It is demonstrated that the Cdase gene encodes for all measurable ceramidase function in Drosophila. BWA, an alkaline ceramidase homologue, does not exhibit ceramidase activity. Bwa genetically interacts with other ceramide-metabolizing enzymes by influencing the flux through the sphingolipid pathway.
Abstract
Ceramidases catalyze the conversion of ceramide to sphingosine. They are acylaminohydrolases that catalyze the deacylation of the amide-linked saturated fatty acid from ceramide to generate sphingosine. They also catalyze the reverse reaction of ceramide biosynthesis using sphingosine and fatty acid. In mammals, different proteins catalyze these reactions while individually exhibiting optimal activity over a narrow pH range and have been accordingly called acid, neutral, and alkaline ceramidases. Several genes encode for variants of alkaline ceramidase in mammals. Brainwashing (Bwa) is the only putative alkaline ceramidase homologue present in Drosophila. In this study we have demonstrated that BWA does not exhibit ceramidase activity and that bwa null mutants display no loss of ceramidase activity. Instead, the neutral ceramidase gene CDase encodes the protein that is responsible for all measurable ceramidase activity in Drosophila. Our studies show strong genetic interaction of Bwa with CDase and the Drosophila ceramide kinase gene (DCERK). We show that, although BWA is unlikely to be a ceramidase, it is a regulator of sphingolipid flux in Drosophila. Bwa exhibits strong genetic interaction with other genes coding for ceramide-metabolizing enzymes. This interaction might partly explain its original identification as a ceramidase.
INTRODUCTION
Most sphingolipids exist as integral components of membranes. Several of them are found to exist in monomeric forms within the cytosolic milieu and in the circulatory system. Membrane sphingolipids, especially those found at the cell surface, can act as antigenic determinants, cell–cell interaction molecules, essential components of glycolipid calyx surrounding the cells, and structural determinants of plasma membrane and perform a wide spectrum of other functions (Karlsson et al., 1968; Merrill et al., 1997; Shayman, 2000). It is believed that several of the sphingolipids are generated within the membrane milieu as second messengers in response to cellular stimuli (Spiegel et al., 1996; Spiegel and Merrill, 1996). Therefore it is not surprising that they have been implicated in a wide array of events ranging from cellular apoptosis to cell division and differentiation. Sphingolipids are hydrophobic compounds derived from the aliphatic amino alcohol sphingosine. Sphingosines are N-acylated to generate ceramides (Merrill et al., 2001). In most eukaryotes, the de novo synthesis of sphingolipids begins with the condensation of serine and a fatty acyl coenzyme A in the endoplasmic reticulum (ER). Reactions leading up to the biosynthesis of ceramide occur in the ER, after which the ceramide is translocated to the Golgi complex, a process that is thought to be mediated mostly by the action of ceramide transfer protein (van Meer and Holthuis, 2000; Hanada et al., 2003; Hanada, 2006). The transfer of ceramide to the Golgi complex ensures the subsequent modification of ceramide at the 2-OH position that results in the generation of complex sphingolipids. Most of the enzymes required for the generation of complex sphingolipids are localized to the post-ER compartments. The variation in the chain length of fatty acids (of sphingosine), n-acylated fatty acids in ceramide, and various substitutions at the 2-OH position are responsible for the existence of more than 300 species of sphingolipids in a mammalian organism (Hannun et al., 2001).
Ceramide is a central component in the sphingolipid metabolic pathway (Hannun and Obeid, 2008). It is a key intermediate in the de novo biosynthetic pathway, first generated in the ER and then transferred to the Golgi complex. Upon its transfer, it is further metabolized in one of many different biosynthetic pathways. It acts as a substrate in the biosynthesis of sphingomyelin, sphingosine and sphingosine 1-phosphate, ceramide 1-phosphate, and a series of glycosphingolipids with increasingly complex carbohydrate substitution at the 2-OH position. It is also an important intermediate in the sphingolipid catabolic pathway. Thus ceramide fulfills the role of a branch point intermediate that could be rerouted through one of several possible metabolic routes. Therefore ceramide can be considered a key component of several regulatory circuits and cycles, such as the sphingomyelin cycle proposed to be operative in the sphingolipid metabolic pathway (Holthuis et al., 2001; Hannun and Obeid, 2008). Among the enzymes that act on ceramide are sphingomyelin synthases, ceramidases, ceramide kinases, and glucosylceramide synthase, which lead to the generation of sphingomyelin, sphingosine, ceramide 1-phosphate, and glucosylceramides, respectively. Ceramide is also a product in the reaction catalyzed by sphingomyelinases, glucocerebrosidase, and so forth. Ceramidases are a class of n-acyl fatty acyl hydrolases that generate fatty acid and sphingosine from ceramide. They are variously classified as acid, neutral, or alkaline ceramidases based on their pH optimum for the ceramidase activity. It is now generally believed that these activities are encoded by separate enzymes (Mao and Obeid, 2008).
Drosophila encodes for a neutral ceramidase homologue, called CDase (Acharya et al., 2003, 2004; Rohrbough et al., 2004; Acharya and Acharya, 2005; Acharya et al., 2008). Using a viable null mutant of Drosophila CDase we now demonstrate that it accounts for all measurable ceramidase activity in the fly. We generated deletion mutants of Brainwashing (Bwa), the putative alkaline ceramidase homologue, and showed that these mutants exhibit no loss in ceramidase activity. Using genetic epistasis experiments and mass spectrometric analysis of sphingolipid metabolites, we show that BWA protein regulates sphingolipid flux and affects ceramide concentrations in the animal without functioning as a ceramidase.
RESULTS
CDase null mutations have no residual ceramidase activity at neutral pH
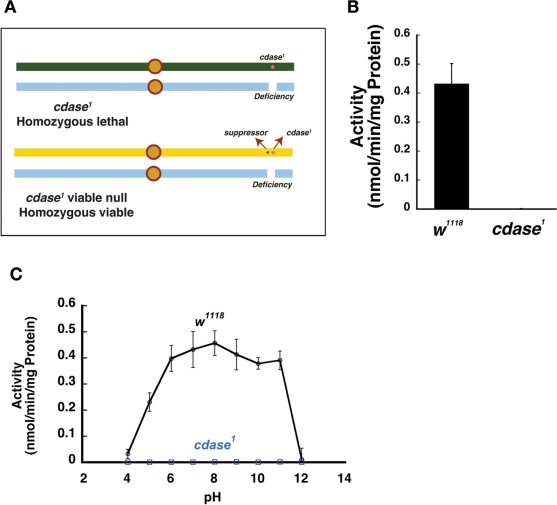
Recently we reported the generation and characterization of a null mutant of Drosophila ceramidase, cdase1 (Acharya et al., 2008). These flies were generated by ethyl methanesulfonate (EMS) mutagenesis and harbor a termination codon at amino acid 641 of the CDase gene. The resulting premature termination led to an extremely unstable protein as examined by Western analysis, and the cdase1 flies are CDase null mutants. The mutant flies were homozygous embryonic lethal (or lethal over a deficiency that uncovers the CDase genomic region as show in Figure 1A) and are rescued by the introduction of a transgene. The presence of a closely linked suppressor mutation on the same chromosome, however, allowed us to recover viable null mutants over the deficiency (suppressor cdase1 or viable null cdase1) (Figure 1A). The identity of this viable suppressor mutation remains unknown. Although the viable null cdase1 flies demonstrated specific phototransduction defects, they were not only viable but also fertile. The availability of adult fertile viable null CDase mutants enabled us to further explore the ceramidase function in Drosophila. Tissue extracts derived from viable null cdase1 mutants demonstrated no residual ceramidase activity at pH 7.0 as described previously (Figure 1B) (Acharya et al., 2008). The control w1118 had a specific activity of 0.43 nmol·min-1·mg-1 protein. The enzymatic activity was restored by the introduction of a genomic copy of the CDase gene into these mutant flies (Acharya et al., 2008). After confirming the lack of neutral ceramidase activity in the cdase1 viable null flies, we decided to undertake biochemical and genetic studies to investigate the presence of other ceramidases and probe the genetic interactions of CDase in Drosophila.
FIGURE 1:
CDase is the major ceramidase active in the pH range 5.0–11.0. (A) A viable null allele of cdase1 was obtained by the presence of suppressor mutation that enabled the recovery of the lethal cdase1 over a deficiency that uncovers the CDase gene region. (B) The viable null allele of cdase1 displays no ceramidase activity. (C) The viable null mutant displays no ceramidase activity in the pH range 5.0–11.0, whereas the wild-type control w1118 demonstrates robust ceramidase activity in that pH range. cdase1 in (B) and (C) refers to the viable null mutant. All activity measurements are an average of at least six measurements, and the error bars denote SD.
CDase accounts for all observable ceramidase activity in Drosophila
Previous studies measuring ceramidase activity in mammalian tissues and tissue culture cells have confirmed the presence of biochemical activity across a wide range of pH values, and the activities at acid and alkaline pH were concluded to be a combined contribution of several ceramidases, each of which demonstrated an optimum enzymatic activity over a narrow pH range (Hassler and Bell, 1993; Merrill et al., 1997; van Echten-Deckert et al. 1997; Luberto and Hannun, 1999). For example, acid ceramidase protein has an optimal pH of ∼4.5 (Li et al., 1999). Alkaline ceramidases demonstrate maximum activity at approximately pH 8.0. Neutral ceramidases (CDase and its homologues) were shown to be active in the neutral pH range. We explored the presence of additional ceramidases that could contribute to the enzymatic activity in Drosophila, then assessed ceramidase activity in tissue extracts from control w1118 and viable null cdase1 mutants prepared in appropriate buffers, the pH values of which ranged from pH 5.0 to pH 11.0. We measured in control w1118 extracts for total enzymatic activity (Figure 1B). We used the viable null cdase1 mutant extracts to measure activity of other ceramidases that could contribute to the enzymatic activity under the experimental conditions. w1118 extracts showed activity across a broad range of pH. Whereas there was minimal activity at pH 4.0, 50% of peak activity was observed at pH 5.0, and activity reached a maximum at approximately pH 8.0. Comparable enzymatic activity was observed even at pH 11.0 before tapering off to zero at approximately pH 12.0 (Figure 1C). Viable null cdase1 mutants, in contrast, showed no enzymatic activity at any of the pH values measured. Also, introduction of two copies of the CDase transgene restored the activity back to the levels observed in w1118 extracts. We have in the past confirmed this activity by overexpressing CDase protein using the Upstream Activating Sequence (UAS)-Gal4 system and driving the expression of CDase using a photoreceptor-specific Gal4 promoter (Acharya et al., 2003). In that study, head extracts were used for the biochemical assays. Also, overexpression of CDase showed increased activity across the whole spectrum of pH from 5.0 to 11.0 (unpublished data). These studies suggest that CDase is the major contributor of biochemical ceramidase activity in Drosophila.
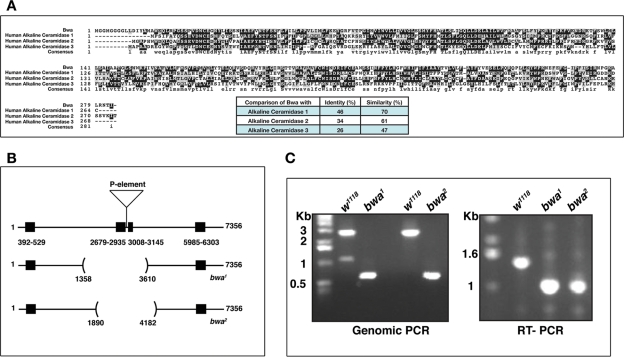
This was a surprising finding because other organisms have more than one protein catalyzing this activity (Hannun et al., 2001). Our results, however, indicate that, in Drosophila, CDase protein shows enzyme activity over a broad pH range, and the results from viable null cdase1 mutants indicates that CDase contributes to most of the observable ceramidase activity. Drosophila does not encode for an acid ceramidase homologue (Yoshimura et al., 2002; Acharya and Acharya, 2005), and a sequence homology search indicated the presence of Bwa as a Drosophila alkaline ceramidase homologue (Boquet et al., 2000; Acharya and Acharya, 2005). Bwa was initially identified as a gene the protein product of which was thought to be involved in the separation of the two hemispheres of the Drosophila brain. A P-element insertion in the intron of this gene was reported as a mutant, and these flies were reported to exhibit fusion of the brain hemispheres. The fusion of the brain was not reliably seen in succeeding generations of the P-insertion flies and was therefore not a true phenotype (Boquet et al., 2000; personal communication). The protein encoded by the Bwa gene shows 46% identity and 70% similarity to the human alkaline ceramidase gene 1 (Figure 2A). We therefore expected BWA protein to demonstrate ceramidase activity as has been reported for members of the alkaline ceramidase family (Mao et al., 2001). Our initial analysis of the cdase1 mutant, however, failed to detect enzymatic activity that could be attributed to the putative alkaline ceramidase homologue, the Bwa gene product. We therefore decided to investigate the role of BWA in ceramidase function in Drosophila in greater detail.
FIGURE 2:
Generation of deletion null mutants of Bwa, the putative alkaline ceramidase homologue in Drosophila. (A) Bwa is the alkaline ceramidase homologue in Drosophila with closest homology to mammalian alkaline ceramidase 1. (B) A diagram demonstrating the extent of genome deletion in the bwa1 and bwa2 null mutants generated by mobilizing the P-element insertion in the second intron of the gene. (C) Left panel – Genomic PCR of the two deletion mutants generated by mobilizing a P-element. RT-PCR analysis confirms the deletion of two exons in the messenger RNA (mRNA) of the mutant flies resulting in truncated mRNA products.
Isolation of a bwa null mutant
The Bwa gene (CG13969) localizes to the left arm of the second chromosome at 38B2–3. The gene spans 7,356 nucleotides and contains four exons (392–529; 2679–2935; 3008–3145; and 5985–6303). The transcript for the gene is expressed abundantly in most stages of development (except in the first and early part of second instar larvae) and in the adults (FlyAtlas; Chintapalli et al., 2007). A previously reported P-element is inserted in the second intron of the gene (Boquet et al., 2000). Reverse transcription–PCR (RT–PCR) of the BWA gene product from these flies yielded full-length transcript, indicating that the P-insertion did not disrupt the gene. We mobilized this P-element and isolated two deletion alleles (Figure 2B). Both bwa alleles delete exons 2 and 3. Whereas bwa1 deletes a 2.358-kb fragment (between nucleotides 1352 and 3610 of the gene), bwa2 deletes a 2.282-kb fragment (between nucleotides 1890 and 4182 of the gene). In both instances, RT–PCR sequencing of the truncated messenger RNAs revealed splicing of the first exon to the fourth resulting in a truncated 47-amino-acid peptide. Thus both the alleles are null for BWA. The initial phenotypic characterization is reported for both mutants, and all biochemical data are from the bwa1 allele.
bwa mutants are viable and show no loss of ceramidase activity
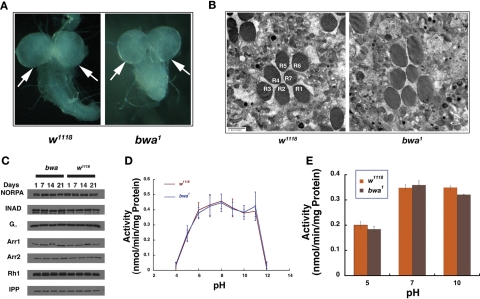
Both bwa mutant alleles resulted in viable and fertile flies that had a normal life span (Supplemental Figure 1A). Because the transcript levels are highest in the male accessory gland, we compared the hatch rate of the mutant embryos to that of control w1118 flies (in Day 3 and Day 10 flies), to examine for fertility defects (FlyAtlas; Chintapalli et al., 2007). The hatch rate of the embryos from both mutant alleles was comparable to wild type at both ages. Also, ∼95% of w1118 embryos reached adulthood, and ∼90% of bwa mutant embryos eclosed as adults, indicating no significant difference in the developmental viability of these mutant flies. We also crossed both 3- and 10-d-old male and female bwa mutant flies to control w1118 female and male flies, respectively. No differences in hatch rates of the embryos were observed in the progeny of these crosses. In addition, as can be seen in Figure 3A, the cerebral hemispheres appeared normal, and no fusion is observed between the two mutant hemispheres in both the alleles, confirming that Bwa gene product is not involved in segregation of the two cerebral hemispheres. Photoreceptors are among one of the most active organs in Drosophila, and mutations in sphingolipid metabolic enzymes, especially cdase1, show degeneration of photoreceptors. Examination of 10-d-old bwa mutant flies showed no obvious degeneration of the photoreceptors as seen in Figure 3B, indicating no endogenous function for the protein in maintaining photoreceptor homeostasis. Control w1118 and bwa mutant head extracts show no difference in the steady-state levels of Rh1, Arr1, Arr2, INAD, NORPA, or Gα, when examined by Western blot analysis (Figure 3C).
FIGURE 3:
Analysis of bwa mutants. (A) bwa null mutants do not display a defect in fusion of the central lobes of the brain. The control and the mutant lobes are both well developed and connected in the middle to other lobes in the developing third instar larvae. (B) Transmission electron micrograph (TEM) comparison of ommatidia from control w1118 and bwa mutant show well-organized rhabdomeres and intact cellular morphology. (C) Phototransduction components are not affected in bwa mutants as observed by Western analysis. 1-, 7-, 14-, and 21-d-old w1118 and bwa fly head extracts were probed for the indicated proteins, and inositol polyphosphate 1-phosphatase (IPP) was used as loading controls. (D) Extracts prepared from bwa mutant display no loss of ceramidase activity across the pH range 5.0–11.0 compared to the control w1118 fly extracts. (E) Extracts prepared from bwa mutant flies show reverse ceramidase activity similar to extracts prepared from control w1118 flies at pH 5.0, 7.0, and 10.0, indicating no loss of reverse ceramidase activity in the mutant flies upon loss of BWA protein.
The human alkaline ceramidase homologue has been reported to have robust ceramidase activity (Mao et al., 2000a, 2000b, 2001, 2003; Xu et al., 2006; Mao and Obeid, 2008), thus we assessed the bwa mutants for loss of ceramidase activity. w1118 was used as a wild-type control, and the activity was compared against viable null cdase1, the null mutant of CDase. As seen in Figure 3D, extracts prepared from bwa mutant did not demonstrate loss of ceramidase activity compared to w1118 extracts from pH 5.0 to 11.0. Viable null cdase1 mutant, in contrast, showed complete loss of ceramidase activity (Figure 1C), indicating that CDase in Drosophila contributes all observable enzymatic activity. The FlyBase records high levels of Bwa transcript in the pupal stages of flies. Therefore we compared ceramidase activity from pupal extracts of wild-type controls and bwa and cdase1 mutants (Supplemental Figure 1B). Pupal extracts showed no loss of ceramidase activity in bwa mutants, and all ceramidase activity could be ascribed to the CDase protein. We also tested the enzymatic activity of extracts from w1118, viable null cdase1, and bwa for reverse ceramidase activity because alkaline ceramidases have been proposed to be efficient reverse ceramidases or acyl-coA–independent ceramide synthases (Mao et al., 2000a, 2000b). In assays performed to measure reverse ceramidase activity, we did not find any difference in activity from control w1118 and bwa mutants at pH 5.0, 7.0, and 10.0, indicating that the BWA did not contribute to any measurable reverse ceramidase activity in Drosophila (Figure 3E).
Sphingolipid profile of bwa mutant and overexpressors
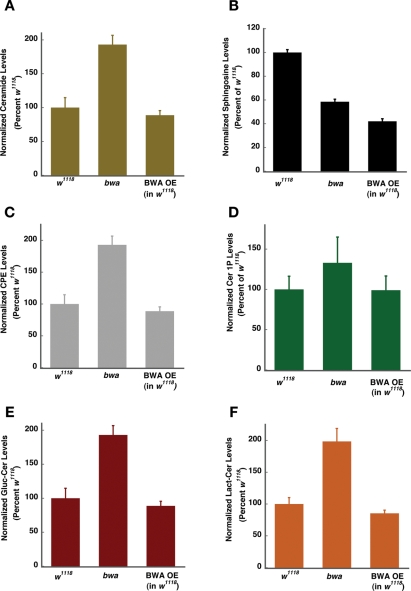
To estimate the sphingolipid content of the control w1118, bwa mutant, and control flies overexpressing the BWA protein, we used ultrafast liquid chromatography coupled with tandem mass spectrometry (UFLC MS/MS) (Table 1). The BWA protein that is overexpressed is functional because it rescues the lethality of the cdase1 mutant and thus would provide the best control to evaluate the opposite effects of loss of BWA protein. As can be seen in Figure 4, A–F, the loss of BWA protein results in accumulation of ceramide, ceramide phosphorylethanolamine (CPE), ceramide 1-phosphate, glucosylceramide, and lactosylceramide. We also observe a significant reduction in sphingosine levels (Figure 4B). The increased ceramide and decreased sphingosine levels in the bwa mutant could arise from a ceramidase-like function of BWA. If BWA were indeed a ceramidase, we would expect that the overexpressor flies would have lower levels of ceramide and a corresponding increase in sphingosine levels compared to controls. Our data reveal that BWA overexpressor extracts show little decrease in the steady-state levels of ceramide. Instead, they show a substantial decrease in the levels of sphingosine, a finding not expected if BWA is a ceramidase (Figure 4, A–F). It is worth noting that sphingosine is not only generated by deacylation of ceramide to sphingosine by ceramidase, but is also an intermediate in the ceramide biosynthetic pathway. The decreased sphingosine observed in the bwa mutant could also result from increased conversion of the sphingosine generated in the de novo biosynthetic pathway into other metabolites such as CPE, ceramide 1-phosphate, lactosylceramide, and glucosylceramide.
Table 1.:
The conditions of SRM for the quantification of sphingosine and ceramide species.
| Compound | Precursor | Product | CV | CE |
|---|---|---|---|---|
| Sphingosine | ||||
| d14:1 | 244.2 | 208.2 | 15 | 15 |
| d16:1 | 272.3 | 236.3 | 15 | 15 |
| d17:1 | 286.3 | 268.2 | 15 | 15 |
| Ceramide | ||||
| d14:1/16:0 | 482.5 | 208.3 | 25 | 25 |
| d14:1/18:1 | 508.5 | 208.3 | 30 | 25 |
| d14:1/18:0 | 510.5 | 208.3 | 30 | 25 |
| d14:1/20:0 | 538.5 | 208.3 | 35 | 30 |
| d14:1/22:0 | 566.5 | 208.3 | 35 | 30 |
| d14:1/24:1 | 592.6 | 208.3 | 40 | 30 |
| d14:1/24:0 | 594.6 | 208.3 | 40 | 30 |
| d16:1/14:0 | 482.5 | 236.3 | 25 | 25 |
| d16:1/16:0 | 510.5 | 236.3 | 25 | 25 |
| d16:1/18:0 | 538.5 | 236.3 | 30 | 25 |
| d16:1/18:1 | 536.5 | 236.3 | 30 | 25 |
| d16:1/20:0 | 566.5 | 236.3 | 35 | 30 |
| d16:1/22:0 | 594.6 | 236.3 | 35 | 30 |
| d16:1/24:0 | 622.6 | 236.3 | 40 | 30 |
| d16:1/24:1 | 620.6 | 236.3 | 40 | 30 |
| d18:1/12:0 | 482.6 | 264.2 | 25 | 25 |
| d18:1/25:0 | 664.8 | 646.7 | 35 | 15 |
| Ceramide 1-phosphate | ||||
| d14:1/14:0 | 534.4 | 208.3 | 30 | 25 |
| d14:1/16:0 | 562.4 | 208.3 | 30 | 25 |
| d14:1/18:1 | 588.5 | 208.3 | 30 | 25 |
| d14:1/18:0 | 590.5 | 208.3 | 30 | 25 |
| d14:1/20:0 | 618.5 | 208.3 | 35 | 25 |
| d14:1/22:0 | 646.5 | 208.3 | 35 | 30 |
| d14:1/24:1 | 672.5 | 208.3 | 40 | 30 |
| d14:1/24:0 | 674.5 | 208.3 | 40 | 30 |
| d16:1/16:0 | 590.5 | 236.3 | 30 | 25 |
| d16:1/18:0 | 618.5 | 236.3 | 30 | 25 |
| d16:1/18:1 | 616.5 | 236.3 | 30 | 25 |
| d16:1/20:0 | 646.5 | 236.3 | 30 | 25 |
| d16:1/22:0 | 674.5 | 236.3 | 35 | 25 |
| d16:1/24:0 | 702.6 | 236.3 | 35 | 30 |
| d16:1/24:1 | 700.6 | 236.3 | 40 | 30 |
| d18:1/12:0 | 562.5 | 264.5 | 30 | 25 |
| Phosphoethanolamine ceramide | ||||
| d14:1/18:0 | 633.5 | 208.3 | 35 | 30 |
| d14:1/20:0 | 661.5 | 208.3 | 35 | 30 |
| d16:1/18:0 | 661.5 | 236.3 | 35 | 30 |
| d14:1/22:0 | 689.6 | 208.3 | 40 | 30 |
| d16:1/20:0 | 689.6 | 236.3 | 35 | 30 |
| d16:1/22:0 | 717.6 | 236.3 | 35 | 30 |
| Glucosyl ceramide | ||||
| d14:1/18:0 | 672.5 | 208.3 | 35 | 30 |
| d14:1/20:0 | 700.6 | 208.3 | 35 | 30 |
| d16:1/18:0 | 700.6 | 236.3 | 40 | 30 |
| d14:1/22:0 | 728.6 | 208.3 | 40 | 30 |
| d16:1/20:0 | 728.6 | 236.3 | 40 | 30 |
| d16:1/22:0 | 756.6 | 236.3 | 40 | 30 |
| d18:1/12:0 | 644.7 | 264.5 | 35 | 30 |
| Lactosyl ceramide | ||||
| d14:1/18:0 | 834.6 | 208.3 | 40 | 30 |
| d14:1/20:0 | 862.6 | 208.3 | 40 | 30 |
| d16:1/18:0 | 862.6 | 236.3 | 40 | 30 |
| d14:1/22:0 | 890.7 | 208.3 | 40 | 30 |
| d16:1/20:0 | 890.7 | 236.3 | 40 | 30 |
| d16:1/22:0 | 918.7 | 236.3 | 45 | 30 |
| d18:1/12:0 | 806.7 | 264.4 | 35 | 35 |
FIGURE 4:
bwa mutants show a general increase in levels of sphingolipid metabolites. (A) Mutant extracts show an approximate twofold increase in ceramide, and the overexpressor extracts do not show a decrease in ceramide levels compared to extracts prepared from control w1118 flies. (B) Lipid extracts prepared from mutant flies show a twofold decrease in sphingosine compared to control, but overexpressor flies show a further decrease in the levels of sphingosine. (C) Mutants show a near twofold increase in the steady-state levels of CPE, whereas the overexpressors show a marginal decrease. (D) Ceramide 1-phosphate (Cer 1P) levels are slightly increased in the mutants and not changed in the overexpressors. (E) Glucosylceramide is increased in the mutant by approximately twofold and minimally affected in the overexpressor flies compared to the control. (F) Lactosylceramide is increased in the mutant animals and not substantially changed in the overexpressor flies.
In summary, extracts from bwa mutants do not show decreased ceramidase activity, overexpression of BWA does not increase the ceramidase activity, and increased expression of BWA does not increase the endogenous sphingosine levels. In bwa mutants, we see a uniform increase in production of a number of metabolites, including ceramide, CPE, ceramide 1-phosphate, glucosylceramide, and lactosylceramide. In contrast, overexpression of the protein does not greatly reduce the steady-state levels of any metabolites except sphingosine, the levels of which are decreased compared to those in wild type and bwa mutants. Thus our data support the hypothesis that BWA is more a regulator of the flux through the sphingolipid biosynthetic pathway than is a ceramidase.
Genetic interaction of Bwa with CDase
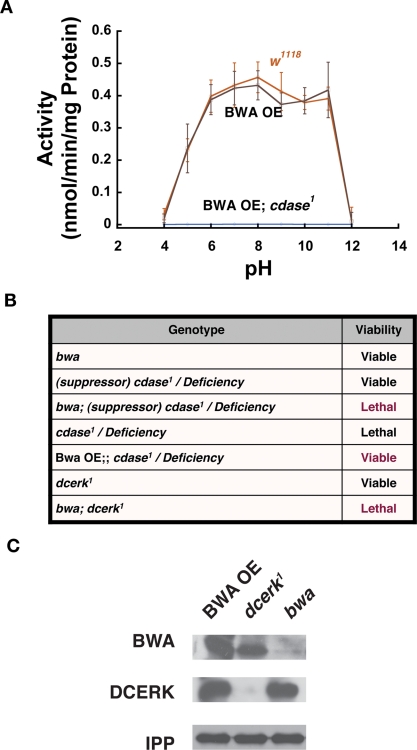
Bwa encodes for a 283-amino-acid protein. We have failed to detect the endogenous BWA protein from wild-type controls in Western blots, and we have raised several antibodies against BWA. These antibodies failed to recognize an endogenous band at the appropriate molecular weight on Western blots of wild-type controls that was missing in bwa mutants. Our results indicated that either the protein was unstable or there was limited expression of the protein in these extracts. The latter scenario is unlikely, however, because a large amount of the transcript has been detected in head and intestine of the flies. We therefore used a gain-of-function strategy to evaluate the protein function in vivo. We overexpressed BWA using the UAS-Gal4 system. This strategy enabled the production and detection of substantially large amounts of the protein at the expected molecular weight (∼30 kDa). The protein was visible on Western analysis of whole fly extracts, when ubiquitously expressed using an Actin-Gal4 driver or in head extracts when BWA was expressed in the photoreceptors using Gmr-Gal4 (an eye-specific Gal4 driver). We examined tissue extracts from BWA overexpressor flies for additional or new ceramidase activity as compared to wild-type control flies. As can be seen in Figure 5A, targeted overexpression of BWA in w1118 controls exhibited the same level of activity as control w1118 flies that were not expressing the protein. In fact, overexpression of the BWA protein does not show an increase in ceramidase activity over the entire pH range from 5.0 to 11.0. These results indicate that BWA protein exhibited no demonstrable ceramidase activity in Drosophila tissue extracts. The extracts from these flies were also compared for reverse ceramidase activity, and no difference was observed (unpublished data).
FIGURE 5:
Bwa genetically interacts with other ceramide-metabolizing flies. (A) BWA overexpressor flies do not show an increase in ceramidase activity at any of the pH over the range 5.0–11.0 when expressed in w1118 control background. Alternatively, cdase1 lethal mutants rescued by overexpression of BWA show no activity in any of the pH over the range 5.0–11.0. The experiments described here and in Figure 3C were all done together. Thus the w1118 activities are the same. All activity measurements are an average of at least six measurements, and the error bars denote SD. (B) Genetic interactions of bwa mutants and BWA overexpressors with CDase and DCERK. bwa and viable null cdase1 allele (denoted here as suppressor, cdase1, for clarity) are individually viable, but the bwa; viable cdase1 (suppressor, cdase1) mutants are homozygous lethal. Similarly, dcerk1 is viable; however, bwa; dcerk1 double mutants are homozygous lethal. The cdase1 allele in the absence of the suppressor mutation is homozygous lethal or lethal over a deficiency. Overexpression of BWA in this background rescues the lethality of cdase1. (C) dcerk1 mutants show an enormous increase in the level and stabilization of BWA protein in their extracts. The first lane contains extracts from BWA overexpressor, the second lane from dcerk1 mutant, and the third lane from the bwa mutant. Equivalent extracts were probed for DCERK, BWA, and also for IPP as loading control. As can be observed, there is increased production/stabilization of BWA protein in dcerk1 mutant. The faint band seen in bwa mutant extract probed for BWA is a nonspecific band.
Because the bwa mutants were viable and fertile and show no other obvious phenotype or compromise in basic function, we probed for genetic links between Bwa and other genes the products of which act on ceramide in Drosophila. Extracts from bwa mutants show an increase in the level of ceramide and other sphingolipids and thus might show epistatic interaction with other ceramide-metabolizing enzymes. CDase converts ceramide to sphingosine, and Drosophila ceramide kinase (DCERK) converts ceramide to ceramide kinase. We have generated null mutants for CDase (cdase1) and a severely hypomorphic allele for ceramide kinase (dcerk1). cdase1 mutants in isolation are homozygous lethal, and dcerk1 mutants are viable but have destabilization of NORPA (eye-specific phospholipase C) in photoreceptors, and retinal cells undergo degeneration (Acharya et al., 2008; Dasgupta et al., 2009). We undertook genetic interaction studies of Bwa with these two genes because both mutants result in accumulation of ceramide. As described earlier, a closely linked unknown suppressor mutation allowed us to recover viable null mutants of cdase1 over a deficiency that uncovers this region (Figure 5B). When the EMS-mutagenized chromosome was backcrossed to wild-type chromosomes to get rid of the suppressor and other incidental mutations (this process is necessary to outcross all other mutations induced by EMS treatment of the chromosome), then cdase1 was homozygous lethal and lethal over the deficiency. We used both these mutant cdase “alleles” in our genetic interaction studies with bwa mutants. We first tested the effect of bwa null allele in the viable suppressor, cdase1/deficiency (viable null) combination. We failed to recover any viable progeny that was homozygous for bwa1 and suppressor, cdase1/deficiency (Figure 5B). Thus the double mutants of bwa1; suppressor, cdase1/deficiency was lethal even in the presence of the suppressor mutation. This result implied a genetic interaction between Bwa and CDase. Thus BWA was playing an essential role in the survival of ceramidase mutant flies in the suppressor genetic background. In parallel experiments, we also expressed BWA ubiquitously using an Act-Gal4 (Gal4 driven under the control of the actin promoter). The overexpression of BWA was sufficient to render even the lethal version (cleaned) of cdase1 viable. Thus targeted overexpression of BWA was capable of rescuing the lethality of cdase1 mutant. These experiments suggest a strong genetic interaction between Bwa and CDase and implicate them in essential overlapping functions in the fly.
Ceramide kinase converts ceramide to ceramide 1-phosphate. We had recently shown that ceramide kinase mutant dcerk1 accumulated ceramide. We therefore decided to test whether Bwa genetically interacted with ceramide kinase mutants. dcerk1 is a severely hypomorphic mutant of DCERK. It was generated by the mobilization of a P-element situated in the promoter region of the DCERK gene (Dasgupta et al., 2009). These mutants are viable and have decreased fertility, but they can be maintained as a homozygous stock at room temperature. Introduction of bwa mutant allele into the dcerk1 mutants results in lethality (Figure 5B). Thus bwa; dcerk1 fail to develop into adult flies, indicating a requirement for BWA protein in the survival of dcerk1 mutant flies. We also observed that BWA protein, which we fail to observe in wild type and other backgrounds, is greatly stabilized and increased in the dcerk1 single mutant flies to levels attained in the photoreceptors of flies expressing Gmr-Gal4–driven UAS-Bwa (Figure 5C).
These studies indicate a strong genetic interaction among Bwa, CDase, and DCERK, and the gain-of-function studies also indicate that transgenic overexpression of BWA results in a functional protein. The rescue of cdase1 lethality provided us with a genetic background to investigate the biochemical activity of BWA in a background bereft of the predominant source of ceramidase activity. We therefore performed ceramidase assay from pH 5.0 to 11.0 in cdase1 mutants rescued by BWA overexpression to see whether we could observe an increased activity over the background observed in cdase1 viable null mutants. As can be seen in Figure 5A, we did not see any activity across the entire pH spectrum.
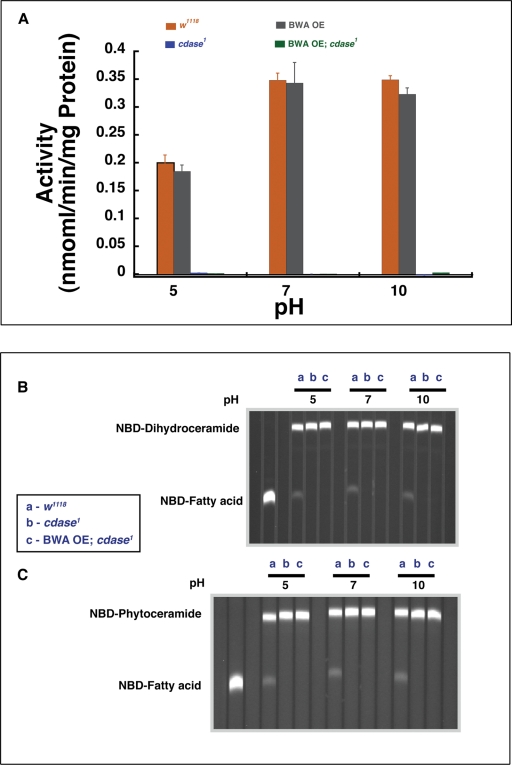
It was earlier reported that human alkaline ceramidase homologues exhibit ceramidase activity but preferred specific substrates, such as long-chain ceramides (d-erythro-C24 ceramide), phytoceramide, or dihydroceramide (Mao et al., 2000a, 2000b, 2001, 2003; Xu et al., 2006; Mao and Obeid, 2008). We therefore tested ceramidase activity using each of these substrates in extracts prepared from w1118, viable null cdase1 mutants, and cdase1 lethal rescued by BWA overexpression. Figure 6A shows ceramidase activity measured at pH 5.0, 7.0, and 10.0 using d-erythro-C24 ceramide as the substrate; in these assays, only wild-type extracts containing CDase enzyme shows activity. Thus CDase contributes to all observable activity on long-chain ceramide. To rule out the requirements for specific lipid-based endogenous cofactors for BWA ceramidase activity, we provided the substrate ceramides in liposomes prepared from lipid extracts of wild-type flies (instead of detergent-based substrate delivery). We performed activity measurements under these conditions in control w1118, cdase1 viable null mutant, and cdase1 lethal mutant rescued by BWA overexpressor flies at pH 5.0, 7.0, and 10.0. Only control w1118 extracts displayed activity under these conditions (unpublished data), ruling out lipid-specific cofactor requirement for ceramidase activity for BWA protein. Figure 6, B and C, shows ceramidase activity against 6-((N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino) hexanoyl)sphingosine (NDB)-dihydroceramide and NBD-phytoceramide. Here again NBD fatty acid generation or phytoceramidase and dihydroceramidase activity is exhibited only by extracts containing CDase and not in BWA overexpressors. (Extracts from w1118 and bwa showed similar activity in using this substrate at pH 5.0, 7.0, and 10.0, indicating that BWA protein did not contribute to any of the ceramidase activity exhibited against this substrate by Drosophila extracts; see Figure 3E.)
FIGURE 6:
BWA overexpressor flies show no ceramidase activity. (A) Extracts from control w1118, viable null cdase1 (cdase1 null with the suppressor mutation), and BWA overexpressors in w1118 (BWA OE), and extracts from cdase1 null lethal flies that were rescued by BWA overexpression (BWA OE; cdase1) flies, were assayed for activity using a long-chain ceramide (d-erythro C-24 ceramide) as substrate. Control w1118 and w1118 overexpressing extracts show the same specific ceramidase activity against the long-chain ceramide, implying that BWA overexpression does not contribute to any observable increase in ceramidase activity at any of the three pHs indicated. Similarly, the viable allele of cdase1 and the lethal mutant of cdase1 rescued by overexpression of BWA show no activity when the assay is performed at the three pHs indicated. Thus BWA protein that is genetically active exhibits no biochemical ceramidase activity. All activity measurements are an average of at least six measurements, and the error bars denote SD. (B) Control extract is able to deacylate NBD-dihydroceramide, and this activity is lost in viable allele of cdase1. The activity is not recovered in lethal mutants of cdase1 rescued by overexpression of BWA. (C) Control extract is able to deacylate NBD-phytoceramide, and this activity is lost in viable allele of cdase1 (suppressor, cdase1). The activity is not recovered in lethal mutants of cdase1 rescued by overexpression of BWA.
In an attempt to evaluate whether the BWA protein could complement the defects of ceramidase activity in yeast, we expressed BWA protein in yeast mutant for either YPC1 or the YDC1 gene product (Supplemental Figure 2A). Δydc1 mutants are thermolabile (Mao et al., 2000b). Expression of BWA did not rescue the heat susceptibility of Δydc1 (Supplemental Figure 2B) or alter the flux through the sphingolipid biosynthetic pathway (Supplemental Figure 2C). These experiments indicate that the Drosophila BWA protein, unlike the mammalian or the yeast protein, does not possess ceramidase activity. It is also noteworthy that although BWA has profound influence on sphingolipid flux in Drosophila, it is unable to affect the metabolic pathway in yeast.
DISCUSSION
Our studies indicate that the neutral ceramidase homologue, CDase, is the only functional ceramidase present in Drosophila. Enzyme activity studies demonstrate robust ceramidase activity in the pH range 5.0–11.0 in Drosophila extracts. All of this activity is lost in extracts of viable null mutant of CDase. Only one other homologue of a putative alkaline ceramidase is encoded by the genome and is named Brainwashing. Previously, in a screen designed to identify mutations in the development and organization of Drosophila brain, the Preat laboratory had identified a P-element insertion into the second intron of CG13969 (Boquet et al., 2000). The authors initially noted that these flies had defective fusion of the cerebral hemispheres. This phenotype was not penetrant, however, in subsequent generations of the flies and did not segregate with the P-insertion. Thus this phenotype was caused by an unlinked secondary mutation in the fly and not by disruption of the CG13969. We used this insertion to generate deletion alleles of Bwa in the current study. These mutants do not have defects in hemisphere fusion. We could not find any observable phenotype in these mutant animals.
Recently it was reported that mutations in the bwa gene, termed as Dacer by the authors in that study, resulted in increased longevity of the mutant flies (Yang et al., 2010). It should be noted in the study that the authors have used a P-element insertion in the fourth exon of the gene as the mutagen. Neither were rescue experiments performed nor was the P-element mobilized to generate defined deletion mutants. In our studies, mere insertion of the P-element does not abrogate the ability to generate transcripts of the gene. We have shown that the insertion of P-element in the second intron of Bwa did not affect the transcription of the gene. Our deletion mutants do not exhibit increased longevity or life span (Supplemental Figure 1A). Also in the study by Yang et al., BWA protein was expressed in Sf9 cells, and the activity was measured from microsomal preparations. Those measurements were taken in the background of endogenous proteins from Sf2 cells. The viable cdase1 null background shows no endogenous ceramidase activity, and in this background endogenous or overexpressed functional BWA exhibits no ceramidase activity. We have also expressed BWA in S2 cells and partially purified it using affinity chromatography. The partially purified protein has also been tested for enzymatic activity and exhibits no ceramidase function (Supplemental Figure 3, A–C). Like the mammalian and the yeast homologues, the protein shows a reticular pattern of expression overlapping with the ER-Golgi markers (Supplemental Figure 3D). Overwhelming evidence from our group indicates that the Drosophila BWA protein is not a ceramidase but is a regulator of the ceramide flux within the Drosophila. Mao and colleagues have extensively reported on the yeast alkaline ceramidase and the mammalian homologues ACER1–3 (Mao et al., 2000a, 2000b, 2001, 2003; Xu et al., 2006; Mao and Obeid, 2008). The proteins catalyze ceramidase reactions.
In reconciling the present study with their extensive work with the yeast and mammalian enzymes it is possible that BWA protein started out as a ceramidase and has generally retained this activity across species and kingdom. In certain clades (perhaps) or special circumstances, such as in Drosophila melanogaster, however, the protein lost its ceramidase activity and in its place acquired a role of monitoring ceramide levels. Further studies in this regard should resolve this matter. An example of such a change is seen in the human caspase-12 gene. Eighty percent of humans do not encode for a functional caspase-12 gene (Fischer et al., 2002; Kachapati et al., 2006; Xue et al., 2006). In these human populations, and unlike in mouse and chimpanzees, the caspase-12 gene has accumulated several mutations rendering it inactive as a caspase protein; hence, it is not involved in ER-mediated apoptosis. It is now believed that caspase-12 gene product is involved in mediating the ill effects of sepsis and is one of the major causes of mortality in children across the world. The pressures to avoid sepsis and to survive are believed to be strong selective forces in the spread of the inactive form of caspase-12 in humans.
A search of the motifs present in the BWA protein using the PRINTS program (http://www.bioinf.manchester.ac.uk/fingerPRINTScan/) indicates that the protein demonstrates motifs with some similarity to those present in channels, bacterial transporters, and translocase proteins (Attwood et al., 2004). Thus BWA might function to assist the movement of ceramides in the membrane environment, and perhaps even regulate the sphingolipid metabolic pathway by facilitating easier movement of ceramides across layers of membranes in its vicinity. Increased mobility might in turn improve the accessibility of ceramide to other enzymes that act on ceramide, thereby expediting its metabolism. It is worth noting that, although the bwa mutant accumulates amounts of ceramide similar to those in cdase1 and dcerk1 mutants, it exhibits neither the lethality observed in the former nor the retinal degeneration observed in the latter. Because each of these enzymes localizes to different compartments of the cell, the pool of ceramides that is at least initially affected in each of these mutant backgrounds will be different. Therefore the effect of ceramide accumulation in each one of these mutants will depend on the physiological function affected by such ceramide accumulation. Accumulation in cdase1 mutants in the outer leaflet and the endolysosomal compartment is detrimental to synaptic signaling in cdase mutants and thus they are embryonic lethal (Rohrbough et al., 2004; Acharya et al., 2008). Accumulation of similar levels of ceramide initiated in the inner membrane of ceramide kinase mutants is detrimental to photoreceptor structure and function but does not compromise embryonic synaptic signaling as to cause death (Dasgupta et al., 2009). In the case of bwa mutants, the accumulation of ceramide (perhaps in other inner membrane compartments) does not affect the development or phototransduction in Drosophila. Thus context-dependent ceramide accumulation is critical for the observed phenotypes.
Thus our experiments demonstrate that all observable ceramidase activity in Drosophila is contributed by CDase protein and no activity is attributable to BWA. Yet we see a strong genetic interaction of Bwa with genes for two of the enzymes that function to reduce ceramide levels in vivo. These studies imply that BWA either exhibits ceramidase activity that is activated only under specific circumstances or could have an as yet unknown biochemical function that would effectively decrease ceramide levels in vivo without biochemical activity as a ceramidase.
MATERIALS AND METHODS
Ceramidase assay
The assay was performed as described previously with slight modifications (Dobrowsky and Kolesnick, 2001). Briefly, [3H]C16- or C24-ceramide and cold C16- or C24-ceramide in chloroform/methanol (2:1 vol/vol) were added to each assay tube and evaporated the solvent (the [3H]C16- or C24-ceramide was obtained from the lipidomics shared resources, Medical University of South Carolina, Charleston, SC). The residue was resuspended in 100 μl of 100 mM buffer (NaOAc, pH 4.0–6.0; HEPES, pH 6.5–8.5; glycine, pH 9.0–11.0; Na2HPO4, pH 11.5–12.5) containing 0.2% Triton X-100 and 5 mM CaCl2. The reaction was initiated by the addition of ∼10 μg of total protein and was incubated for 1–2 h at 37°C. (Drosophila extracts are prepared by homogenization in the corresponding buffer containing protease inhibitors. The lysate is spun at 5000 × g for 10 min at 4°C. The supernatant is collected, and the protein concentration is determined.) The reaction was terminated by the addition of 2 ml of isopropyl alcohol/heptane/2N NaOH (78:20:2). Cold palmitic acid (50 μg) was added as carrier lipid, and phase partitioning was induced by the addition of 1.2 ml of heptane and 1 ml of water. After centrifugation, the upper phase was discarded. The lower phase was washed with two 1-ml aliquots of heptane, and the upper phase was discarded after each centrifugation. The lower phase was acidified with 1 ml of 1N H2SO4 to protonate the released fatty acid. Heptane (2 ml) was added, vortexed, and centrifuged, and 1.5 ml of the upper phase was transferred to a scintillation vial and counted. The activity was calculated. Most assays were done in duplicate and repeated at least three times.
It is possible that BWA activity requires cofactors that might not be supplied in the regular assay. To verify this possibility, we designed an assay in which the substrate (ceramide) was provided in liposomes prepared from Drosophila total lipid extract. Briefly, 10–15 flies were washed with phosphate-buffered saline (PBS) three times and homogenized in the assay buffer. The homogenate was mixed with equal volumes of chloroform methanol (2:1) and vortexed. The organic phase was collected and evaporated under nitrogen. Radiolabeled ceramide was added to the total lipid extract and suspended in assay buffer. This suspension was sonicated in a water bath sonicator to prepare liposomes. These liposomes were used as substrate for the ceramidase assay as described earlier in this article.
Reverse ceramidase assay
The assay was performed as described with slight modifications (El Bawab et al., 2001). Briefly, the substrates ([3H]palmitic acid, cold palmitic acid, and sphingosine) were dried and then resuspended in 100 μl of 100 mM buffer (NaOAc, pH 5.0; HEPES, pH 7.0; glycine, pH 10.0), containing 0.2% Triton X-100; 20 μg of fly total protein was used for each reverse action. The enzymatic reaction was carried out at 37°C for 1–2 h. The reaction was terminated by adding 2 ml of isopropyl alcohol/heptane/2N NaOH (78:20:2), followed by 1 ml of water and 1 ml of heptane. The upper phase was collected after centrifugation. The lower phase was washed with 2 ml of heptane, and the upper phase was collected again and counted in a scintillation counter. Most assays were done in duplicate and repeated three times.
Expression and purification of bwa protein in S2 cells
Drosophila Bwa was cloned into pRmHa-C vector in frame with the Flag and Histidine tag and expressed in S2 cells using standard techniques. Protein expression in these stable cell lines was induced by adding copper sulfate (0.5 mM) to the medium. The cells were washed and lysed in 20 mM sodium phosphate buffer, pH 7.4, containing protease inhibitors. Nuclei and other cellular debris were removed by low-speed centrifugation. A membrane pellet was prepared from the supernatant by ultracentrifugation at 110,000 × g for 1 h.
The membrane pellet was lysed in buffer A (5 mM imidazole, 500 mM sodium chloride in 20 mM phosphate buffer) with 1% Triton X-100. BWA was purified using nickel affinity chromatography. Briefly, the lysate was loaded on to a Histrap column equilibrated with buffer A, and the column was washed with 5 column volumes of the same buffer. The unbound proteins were washed off by increasing the concentration of imidazole to 200 mM in buffer A. Finally the bound BWA protein was eluted with buffer B (400 mM imidazole, 500 mM sodium chloride in 20 mM phosphate buffer), and 200-μl fractions were collected. The fractions were tested on 12% SDS–PAGE to check for purity. The fraction with the highest purity was used in the ceramidase assay.
Phytoceramidase and dihydroceramidase activities
The activity was measured using C12-NBD-phytoceramide or NBD-C12-dihydroceramide (from Matreya, Pleasant Gap, PA) as substrates. The solvent was evaporated and resuspended in 100 μl of 100 mM buffer (NaOAc, pH 5.0; HEPES, pH 7.0; glycine, pH 10.0) containing 0.2% Triton X-100 and 2.5 mM CaCl2. After incubation at 37°C for 60 min, NBD-C12–fatty acid was released. The reactions were stopped by boiling for 5 min and dried under a SpeedVac. Lipids were dissolved in 35 μl of chloroform/methanol (2:1), and 25 μl was spotted onto a thin-layer chromatography (TLC) plate. The released NBD-fatty acid was separated from NBD-ceramide by TLC in a solvent of chloroform/methanol/25% ammonium hydroxide (90:30:0.5). The TLC plate was scanned using a PhosphorImager system set at the fluorescence mode. The NBD-fatty acid product was identified by comparison with NBD-C12 (Matreya, Pleasant Gap, PA).
Generation of bwa mutants
The bwa null deletion mutants were generated by mobilizing a P-element described previously (Boquet et al., 2000). The P-element was inserted into the second intron of Bwa and was mobilized using standard P-element mutagenesis screen. Approximately 500 excision lines were generated and screened by PCR for potential deletion alleles. Two lines, bwa1 and bwa2, were identified in the screen by genomic PCR, RT-PCR, and sequencing of the RT-PCR products. Oligos were used for genomic PCR of the two mutants: bwa1 – forward primer – ATACGCTCGCGATTTCATTT, reverse primer – TCGAAACTGTCCCGAAAAAC; bwa2 – forward primer – TCCCCAACACGATGACAATA, reverse primer – TGGCCACCGGTATAACACTT. For RT-PCR the following primers were used: forward primer – CTAAGATTGCAAGGAAGGGCG, reverse primer – TTTAGGCCCAAGTGTTCAGG. Both mutants had deletions that resulted in the generation of a truncated peptide containing 47 amino acids of the first exon
MGGMGGGGLLDIYAMAWEHLRPGSSPVDWCEGNYLISSNIAEFVNTS stop.
Antibodies
Antibodies used in this study were raised using standard protocol in rabbit against the following peptide from the C terminus of the protein: C G I P F I S I R N P G K A L R N T I.
Sphingolipid measurements
Lipids were extracted from fly heads following the protocol described previously. Liquid chromatography was performed on an ACQUITY UPLC system (Waters Co., Milford, MA). The sphingolipids were separated using an ACQUITY UPLC BEH Shield RP18 column (100 mm × 2.1 mm i.d.) maintained at 60°C. The mobile phase was composed of 5 mM HCOONH4 in water containing 0.2% formic acid (solvent A) and 5 mM HCOONH4 in methanol containing 0.2% formic acid (solvent B) with a flow rate of 0.3 ml/min. The column was eluted with isocratic elution 20% of solvent A for 5 min, followed by a linear gradient from 80% to 99% solvent B in 6 min. The column temperature was 60°C.
Mass spectrometric detection was carried out on a TQD mass spectrometer (Waters Co.) with the electrospray ionization (ESI) source set in positive mode. Quantification was performed using selected reaction monitoring (SRM) of the transitions with a can time of 0.014−0.036 s per transition. Parameters used are as follows: capillary voltage 2.0 kV, source temperature 120°C, desolvation temperature 450°C. Nitrogen was used as for desolvation and as cone gas with flow rates of 1000 and 50 l/h, respectively.
Expression of Drosophila bwa protein in the yeast
Δypc1, Δydc1, and wild-type Mat A type (BY4741) Saccharomyces cerevisiae were obtained from Open Biosystems (Huntsville, AL). Cloning and expression of Drosophila Bwa gene was essentially similar to that described previously (Mao et al., 2001). Briefly, the Drosophila Bwa cDNA was cloned in to pYES2 vector in frame with the V5 tag. The three yeast strains were transformed with the construct by using lithium acetate. The cells were plated on Synthetic Complete medium lacking uracil (SC –Ura) and incubated for 2 d at 30°C. For the induction of the protein, the cells were grown in SC –Ura supplemented with 2% galactose for 4 h, and the protein expression was verified by immunoblotting using anti-V5 antibody.
Heat stress study
Exponentially growing cells (5.2 × 105 total in 100 μl) were incubated at room temperature (untreated) or at 45°C for 50 min (heat stress). The untreated cells were diluted 500 times, and 5 μl was plated on to Yeast Proteome Database (YPD) plates. For the heat-stressed cells, 10 μl was directly plated on the plates. The plates were incubated at 30°C for 2 d, and the colonies were counted using a Bio-Rad (Hercules, CA) gel documentation system.
Labeling of yeast cells
A single isolated yeast colony was inoculated into SC –Ura supplemented with 2% glucose and cultured overnight. The overnight culture was inoculated into 1 ml of fresh medium, and the cells were grown to an optical density of 0.6–0.7. The culture medium was then supplemented with 30 μCi of [3H]palmitic acid, and the cells were further incubated at 30°C for an additional 4 h. Total lipids were extracted, deacylated by monomethylamine, and resolved by TLC by using the solvent system containing chloroform, methanol, and 4.2 N ammonium hydroxide in the ratio of 9:7:2. For the cells transfected with pYES2 Bwa construct, the medium contained galactose instead of glucose during the induction and labeling period.
Supplementary Material
Acknowledgments
We thank Shyam Sharan and Ira Daar for critical reading of the manuscript. Jun Yonekubo (Nihon Waters K.K.) is thanked for his helpful discussion and support for the experiments. This work was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health, Department of Health and Human Services. Work in Usha Acharya’s laboratory was supported by a grant from the National Institutes of Health (R01EY16469). We thank Alicja Biewalska, Director of the Shared Lipidomic Resources at the Medical University of South Carolina, for providing us with [3H]C16- and C24-ceramide.
Abbreviations used:
- CPE
ceramide phosphoryletanolamine
- DCERK
Drosophila ceramide kinase
- EMS
ethyl methanesulfonate
- ER
endoplasmic reticulum
- IPP
inositol polyphosphate 1-phosphatase
- NDB
6-((N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino) hexanoyl)sphingosine
- RT–PCR
reverse transcription–PCR
- SC –Ura
Synthetic Complete medium lacking uracil
- SRM
selected reaction monitoring
- TEM
transmission electron micrograph
- TLC
thin-layer chromatography
- UAS
Upstream Activating Sequence
- UFLC MS/MS
ultrafast liquid chromatrography coupled with tandem mass specrometry
- YPD
Yeast Proteome Database.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-05-0453) on December 9, 2010.
REFERENCES
- Acharya JK, et al. Cell-nonautonomous function of ceramidase in photoreceptor homeostasis. Neuron. 2008;57:69–79. doi: 10.1016/j.neuron.2007.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya U, Acharya JK. Enzymes of sphingolipid metabolism in Drosophila melanogaster. Cell Mol Life Sci. 2005;62:128–142. doi: 10.1007/s00018-004-4254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya U, Mowen MB, Nagashima K, Acharya JK. Ceramidase expression facilitates membrane turnover and endocytosis of rhodopsin in photoreceptors. Proc Natl Acad Sci USA. 2004;101:1922–1926. doi: 10.1073/pnas.0308693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya U, Patel S, Koundakjian E, Nagashima K, Han X, Acharya JK. Modulating sphingolipid biosynthetic pathway rescues photoreceptor degeneration. Science. 2003;299:1740–1743. doi: 10.1126/science.1080549. [DOI] [PubMed] [Google Scholar]
- Attwood TK, Bradley P, Gaulton A, Maudling N, Mitchell AL. lutionary applications: Encyclopaedia of Genetics, Genomics, Proteomics and Bioinformatics. In: Dunn LJM, editor. P Little, and A Subramaniam, New York: John Wiley & Sons; [Google Scholar]
- Boquet I, Hitier R, Dumas M, Chaminade M, Preat T. Central brain postembryonic development in Drosophila: implication of genes expressed at the interhemispheric junction. J Neurobiol. 2000;42:33–48. [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Dasgupta U, et al. Ceramide kinase regulates phospholipase C and phosphatidylinositol 4, 5, bisphosphate in phototransduction. Proc Natl Acad Sci USA. 2009;106:20063–20068. doi: 10.1073/pnas.0911028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowsky RT, Kolesnick RN. Analysis of sphingomyelin and ceramide levels and the enzymes regulating their metabolism in response to cell stress. Methods Cell Biol. 2001;66:135–165. doi: 10.1016/s0091-679x(01)66007-2. [DOI] [PubMed] [Google Scholar]
- El Bawab S, Birbes H, Roddy P, Szulc ZM, Bielawska A, Hannun YA. Biochemical characterization of the reverse activity of rat brain ceramidase. A CoA-independent and fumonisin B1-insensitive ceramide synthase. J Biol Chem. 2001;276:16758–16766. doi: 10.1074/jbc.M009331200. [DOI] [PubMed] [Google Scholar]
- Fischer H, Koenig U, Eckhart L, Tschachler E. Human caspase 12 has acquired deleterious mutations. Biochem Biophys Res Commun. 2002;293:722–726. doi: 10.1016/S0006-291X(02)00289-9. [DOI] [PubMed] [Google Scholar]
- Hanada K. Discovery of the molecular machinery CERT for endoplasmic reticulum-to-Golgi trafficking of ceramide. Mol Cell Biochem. 2006;286:23–31. doi: 10.1007/s11010-005-9044-z. [DOI] [PubMed] [Google Scholar]
- Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Luberto C, Argraves KM. Enzymes of sphingolipid metabolism: from modular to integrative signaling. Biochemistry. 2001;40:4893–4903. doi: 10.1021/bi002836k. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- Hassler DF, Bell RM. Ceramidases: enzymology and metabolic roles. Adv Lipid Res. 1993;26:49–57. [PubMed] [Google Scholar]
- Holthuis JC, Pomorski T, Raggers RJ, Sprong H, Van Meer G. The organizing potential of sphingolipids in intracellular membrane transport. Physiol Rev. 2001;81:1689–1723. doi: 10.1152/physrev.2001.81.4.1689. [DOI] [PubMed] [Google Scholar]
- Kachapati K, O’Brien TR, Bergeron J, Zhang M, Dean M. Population distribution of the functional caspase-12 allele. Hum Mutat. 2006;27:975. doi: 10.1002/humu.9448. [DOI] [PubMed] [Google Scholar]
- Karlsson KA, Samuelsson BE, Steen GO. Structure and function of sphingolipids. 2. Differences in sphingolipid concentration, especially concerning sulfatides, between some regions of bovine kidney. Acta Chem Scand. 1968;22:2723–2724. doi: 10.3891/acta.chem.scand.22-2723. [DOI] [PubMed] [Google Scholar]
- Li CM, et al. The human acid ceramidase gene (ASAH): structure, chromosomal location, mutation analysis, and expression. Genomics. 1999;62:223–231. doi: 10.1006/geno.1999.5940. [DOI] [PubMed] [Google Scholar]
- Luberto C, Hannun YA. Sphingolipid metabolism in the regulation of bioactive molecules. Lipids. 1999;34 (suppl):5–11. doi: 10.1007/BF02562221. [DOI] [PubMed] [Google Scholar]
- Mao C, Obeid LM. Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochim Biophys Acta. 2008;1781:424–434. doi: 10.1016/j.bbalip.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Xu R, Bielawska A, Obeid LM. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. J Biol Chem. 2000a;275:6876–6884. doi: 10.1074/jbc.275.10.6876. [DOI] [PubMed] [Google Scholar]
- Mao C, Xu R, Bielawska A, Szulc ZM, Obeid LM. Cloning and characterization of a Saccharomyces cerevisiae alkaline ceramidase with specificity for dihydroceramide. J Biol Chem. 2000b;275:31369–31378. doi: 10.1074/jbc.M003683200. [DOI] [PubMed] [Google Scholar]
- Mao C, Xu R, Szulc ZM, Bielawska A, Galadari SH, Obeid LM. Cloning and characterization of a novel human alkaline ceramidase. A mammalian enzyme that hydrolyzes phytoceramide. J Biol Chem. 2001;276:26577–26588. doi: 10.1074/jbc.M102818200. [DOI] [PubMed] [Google Scholar]
- Mao C, Xu R, Szulc ZM, Bielawski J, Becker KP, Bielawska A, Galadari SH, Hu W, Obeid LM. Cloning and characterization of a mouse endoplasmic reticulum alkaline ceramidase: an enzyme that preferentially regulates metabolism of very long chain ceramides. J Biol Chem. 2003;278:31184–31191. doi: 10.1074/jbc.M303875200. [DOI] [PubMed] [Google Scholar]
- Merrill AH, Jr, Schmelz EM, Dillehay DL, Spiegel S, Shayman JA, Schroeder JJ, Riley RT, Voss KA, Wang E. Sphingolipids—the enigmatic lipid class: biochemistry, physiology, and pathophysiology. Toxicol Appl Pharmacol. 1997;142:208–225. doi: 10.1006/taap.1996.8029. [DOI] [PubMed] [Google Scholar]
- Merrill AH, Jr, Sullards MC, Wang E, Voss KA, Riley RT. Sphingolipid metabolism: roles in signal transduction and disruption by fumonisins. Environ Health Perspect. 2001;109(suppl 2):283–289. doi: 10.1289/ehp.01109s2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrbough J, Rushton E, Palanker L, Woodruff E, Matthies HJ, Acharya U, Acharya JK, Broadie K. Ceramidase regulates synaptic vesicle exocytosis and trafficking. J Neurosci. 2004;24:7789–7803. doi: 10.1523/JNEUROSCI.1146-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayman JA. Sphingolipids. Kidney Int. 2000;58:11–26. doi: 10.1046/j.1523-1755.2000.00136.x. [DOI] [PubMed] [Google Scholar]
- Spiegel S, Foster D, Kolesnick R. Signal transduction through lipid second messengers. Curr Opin Cell Biol. 1996;8:159–167. doi: 10.1016/s0955-0674(96)80061-5. [DOI] [PubMed] [Google Scholar]
- Spiegel S, Merrill AH., Jr Sphingolipid metabolism and cell growth regulation. FASEB J. 1996;10:1388–1397. doi: 10.1096/fasebj.10.12.8903509. [DOI] [PubMed] [Google Scholar]
- van Echten-Deckert G, Klein A, Linke T, Heinemann T, Weisgerber J, Sandhoff K. Turnover of endogenous ceramide in cultured normal and Farber fibroblasts. J Lipid Res. 1997;38:2569–2579. [PubMed] [Google Scholar]
- van Meer G, Holthuis JC. Sphingolipid transport in eukaryotic cells. Biochim Biophys Acta. 2000;1486:145–170. doi: 10.1016/s1388-1981(00)00054-8. [DOI] [PubMed] [Google Scholar]
- Xu R, Jin J, Hu W, Sun W, Bielawski J, Szulc Z, Taha T, Obeid LM, Mao C. Golgi alkaline ceramidase regulates cell proliferation and survival by controlling levels of sphingosine and S1P. FASEB J. 2006;20:1813–1825. doi: 10.1096/fj.05-5689com. [DOI] [PubMed] [Google Scholar]
- Xue Y, et al. Spread of an inactive form of caspase-12 in humans is due to recent positive selection. Am J Hum Genet. 2006;78:659–670. doi: 10.1086/503116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q. Role of Drosophila alkaline ceramidase (Dacer) in Drosophila development and longevity. Cell Mol Life Sci. 2010;67:1477–1490. doi: 10.1007/s00018-010-0260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y, Okino N, Tani M, Ito M. Molecular cloning and characterization of a secretory neutral ceramidase of Drosophila melanogaster. J Biochem. 2002;132:229–236. doi: 10.1093/oxfordjournals.jbchem.a003215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.