p)::candA-C::ceyfp::niaD(t); ΔcandA-C::ble; ΔwA; ΔnkuA::argB | pME3742 in AGB553 |
| AGB558 | niiA(p)::neyfp::niiA(t); niaD(p)::candA-C::ceyfp::niaD(t); ΔwA; ΔcandA-N::natR; ΔcandA-C::ble; pyrG89/AfpyrG; pyroA4; ΔnkuA::argB | pME3740 in AGB554 |
| AGB559 | niiA(p)::neyfp::culA::niiA(t); pyrG89/AfpyrG; pyroA4; ΔwA; niaD(p)::candA-C::ceyfp::niaD(t); ΔcandA-C::ble; ΔnkuA::argB; ΔcandA-N::natR | pME3741 in AGB554 |
| AGB560 | niiA(p)::neyfp::culAK710R::niiA(t); pyrG89/AfpyrG; pyroA4; niaD(p)::candA-C::ceyfp::niaD(t); ΔcandA-C::ble; ΔwA; ΔnkuA::argB; ΔcandA-N::natR | pME3742 in AGB554 |
| AGB561 | niiA(p)::neyfp::culA::niiA(t); pyrG89/AfpyrG; pyroA4; ΔwA; niaD(p)::candA-N::C::ceyfp::niaD(t); ΔcandA-C::ble; ΔnkuA::argB; ΔcandA-N::natR | pME3743 in AGB554 |
| AGB562 | niiA(p)::neyfp::culAK710R::niiA(t); pyrG89/AfpyrG; pyroA4; niaD(p)::candA-N::C::ceyfp::niaD(t); ΔcandA-C::ble; ΔwA; ΔnkuA::argB; ΔcandA-N::natR | pME3744 in AGB554 |
| AGB563 | niiA(p)::neyfp::niiA(t); pyrG89/AfpyrG; pyroA4; niaD(p)::candA-N::C::ceyfp::niaD(t); ΔcandA-C::ble; ΔwA; ΔnkuA::argB; ΔcandA-N::natR | pME3745 in AGB554 |
| AGB564 | ΔcandA-N::natR; pyrG89; pyroA4; ΔnkuA::argB | pME3601 in TNO2A3 |
| AGB565 | niiA(p)::ceyfp::niiA(t); pyrG89/AfpyrG; pyroA4; niaD(p)::neyfp::RKRRR::candA-N::niaD(t); ΔwA; ΔnkuA::argB; ΔcandA-N::natR | pME3749 inAGB564 |
| AGB566 | niiA(p)::ceyfp::niiA(t); pyrG89/AfpyrG; pyroA4; niaD(p)::neyfp::RKRRR::candA-N::niaD(t); ΔcandA-C::ble; ΔwA; ΔnkuA::argB; ΔcandA-N::natR | pME3749 in AGB554 |
| AGB567 | niiA(p)::culAK710R::ceyfp::niiA(t); pyrG89/AfpyrG; pyroA4; niaD(p)::neyfp::RKRRR::candA-N::niaD(t); ΔwA; ΔnkuA::argB; ΔcandA-N::natR | pME3748 in AGB564 |
| AGB568 | niiA(p)::culAK710R::ceyfp::niiA(t); pyrG89/AfpyrG; pyroA4; niaD(p)::neyfp::RKRRR::candA-N::niaD(t); ΔcandA-C::ble; ΔwA; ΔnkuA::argB; ΔcandA-N::natR | pME3748 in AGB554 |
| AGB569 | niiA(p)::culA::ceyfp::niiA(t); pyrG89/AfpyrG; pyroA4; niaD(p)::neyfp::RKRRR::candA-N::niaD(t); ΔwA; ΔnkuA::argB; ΔcandA-N::natR | pME3747 in AGB564 |
| AGB570 | niiA(p)::culA::ceyfp::niiA(t); pyrG89/AfpyrG; pyroA4; niaD(p)::neyfp::RKRRR::candA-N::niaD(t); ΔcandA-C::ble; ΔwA; ΔnkuA::argB; ΔcandA-N::natR | pME3747 in AGB554 |
| AGB571 | alcA(p)::mrfp::RKRRR::candA-N::his2A(t)::ptrA; pyroA4; pyrG89; ΔcandA-N::pyr-4; ΔcandA-C::ble | pME3750 in AGB268 |
| AGB572 | alcA(p)::mrfp::RKRRR::candA-N::his2A(t)::ptrA; pyroA4; pyrG89; ΔcandA-N::pyr-4 | pME3750 in AGB264 |
a Fungal Genetics Stock Center (University of Missouri, Kansas City, MO).
The A. nidulans BAC library (obtained from Clemson University, Clemson, SC) was screened for candA-C using a gene-specific probe, amplified from genomic wild-type DNA. Probes used for BAC filter hybridization were [α-32P]-dATP labeled with the HexaLabel DNA Labeling Kit (MBI Fermentas GmbH, St. Leon-Rot, Germany) and detection was performed using the BioMaxMS film (Kodak Molecular Imaging, New Haven, CT). A 9.3-kb HpaI fragment from BAC library clone E19, plate 9, containing the complete candA-C coding region was cloned into pBluescript II SK+ (MBI Fermentas GmbH) cut with EcoRV yielding plasmid pME3609.
Plasmid construction
Details are given in the Supplemental Material.
Yeast two-hybrid analyses
The yeast two-hybrid test was described (Golemis and Brent, 1996). Details are given in the Supplemental Material.
Protein isolation and analysis
Protein isolation has been described (Busch et al., 2007). Mouse anti-GFP (Clontech-Takara Bio Europe, Saint-Germain-en-Laye, France), rabbit anti-RFP (Abcam, Cambridge, UK), and mouse anti–α-tubulin (Sigma-Aldrich, St. Louis, MO) antibodies were used as primary antibodies. Horseradish peroxidase-coupled goat anti–mouse IgG (Jackson Immuno Research Laboratories, Westgrove, PA) or goat anti-rabbit IgG (Invitrogen, Eugene, OR), respectively, were used as secondary antibodies.
In silico analyses
Basic Local Alignment Search Tool searches were performed using data from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). Clustal W (http://npsa-pbil.ibcp.fr/) and MultAlin (Corpet, 1988) (http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html) were used for alignments.
DNA isolation and hybridization
Isolation of genomic DNA of A. nidulans was described (Busch et al., 2007). Southern analysis was performed with the Gene Images Random-Prime DNA labeling kit and the Gene Images CDP-Star Detection Kit (GE Healthcare Life Sciences, Munich, Germany). For signal detection, Amersham Hyperfilm ECL was used (GE Healthcare Life Sciences, Munich, Germany).
Microscopy
A Kappa PS30 digital camera and ImageBase software (KAPPA opto-electronics GmbH, Gleichen, Germany) was used in combination with an Olympus SZX12 binocular (Olympus, Hamburg, Germany) or a ZEISS Axiolab (ZEISS AG, Oberkochen, Germany) light microscope. Fluorescence microscopy was conducted as described (Helmstaedt et al., 2008).
Chemical analysis
1H NMR spectra were recorded on Varian Inova 600 and Unity 300 spectrometers, respectively, at 600 and 300 MHz at 298 K (VARIAN Inova, Palo Alto, CA). Chemical shifts in CD3OD are reported as δ values (ppm) relative to CH3OH (δ = 3.30) as an internal reference unless stated otherwise. 13C NMR spectra were recorded at 150.8 MHz. Chemical shifts in CD3OD are reported as δ values relative to CD3OD (δ = 49.0); the multiplicity of the signals was determined by the heteronuclear singular quantum coherence (HSQC; edited) technique. Two-dimensional NMR spectra included H,H-correlated spectroscopy (H,H-COSY; gCOSY pulse sequence), HSQC (gHSQC pulse sequence), and heteronuclear multiple bond connectivity (HMBC; gHMBC pulse sequence). Electron impact MS was used with an ionizing voltage of 70 eV (Finnigan MAT 95). UV spectra were obtained in methanol on a Varian Cary 3E (VARIAN, Palo Alto, CA). Infrared spectra were recorded on a Perkin-Elmer Fourier transform IR 1600 spectrometer as KBr pellets (Perkin-Elmer, Waltham, MA). Solvents for extraction and chromatography were of technical grade and distilled before use. Thin-layer chromatography (TLC) was carried out: type A, on silica gel 60 F254 plates/0.2 mm (Merck, Darmstadt, Germany) using CHCl3:MeOH (9:1) as solvent; or type B, on RP-18 F254S: plates/0.2 mm (Merck) using MeOH:H2O (7:3) as solvent. Silica gel 60/0.040–0.063 mm (Machery & Nagel, Düren, Germany) and Sephadex LH-20 (Pharmacia, Germany) were used for column chromatography. Flash column chromatography was performed using silica gel 60/0.025–0.040 mm (Machery & Nagel). Medium-pressure liquid chromatography was performed with Knauer Wellchrom Maxi-Star K 1000 pumps (Knauer, Berlin, Germany) using a Merck LiChroprep RP-18/0.040–0.063 mm column (B, 310–25). HPLC was carried out on JASCO HPLC systems with PU-2080 Plus or PU-1587 pumps (JASCO, Gross-Umstadt, Germany), respectively, using column A (Macherey & Nagel, Superspher-100 RP-18 endc., 4 μm, 100 × 2 mm, flow rate 3.0 ml min−1), column B (Macherey & Nagel, Superspher-100 RP-18 endc., 4 μm, 100 × 20 mm, flow rate 18.0 ml min−1), or column C (JASCO Kromasil, 100 C-18, 5 μm, 250 × 8 mm, flow rate 2.5 ml min−1). HPLC/MS/diode array detection (DAD) analysis was carried out using Flux Instruments Rheos 4000, PDA detektor (Finnigan Surveyor), and MS-LC-Q detektor (Finnigan) with Xcalibur 1.3 software (Finnigan).
Metabolite analysis and isolation
In general, 2-l minimal medium cultures containing 10 mM NH4CL as a nitrogen source grown in the light or in the dark were used for metabolite analysis and isolation. The culture broth was separated from the mycelium by filtration using a Miracloth filter (Calbiochem, Merck Biosciences, Nottingham, UK). The mycelium was extracted with MeOH:acetone (3:2, 3 × 1 l). The culture filtrate was adjusted to pH = 5.0 and extracted with ethyl acetate (3 × 2 l), and the solvent was removed by evaporation to yield the crude residue. The crude extracts were dissolved each in 1 ml of MeOH and analyzed by TLC. For HPLC/DAD/MS analysis, the crude extracts (5 mg/ml) and pure compounds (1 mg/ml) were dissolved in MeOH. Analytical HPLC was performed to investigate crude extracts, fractions, and pure compounds. Preparative purification of compounds proceeded from subjection to silica gel chromatography (50 × 2 cm, cyclohexane:ethyl acetate:methanol (5:10:1), which yielded diorcinol). Subsequent size exclusion chromatography (100 × 2.5 cm, Sephadex LH-20, acetone) and reverse phase HPLC gave the other compounds. The metabolites orcinol (up to 1.3 mg), diorcinol (up to 16 mg/l), cordyol C (up to 2.4 mg/l), violaceol I (up to 0.8 mg/l), and violaceol II (up to 2.0 mg/l) were obtained as pure compounds. Additionally, fractions with mixtures of known and presumably unknown metabolites were obtained from all chromatographic steps.
HPLC programs included solution A, H2O; solution B, acetonitrile; and solution C, methanol. HPLC columns used were A) JASCO Kromasil 100 C-18, 5 μm, 250 × 8 mm; B) Machery-Nagel Superspher-100, RP-18 endc., 4 μm, 100 × 2 mm; and C) Machery-Nagel Supersphere-100 RP18 endc., 4 μl, 100 × 20 mm. Isolation of orcinol and cordyol was 20% to 100% B in 25 min, 5 min 100% B, 100% B to 20% B in 2 min, 20% B 8 min, column A. The isolation of violaceol I and violaceol II was 20% to 60% B (A, B with 0.05% HCOOH) in 25 min, 60% to 100% B in 2 min, 3 min 100% B, 100% B to 20% B in 5 min, and 20% B 5 min, column C (Further information on the metabolites chemical characteristics are given in the Supplemental Material; HPLC chromatograms of A. nidulans culture extracts are given in Figures S1 and S2.)
Supplementary Material
Acknowledgments
We thank K. Hofmann and Ö. Bayram for helpful discussions, G. Heinrich and A. Herrmann for excellent technical assistance, and S. Krappmann and M. Kress for plasmids. This work was supported by the Deutsche Forschungsgemeinschaft, the VW Vorab, and the Fonds der Chemischen Industrie.
Abbreviations used:
- BiFC
biomolecular fluorescence complementation
- CRL
cullin RING ligase
- CTD
C-terminal domain
- DAD
diode array detection
- HPLC/MS/UV
high-performance liquid chromatography/mass spectrometry/UV
- NTD
N-terminal domain
- WHB
winged helix binding
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-08-0732) on November 30, 2010.
REFERENCES
- Alonso-Peral MM, Candela H, del Pozo JC, Martinez-Laborda A, Ponce MR, Micol JL. The HVE/CAND1 gene is required for the early patterning of leaf venation in Arabidopsis. Development. 2006;133:3755–3766. doi: 10.1242/dev.02554. [DOI] [PubMed] [Google Scholar]
- Aoki T, Okada N, Ishida M, Yogosawa S, Makino Y, Tamura TA. TIP120B: A novel TIP120-family protein that is expressed specifically in muscle tissues. Biochem Biophys Res Commun. 1999;261:911–916. doi: 10.1006/bbrc.1999.1147. [DOI] [PubMed] [Google Scholar]
- Bornstein G, Ganoth D, Hershko A. Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proc Natl Acad Sci USA. 2006;103:11515–11520. doi: 10.1073/pnas.0603921103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: Global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch S, Eckert SE, Krappmann S, Braus GH. The COP9 signalosome is an essential regulator of development in the filamentous fungus Aspergillus nidulans. Mol Microbiol. 2003;49:717–730. doi: 10.1046/j.1365-2958.2003.03612.x. [DOI] [PubMed] [Google Scholar]
- Busch S, Schwier EU, Nahlik K, Bayram O, Helmstaedt K, Draht OW, Krappmann S, Valerius O, Lipscomb WN, Braus GH. An eight-subunit COP9 signalosome with an intact JAMM motif is required for fungal fruit body formation. Proc Natl Acad Sci USA. 2007;104:8089–8094. doi: 10.1073/pnas.0702108104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussink HJ, Osmani SA. A cyclin-dependent kinase family member (PHOA) is required to link developmental fate to environmental conditions in Aspergillus nidulans. EMBO J. 1998;17:3990–4003. doi: 10.1093/emboj/17.14.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle LA, Meinke DW. A FUSCA gene of Arabidopsis encodes a novel protein essential for plant development. Plant Cell. 1994;6:25–41. doi: 10.1105/tpc.6.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis. Plant Physiol. 2004;135:1020–1026. doi: 10.1104/pp.104.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew EH, Poobalasingam T, Hawkey CJ, Hagen T. Characterization of cullin-based E3 ubiquitin ligases in intact mammalian cells—evidence for cullin dimerization. Cell Signal. 2007;19:1071–1080. doi: 10.1016/j.cellsig.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Chuang HW, Zhang W, Gray WM. Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCF(TIR1) ubiquitin ligase. Plant Cell. 2004;16:1883–1897. doi: 10.1105/tpc.021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutterbuck AJ. Aspergillus nidulans. In: Handbook of Genetics. In: King RC, editor. New York: Plenum; 1974. pp. 447–510. [Google Scholar]
- Cope GA, Deshaies RJ. COP9 signalosome: A multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–671. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- Cope GA, Deshaies RJ. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 2006;7:1. doi: 10.1186/1471-2091-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubiel W. Resolving the CSN and CAND1 paradoxes. Mol Cell. 2009;35:547–549. doi: 10.1016/j.molcel.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: Conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert SE, Kübler E, Hoffmann B, Braus GH. The tryptophan synthase-encoding trpB gene of Aspergillus nidulans is regulated by the cross-pathway control system. Mol Gen Genet. 2000;263:867–876. doi: 10.1007/s004380000250. [DOI] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Feng S, Shen Y, Sullivan JA, Rubio V, Xiong Y, Sun TP, Deng XW. Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell. 2004;16:1870–1882. doi: 10.1105/tpc.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freilich S, Oron E, Kapp Y, Nevo-Caspi Y, Orgad S, Segal D, Chamovitz DA. The COP9 signalosome is essential for development of Drosophila melanogaster. Curr Biol. 1999;9:1187–1190. doi: 10.1016/S0960-9822(00)80023-8. [DOI] [PubMed] [Google Scholar]
- Galagan JE, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Golemis E, Brent R. Protein interaction studies. In: Current Protocols in Molecular Biology. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidmann JG, Smith AJ, Struhl K, editors. New York: Wiley; 1996. pp. 429–454. [Google Scholar]
- Guthrie C, Fink G. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- Helmstaedt K, Laubinger K, Vosskuhl K, Bayram O, Busch S, Hoppert M, Valerius O, Seiler S, Braus GH. The nuclear migration protein NUDF/LIS1 forms a complex with NUDC and BNFA at spindle pole bodies. Eukaryot Cell. 2008;7:1041–1052. doi: 10.1128/EC.00071-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- Käfer E. Origins of translocations in Aspergillus nidulans. Genetics. 1965;52:217–232. doi: 10.1093/genetics/52.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Kawakami T, et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20:4003–4012. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann S, Jung N, Medic B, Busch S, Prade RA, Braus GH. The Aspergillus nidulans F-box protein GrrA links SCF activity to meiosis. Mol Microbiol. 2006;61:76–88. doi: 10.1111/j.1365-2958.2006.05215.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell. 2002;10:1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- Lo SC, Hannink M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol Cell Biol. 2006;26:1235–1244. doi: 10.1128/MCB.26.4.1235-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ. Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen K, Schaefer L, Menon S, Deng XW, Miller JB, Wei N. Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol Cell Biol. 2003;23:6790–6797. doi: 10.1128/MCB.23.19.6790-6797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Tsuge T, Dohmae N, Takio K, Wei N. Association of SAP130/SF3b-3 with Cullin-RING ubiquitin ligase complexes and its regulation by the COP9 signalosome. BMC Biochem. 2008;9:1. doi: 10.1186/1471-2091-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min KW, Kwon MJ, Park HS, Park Y, Yoon SK, Yoon JB. CAND1 enhances deneddylation of CUL1 by COP9 signalosome. Biochem Biophys Res Commun. 2005;334:867–874. doi: 10.1016/j.bbrc.2005.06.188. [DOI] [PubMed] [Google Scholar]
- Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshikawa K, Matsumoto M, Yada M, Kamura T, Hatakeyama S, Nakayama KI. Preferential interaction of TIP120A with Cul1 that is not modified by NEDD8 and not associated with Skp1. Biochem Biophys Res Commun. 2003;303:1209–1216. doi: 10.1016/s0006-291x(03)00501-1. [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: Building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pintard L, Willems A, Peter M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 2004;23:1681–1687. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata E, Yamaguchi Y, Miyauchi Y, Iwai K, Chiba T, Saeki Y, Matsuda N, Tanaka K, Kato K. Direct interactions between NEDD8 and ubiquitin E2 conjugating enzymes upregulate cullin-based E3 ligase activity. Nat Struct Mol Biol. 2007;14:167–168. doi: 10.1038/nsmb1191. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. New York: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Schmidt MW, McQuary PR, Wee S, Hofmann K, Wolf DA. F-box-directed CRL complex assembly and regulation by the CSN and CAND1. Mol Cell. 2009;35:586–597. doi: 10.1016/j.molcel.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeckh V, Scherlach K, Nutzmann HW, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siergiejuk E, Scott DC, Schulman BA, Hofmann K, Kurz T, Peter M. Cullin neddylation and substrate-adaptors counteract SCF inhibition by the CAND1-like protein Lag2 in Saccharomyces cerevisiae. EMBO J. 2009;28:3845–3856. doi: 10.1038/emboj.2009.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- Wee S, Geyer RK, Toda T, Wolf DA. CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat Cell Biol. 2005;7:387–391. doi: 10.1038/ncb1241. [DOI] [PubMed] [Google Scholar]
- Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: Destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- Wu JT, Chan YR, Chien CT. Protection of cullin-RING E3 ligases by CSN-UBP12. Trends Cell Biol. 2006;16:362–369. doi: 10.1016/j.tcb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Yogosawa S, Kayukawa K, Kawata T, Makino Y, Inoue S, Okuda A, Muramatsu M, Tamura T. Induced expression, localization, and chromosome mapping of a gene for the TBP-interacting protein 120A. Biochem Biophys Res Commun. 1999;266:123–128. doi: 10.1006/bbrc.1999.1773. [DOI] [PubMed] [Google Scholar]
- Yogosawa S, Makino Y, Yoshida T, Kishimoto T, Muramatsu M, Tamura T. Molecular cloning of a novel 120-kDa TBP-interacting protein. Biochem Biophys Res Commun. 1996;229:612–617. doi: 10.1006/bbrc.1996.1852. [DOI] [PubMed] [Google Scholar]
- Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell. 2002a;10:1519–1526. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002b;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.