Analyses of 32 meiotic genes from fission yeast with respect to nascent transcription, RNA processing/accumulation, and effects of surveillance factor mutants reveal that the vast majority are “on” in proliferating cells and less than one-third show a transcriptional peak during meiosis, highlighting the important contribution of RNA-level regulation.
Abstract
To determine the relative importance of transcriptional regulation versus RNA processing and turnover during the transition from proliferation to meiotic differentiation in the fission yeast Schizosaccharomyces pombe, we analyzed temporal profiles and effects of RNA surveillance factor mutants on expression of 32 meiotic genes. A comparison of nascent transcription with steady-state RNA accumulation reveals that the vast majority of these genes show a lag between maximal RNA synthesis and peak RNA accumulation. During meiosis, total RNA levels parallel 3′ processing, which occurs in multiple, temporally distinct waves that peak from 3 to 6 h after meiotic induction. Most early genes and one middle gene, mei4, share a regulatory mechanism in which a specialized RNA surveillance factor targets newly synthesized transcripts for destruction. Mei4p, a member of the forkhead transcription factor family, in turn regulates a host of downstream genes. Remarkably, a spike in transcription is observed for less than one-third of the genes surveyed, and even these show evidence of RNA-level regulation. In aggregate, our findings lead us to propose that a regulatory cascade driven by changes in processing and stability of newly synthesized transcripts operates alongside the well-known transcriptional cascade as fission yeast cells enter meiosis.
INTRODUCTION
Although genome-wide studies in diverse eukaryotes have revealed extensive changes in transcript abundance during meiotic development (Reinke et al., 2000; Wellmer et al., 2006; Chalmel et al., 2007), the underlying molecular mechanisms remain largely unknown. In the fission yeast Schizosaccharomyces pombe, the levels of >800 transcripts increase by a factor of four or more in three successive waves that coincide with major meiotic transitions (Mata et al., 2002). Among these mRNAs, ∼100 encoding proteins that function in early events (DNA synthesis and recombination) peak 2–3 h after meiotic induction; 561 mRNAs, including many encoding proteins with roles in cell cycle regulation and cell division, peak at 4–6 h; and 133 mRNAs, mainly encoding spore formation and stress response proteins, peak at 6–9 h. These changes, which extended previous genetic and subtractive hybridization screens for meiosis-specific transcripts (e.g., Watanabe et al., 2001), have been attributed to regulation at both the RNA level, in other words activation of primary transcript processing (Kishida et al., 1994; Averbeck et al., 2005; Malapeira et al., 2005; McPheeters et al., 2009) and inactivation of a selective turnover pathway (Harigaya et al., 2006; Yamanaka et al., 2010), and the DNA level, in other words increased transcription of the cognate genes (e.g., Sugiyama et al., 1994; Ding and Smith, 1998; Mata et al., 2002; Cunliffe et al., 2004; Mata et al., 2007).
Consistent with an important role for transcriptional control, the vast majority of middle meiotic genes are regulated by Mei4p, one of four forkhead transcription factors in fission yeast (Mata et al., 2007). Among these is a gene encoding a cyclin expressed during mid-meiosis, rem1, which contains upstream motifs recognized by members of the forkhead family (Moldón et al., 2008). Intriguingly, this gene is also regulated at the level of splicing, positively by Mei4p, which is produced only during midmeiosis, and negatively by a forkhead protein expressed in both proliferating and differentiating cells, Fkh2p (Moldón et al., 2008). Although trans-acting regulatory factors have not been identified, removal of the mes1 intron is also controlled by an upstream region (Shimoseki and Shimoda, 2001) that contains forkhead recognition motifs. Also consistent with a shared regulatory mechanism, the timing of increased mes1 and rem1 mRNA accumulation during meiosis are similar (Mata et al., 2002). As transcription and splicing of these two transcripts are most likely coupled, the individual contribution of each process to the microarray peaks ∼5 h after meiotic induction cannot be readily assessed.
A third example of meiosis-specific splicing for which mechanistic information is available, crs1 (cyclin regulated by splicing), also contains a splicing regulatory element located outside the coding region (Averbeck et al., 2005; McPheeters et al., 2009); however, in contrast to rem1, it resides downstream. Although the crs1 gene is maximally transcribed in proliferating and early meiotic cells, the RNA accumulates only after concurrent activation of polyadenylation and splicing, which takes place ∼3 h after meiotic induction (McPheeters et al., 2009). These findings converged with the avenue of investigation that uncovered the YTH family RNA binding protein Mmi1p, which binds determinants of selective removal (DSRs) to target certain meiotic transcripts, including crs1, for destruction in mitotically growing cells (Harigaya et al., 2006). Further investigation revealed that 1) temperature-sensitive mmi1 mutations activate 3′ RNA processing and splicing of crs1 in nonmeiotic cells, and 2) the decision to retain the newly synthesized RNA in the nucleus for destruction rather than to process and export it is mediated by a bipartite regulatory element consisting of a DSR and a noncanonical polyadenylation signal (McPheeters et al., 2009). The cotranscriptional regulatory mechanism proposed for crs1 is consistent with the current view that splicing and other RNA processing reactions, which were traditionally designated “posttranscriptional” events, are in fact coupled to RNA synthesis in vivo (reviewed in Perales and Bentley, 2009). In this study, we surveyed gene expression parameters and the impact of mutations in RNA surveillance factors for a diverse array of meiotic genes to assess the contributions of known control mechanisms as well as to potentially uncover new ones. Based on our findings, we propose that the transition from proliferation to differentiation is driven in part by an RNA-level gene regulatory cascade that operates in parallel with, and independent from, previously described meiotic gene expression programs.
RESULTS
Whereas entry into meiosis is normally blocked by the Pat1p kinase via inhibitory phosphorylation of Mei2p, even haploid fission yeast strains harboring the temperature-sensitive pat1–114 allele initiate meiosis when shifted to the restrictive temperature (Iino and Yamamoto, 1985). Similar to mutating Mei2p to a nonphosphorylatable form (Watanabe et al., 1997) or inactivating Tor2p (Álvarez and Moreno, 2006), the pat1ts mutant enters the meiotic differentiation pathway regardless of nutritional conditions (Bähler et al., 1991). We have taken advantage of this mutant’s synchronous developmental program, which operates alongside the ste11-encoded transcription factor induced by nitrogen starvation (Mata and Bähler, 2006), to analyze expression over a meiotic time course for 32 meiotic genes and two control genes (Table 1). The meiotic genes, which were selected primarily on the basis of a dramatic increase (>25-fold) in steady-state RNA levels (column 6) following the transition to meiotic differentiation as judged by microarray analysis (Mata et al., 2002), encode products with a wide array of biological functions (column 5). Ten genes the transcripts of which undergo meiosis-specific splicing (Averbeck et al., 2005) were included in the survey even though two of these (mug93 and SPAPB8E5.10; rows 14 and 31) increase <25-fold. With the exception of meu6 and atg8 (rows 12 and 30), the newly selected genes lack introns (column 4), thus allowing us to assess the role of 3′ processing independent of splicing.
Table 1.:
Salient characteristics of meiotic and vegetative genes included in the survey. Row numbers in column 1 correspond to those in Figures 1, 2, and 4. For genes identifi ed experimentally, column 2 lists the name assigned in the first study, whereas column 3 lists all 34 systematic names (which are derived from the genome sequencing project, with SPA designating a location on chromosome 1, SPB chromosome 2, and SPC chromosome 3; Wood et al., 2002). Column 4 lists the number of introns, and column 5 lists the known or predicted function of the gene product. Information for both column 4 and column 5 is taken from the Sanger Center Web site (http://old.genedb.org/genedb/pombe/). Column 6 shows the fold increase in mRNA accumulation determined by microarray hybridization, and column 7 shows the classifi cation of each gene as early (E), middle (M), late (L), or vegetative (V) in the same study (Mata et al., 2002). The genes are listed in order of the time during meiosis at which RNA accumulation determined by microarray analysis reaches 20% of its maximal value, listed at left in the fi nal column. Listed at right in the fi nal column is the time of maximal accumulation. The difference between these two time points provides an indication of the ascending slope of each peak. For comparison, row 0 shows the same information for crs1, the subject of our previous study (McPheeters et al., 2009).
| 1. | 2. Gene | 3. Systematic | 4. # | 6. Fold | 7. Ma | 8. Hr at: | ||
|---|---|---|---|---|---|---|---|---|
| Row | name | name | int | 5. Product function | increase | class | 20% | Peak |
| 0 | crs1 | SPBC2.09c | 4 | Meiotic cyclin regulated by splicing | 17.7 | E | 1 | 3 |
| 1 | meu13 | SPAC222.15 | 4 | HOP2 recombination protein ortholog | 27.3 | E | 1 | 1–2 |
| 2 | rec8 | SPBC29A10.14 | 4 | Meiotic cohesin complex subunit | 34.6 | E | 1 | 3 |
| 3 | ght3 | SPAC1F8.01 | 0 | Hexose transporter | 34.7 | E | 2 | 4 |
| 4 | fbp1 | SPBC1198.14c | 0 | Fructose-1,6-bisphosphatase | 79.8 | E | 3 | 5 |
| 5 | mei4 | SPBC32H8.11 | 0 | Meiotic forkhead transcription factor | 54.9 | M | 4 | 5 |
| 6 | SPAC1610.02c | 0 | Mitochondrial ribosomal protein subunit L1 | 64.0 | M | 4 | 5 | |
| 7 | SPAPB17E12.09 | 1 | Sequence orphan | 77.9 | M | 4 | 5 | |
| 8 | meu21 | SPAC24C9.07c | 0 | 1,3-β-glucan synthase subunit Bgs2 | 115.9 | M | 4 | 5 |
| 9 | mug131 | SPBC1861.06c | 0 | S. pombe specific UPF0300 family protein 4 | 171.4 | M | 4 | 5 |
| 10 | mug125 | SPAC1A6.08c | 0 | Sequence orphan | 49 | M | 4 | 5 |
| 11 | mcp4 | SPBC16E9.08 | 0 | Pros-pore membrane protein | 62.0 | M | 4 | 5 |
| 12 | meu6 | SPBC428.07 | 2 | Meiotic chromosome segregation protein | 279.0 | M | 4 | 7 |
| 13 | meu4 | SPAC1F8.05 | 0 | Sequence orphan | 570.0 | M | 4 | 8 |
| 14 | mug93 | SPBC32H8.06 | 2 | TPR repeat protein | 4.9 | M | 5 | 5 |
| 15 | mug132 | SPAC11G7.06c | 0 | S. pombe specific UPF0300 family protein 3 | 60.3 | M | 5 | 5 |
| 16 | mfr1 | SPBC1198.12 | 1 | Fizzy-related protein Mfr1 | 91.8 | M | 5 | 5 |
| 17 | mug97 | SPBC146.11c | 0 | Meiotic chromosome segregation | 185.2 | M | 5 | 5 |
| 18 | SPAC10F6.15 | 0 | S. pombe specific UPF0300 family protein 1 | 242.8 | M | 5 | 5 | |
| 19 | mug137 | SPCC1919.11 | 6 | BAR adaptor/SH3 protein | 256.2 | M | 5 | 5 |
| 20 | spo3 | SPAC607.10 | 0 | Sporulation protein | 364.0 | M | 5 | 5 |
| 21 | meu27 | SPCC1259.14c | 0 | S. pombe specific UPF0300 family protein 5 | 120.3 | M | 5 | 5 |
| 22 | spo6 | SPBC1778.04 | 3 | Spo4-Spo6 kinase complex regulatory subunit | 447.8 | M | 5 | 5 |
| 23 | mpf1 | SPAC4G9.05 | 2 | Meiotic PUF family protein | 92.8 | M | 5 | 6 |
| 24 | mug142 | SPBC3H7.09 | 0 | Palmitoyl transferase | 114.7 | M | 5 | 6 |
| 25 | meu25 | SPBC27.03 | 0 | Sequence orphan | 331.4 | M | 5 | 6 |
| 26 | mde7 | SPCC320.07c | 0 | RNA-binding protein | 112.8 | M | 5 | 6 |
| 27 | SPAC1039.11c | 0 | α-glucosidase | 338.1 | M | 5 | 6 | |
| 28 | mug24 | SPCC74.09 | 0 | RNA-binding (RRM) | 414.5 | M | 5 | 6 |
| 29 | meu7 | SPBC16A3.13 | 0 | α-amylase homolog | 507.7 | M | 5 | 6 |
| 30 | atg8 | SPBP8B7.24c | 2 | Autophagy-associated protein | 27.4 | M | 5 | 7 |
| 31 | SPAPB8E5.10 | 2 | Conserved hypothetical | 17.1 | L | 5 | 10 | |
| 32 | SPBP19A11.02c | 0 | GPI anchored ER protein | 114.6 | L | 6 | 8 | |
| 33 | act1 | SPBC32H8.12c | 0 | Actin | NA | V | NA | |
| 34 | SPAC6F6.11c | 4 | Pyridoxine-pyridoxal-pyridoxamine kinase | NA | V | NA | ||
Accumulation of meiotic mRNAs does not parallel changes in transcription
To determine whether the discrepancy between the profiles of RNA synthesis and accumulation during exit from the mitotic cell cycle into meiotic differentiation observed for crs1 (McPheeters et al., 2009) is common or exceptional among meiotic transcripts, we compared these parameters for each of the meiotic and control genes listed in Table 1. Nascent RNA synthesis was monitored by transcriptional run-on (TRO) assays (Hansen et al., 1998; McPheeters et al., 2009). Upon permeabilization of the cells, RNA synthesis stalls due to acute loss of ribonucleoside tri-phosphates (rNTPs) and is restarted via the addition of a buffer containing [α-32P]UTP, followed by a brief (6 min) incubation to allow labeling of transcripts undergoing active synthesis. Measurement of total 32P incorporation over a meiotic time course (Supplemental Figure S1), revealed a gradual decline in the TRO signal from 1 to 4 h after meiotic induction, reaching essentially background levels at the 5-h time point.
To analyze the patterns of nascent RNA synthesis for individual genes, labeled RNAs were hybridized to immobilized PCR fragments located near the 3′ ends of the coding regions, ensuring that the results reflect full-length transcripts (Figure 1A). TRO data for an independent pat1–114–induced meiotic time course look similar (Supplemental Figure S2), and quantitation of the hybridization signals for these and a third TRO experiment (Supplemental Figure S3) indicates that, although transcription of most early, many middle, and both vegetative genes declines to near background levels by 4–5 h after meiotic induction, similar to total 32P incorporation (Supplemental Figure S1), the transitions are more abrupt, with relatively constant signals through the 3-h time point followed by a precipitous decline. Remarkably, the peak transcription of many meiotic genes exceeds that of the highly expressed act1 gene (compare Supplemental Figure S3, panels 5, 12, 14, 16, 22, 28, 30, and 33), with mei4 showing by far the strongest TRO signal in mitotically growing cells of any gene analyzed (Supplemental Figure S3, panel 5).
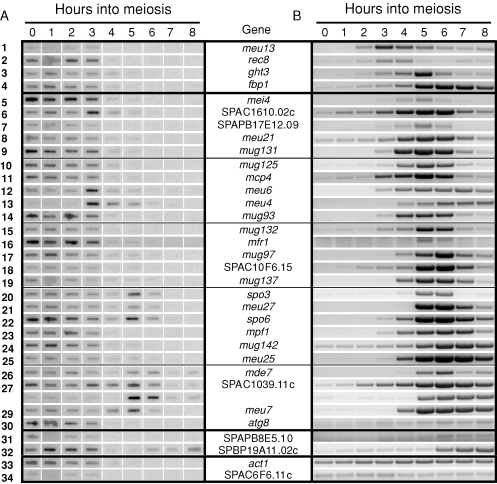
FIGURE 1:
Comparison of nascent transcription with steady-state RNA accumulation for 32 meiotic and 2 vegetative genes. Row numbers correspond to those in Table 1. (A) Analyses of nascent transcription over a meiotic time course using TRO assays. Cells were harvested at the times indicated after meiotic induction and processed as described previously (McPheeters et al., 2009). Newly synthesized 32P-labeled RNAs were detected by hybridization to filter-bound PCR fragments of ∼500 bp corresponding to sequences near the 3′ end of each ORF. The decreased background in the 5- to 8-h TRO assays reflects the lower overall incorporation of label during late meiosis (see Supplemental Figure S1). (B) Semiquantitative RT-PCR analyses of RNA accumulation over a meiotic time course. Total RNA was extracted at the times indicated after the temperature shift, and steady-state RNA levels were assayed by RT-PCR with the same primers used to generate PCR products for the TRO experiment in A; products were visualized negatively after ethidium bromide staining. Reactions were carried out for 21 cycles with three exceptions: for meu4 (row 13) and atg8 (row 23), the cycle number was reduced to 12 and 16, respectively, whereas the low abundance of the SPAC6F6.11c transcript (row 34) necessitated an increase to 28 cycles. Thick lines separate the early, middle, late, and vegetative genes (see Table 1), and thin lines separate every fifth middle meiotic gene for alignment purposes.
Unexpectedly, the profile of RNA accumulation over a meiotic time course is dramatically different from the TRO data for the vast majority of genes surveyed (Figure 1, panel A vs. B). The most striking disparities are observed for the early meiotic genes meu13 and rec8, for which transcription is maximal during mitotic growth and early meiosis. RNA levels, however, do not peak until 3 h after meiotic induction (Figure 1, rows 1 and 2). Transcription of ght3 is likewise maximal in vegetative and early meiotic cells, but RNA accumulation peaks even later, at the 4- to 5-h time points (Figure 1B, row 3) when RNA synthesis has begun to decline (Figure 1A and Supplemental Figure S2, row 3). For fbp1, the transcription and accumulation profiles (Figure 1, row 4) appear as overlapping mirror images, with RNA levels increasing most dramatically in the interval (4–6 h) where new synthesis is detectable but declining (Figure 1A and Supplemental Figure S2, row 4).
In contrast to the early genes, middle meiotic transcripts show fairly uniform peaks 5–6 h after meiotic induction (Figure 1B) despite diverse synthesis patterns. For the largest group (16/26), maximal RNA synthesis occurs in vegetative and early meiotic cells (Figure 1A and Supplemental Figure S2, rows 5, 7–11, 14–19, 23–25, and 30), similar to the early meiotic genes. Notably, a similar transcription profile is observed for SPAPB8E5.10 (Figure 1, row 31), which was classified as a late gene based on microarray data (Mata et al., 2002). The accumulation of this RNA in the apparent absence of ongoing transcription (Figure 1, row 31) most likely reflects the lower sensitivity of the TRO hybridization assay compared to reverse transcription (RT)-PCR. Although we cannot completely exclude the possibility that the efficiency of 32P incorporation decreases at late time points in the TRO experiments, we consider this less likely in that seven middle genes (spo3, meu27, spo6, mde7, SPAC1039.11c, mug24, and meu7) show robust transcription 5–6 h after meiotic induction (Figure 1A and Supplemental Figures S2 and S3, rows 20–22, 26–29), and significant incorporation is seen for a late gene at even later time points (up to 8 h; Figure 1A and Supplemental Figures S2 and S3, row 32).
The late meiotic gene SPBP19A11.02c shows a unique temporal pattern of expression: Although transcribed in proliferating, early, and late meiotic cells with a lull in mid-meiosis, the RNA accumulates only at late time points (Figure 1 and Supplemental Figures S2 and S3, row 32). Also intriguing are three middle genes that show a spike in transcription at 3–4 h but peak RNA accumulation at 5–6 h (Figure 1A and Supplemental Figure S2, rows 6, 12, and 13). For all of the transcriptionally induced genes except meu4 (Figure 1A, row 13), RNA synthesis is readily detectable in growing cells. Finally, the levels of both control RNA polymerase II transcripts, which encode actin and a predicted pyridoxal kinase, remain fairly constant throughout meiosis despite a precipitous decline in transcription (Figure 1 and Supplemental Figures S2 and S3, rows 33 and 34). To provide a more quantitative picture of RNA accumulation, we performed real-time RT-PCR (qRT-PCR) analyses on a representative set of meiotic transcripts (Supplemental Figure S4). Importantly, these RNAs show the same temporal patterns of accumulation as the semiquantitative assays and are also similar to the pattern determined from microarray analysis (Table 1, last column), which likewise measures steady-state RNA levels.
The surprising discrepancy between the profiles of RNA synthesis and accumulation prompted us to seek independent evidence that our assays provide an accurate picture of molecular events during the transition from growth to differentiation. To this end, we took advantage of a study by Bähler and colleagues (Lackner et al., 2007) in which gene expression parameters, including promoter-associated RNA polymerase II and mRNA accumulation, were measured in mitotically growing cells (Table 2). Although seven of the genes in our survey were not analyzed, the available data bolster the discovery reported here that most meiotic genes are transcribed in vegetative cells but the transcripts do not accumulate (Figure 1 and Supplemental Figures S2 and S3). Specifically, 18 of the 25 meiotic genes analyzed show a ratio of mRNA accumulation/Pol II occupancy fourfold lower than either control gene and, for three others, the ratio is >2 (Table 2, column 6), as predicted for active transcription accompanied by nuclear RNA turnover. Conversely, meiotic genes for which we could detect RNA accumulation at the 0 time point clustered near the bottom of the ranking, just above the vegetative genes (column 3).
Table 2.:
Comparison of gene expression measurements reported here (Figure 1) with published data (Lackner et al., 2007)a. The results of analyzing mRNA levels (column 4), RNA polymerase II occupancy (column 5), and mRNA/Poll II occupancy (column 6) were available for 25 of the 32 meiotic genes in our survey as well as both control genes. These genes are listed in columns 1 and 2 in order of their rank in the mRNA/Pol II list (column 7). In column 3, indicating whether RNA detectably accumulates at 0 h (Figure 1B) and column 8, indicating whether a DSR is present in the gene, “yes” (Y) answers are highlighted in gray
| 1. Gene name | 2. Systematic name | 3. 0 h accumulation | 4. mRNA levela | 5. Pol II occupancya | 6. mRNA/Pol IIa | 7. Rank | 8. DSR? |
|---|---|---|---|---|---|---|---|
| rec8 | SPBC29A10.14 | N | 36.90 | 1.20 | 30.85 | 19 | Y |
| mug137 | SPCC1919.11 | N | 27.00 | 0.66 | 41.00 | 30 | N |
| spo6 | SPBC1778.04 | N | 86.50 | 1.87 | 46.33 | 35 | N |
| mug131 | SPBC1861.06c | N | 42.70 | 0.90 | 47.26 | 37 | N |
| crs1 | SPBC2.09c | N | 98.05 | 1.70 | 57.73 | 53 | Y |
| SPAPB8E5.10 | N | 74.10 | 1.24 | 59.73 | 56 | N | |
| mei4 | SPBC32H8.11 | N | 114.65 | 1.47 | 78.21 | 78 | Y |
| mug97 | SPBC146.11c | N | 144.6 | 1.49 | 96.82 | 94 | N |
| meu13 | SPAC222.15 | N | 47.50 | 0.48 | 99.69 | 100 | N |
| mpf1 | SPAC4G9.05 | N | 75.35 | 0.52 | 145.46 | 140 | N |
| meu7 | SPBC16A3.13 | N | 124.55 | 0.84 | 148.01 | 142 | N |
| meu6 | SPBC428.07 | N | 110.60 | 0.73 | 151.20 | 147 | N |
| mug24 | SPCC74.09 | N | 79.45 | 0.44 | 179.95 | 165 | N |
| mcp4 | SPBC16E9.08 | N | 168.90 | 0.63 | 268.31 | 242 | N |
| meu25 | SPBC27.03 | N | 174.05 | 0.61 | 287.69 | 263 | N |
| ght3 | SPAC1F8.01 | N | 119.55 | 0.35 | 341.57 | 312 | N |
| fbp1 | SPBC1198.14c | N | 344.85 | 0.81 | 425.22 | 390 | N |
| mug125 | SPAC1A6.08c | N | 239.25 | 0.65 | 451.15 | 418 | N |
| mug132 | SPAC11G7.06c | N | 224.80 | 0.48 | 465.91 | 434 | N |
| meu21 | SPAC24C9.07c | Y | 558.15 | 0.91 | 615.04 | 613 | N |
| atg8 | SPBP8B7.24c | Y | 909.20 | 1.32 | 688.53 | 718 | N |
| SPBP19A11.02c | Y | 1480.10 | 1.89 | 782.50 | 847 | N | |
| mug142 | SPBC3H7.09 | Y | 963.25 | 0.97 | 998.19 | 1220 | N |
| spo3 | SPAC607.10 | N | 851.80 | 0.84 | 1009.24 | 1243 | N |
| SPAC1039.11c | Y | 1299.55 | 0.57 | 2285.93 | 3059 | N | |
| SPAC1610.02c | Y | 2541.95 | 0.98 | 2599.13 | 3377 | N | |
| act1 | SPBC32H8.12c | Y | 12,327.213 | 6.90 | 1786.94 | 2452 | N |
| SPAC6F6.11c | Y | 2191.80 | 0.62 | 3555.23 | 4048 | N |
Polyadenylation at multiple sites is a common feature of meiotic mRNAs in S. pombe
As the crs1 gene contained a regulatory polyadenylation signal that collaborates with the DSR to silence expression in mitotically growing cells (McPheeters et al., 2009), we next determined whether other meiotic transcripts are polyadenylated at two or more sites. Using RT-PCR with oligo(dT) as the 3′ primer and gene-specific 5′ primers (Figure 2), we found that all four early transcripts in our survey are polyadenylated at two sites; for meu13, rec8, and ght3, both are activated at similar times during meiosis (rows 1–3), whereas the fbp1 polyadenylation signals are differentially used, with the proximal site used only during late meiosis (Figure 2, row 4). The profile of poly(A)+ mRNA accumulation generally mirrors total RNA accumulation (compare Figure 2 with Figure 1B), as expected given that RNA polyadenylation failure precludes export to the cytoplasm (Hilleren et al., 2001). The presence of multiple sites of 3′ processing is not limited to early meiotic genes, as 10 of the 26 middle and both of the late transcripts are polyadenylated at two or more positions. Although one of the two control genes is also polyadenylated at two positions (Figure 2, row 34), analysis of 29 additional constitutively expressed transcripts indicates that multiple 3′ processing sites are less common among vegetative than meiotic genes (unpublished observations).
FIGURE 2:
Analyses of polyadenylation over a meiotic time course. Total RNA was extracted at the times indicated after the temperature shift and the presence of a poly(A) tail was assessed by RT-PCR using a 5′ primer near the 3′ end of the ORF and oligo(dT) as the 3′ primer, as described previously (McPheeters et al., 2009). D and P next to the row number indicate poly(A) sites distal and proximal to the ORF, respectively. The cycle number was 28 except in the case of meu4 (row 13), for which 12 cycles gave a strong signal, consistent with qRT-PCR data (Supplemental Figure S4C). For genes with two closely spaced cleavage/polyadenylation sites, both are displayed on the same gel slice; two separate slices are shown for more widely separated sites. Thick lines separate the early, middle, late, and vegetative genes (see Table 1). The position(s) of poly(A) tail addition (far right) were determined by sequencing of the indicated cDNAs and are expressed as distance (in nt) from the translational stop codon to the cleavage/polyadenylation site.
To map the sites of poly(A) tail addition, we excised each band from gels such as those in Figure 2 and subjected the cDNAs to sequence analysis. For meu13, rec8, and ght3, the 3′ untranslated region (UTR) lengths (148/110, 183/154, and 190/147, respectively; Figure 2, rows 1–3) cluster around the average for S. pombe (169 nucleotides [nt], Wilhelm et al., 2008), and the sites of polyadenylation are in close proximity (38, 29, and 43 nt, respectively), similar to crs1 (McPheeters et al., 2009). Polyadenylation at the meiosis-specific proximal site in fbp1 yields a highly truncated 3′ UTR (210 vs. 412 nt; Figure 2, row 4). For the middle meiotic genes, 3′ UTR lengths vary over a wide range, from 42 nt for the proximal site in spo3 (Figure 2, row 20) to 592 nt for the distal site in mpf1 (row 23), whereas both late genes have shorter than average 3′ UTRs (Figure 2, rows 31 and 32). Interestingly, the longest 3′ UTR (664 nt) is found in one of the control genes, act1 (Figure 2, row 33). Overall, 3′ UTR length does not correlate strongly with the presence of multiple polyadenylation signals or any other parameter we measured.
Nitrogen starvation has a more dramatic effect on vegetative than meiotic genes
As in our previous studies (Averbeck et al., 2005; McPheeters et al., 2009), the foregoing experiments did not impose nitrogen starvation prior to inducing meiosis by shifting the pat1–114 mutant to the nonpermissive temperature. As this protocol differs from other studies that profiled meiotic gene expression, we analyzed accumulation and 3′ processing for representative transcripts following preincubation in nitrogen-free medium to synchronize cells in G1 (Mata et al., 2002). Unexpectedly, the most striking difference was observed in the open reading frame (ORF) RNA detection assay for the control SPAC6F6 transcript (Figure 3G, top), which, rather than accumulating continuously (Figure 1B, row 34), showed diminished accumulation in early and late meiosis. Interestingly, the polyadenylation assay shows continuous accumulation (Figure 3G, bottom), similar to the assays from unstarved cells (Figure 2, row 34), consistent with the microarray study that included preincubation in nitrogen-free medium but analyzed mainly oligo(dT)-primed cDNA (Mata et al., 2002). As transcription of vegetative genes, as well as a subset of the middle and one of the late meiotic transcripts, is below detection limits in late meiosis, these results suggest the existence of a stabilization mechanism, perhaps driven by enhanced efficiency of 3′ processing.
FIGURE 3:
Effect of including a nitrogen starvation step prior to shifting pat1–114 cells to the nonpermissive temperature. (Top) Accumulation of total RNA for representative early, middle, and vegetative transcripts using the same RT-PCR primers as in Figure 1B, which amplify the the 3′ ends of the coding regions. (Bottom) Accumulation of poly(A)+ RNA was analyzed for the same transcripts using RT-PCR with oligo(dT) as the 3′ primer, as in Figure 2.
In contrast to the control gene, the ORF detection and polyadenylated RNA profiles from the nitrogen-starved time course were similar for all three early and four middle meiotic genes (Figure 3, A–F, compare top and bottom panels). There was, however, a difference relative to the unstarved cultures: In each case, the peak was delayed by ∼1 h (compare Figure 2A of McPheeters et al., 2009, for crs1; Figures 1B and 2, rows 2, 4, 5, 22, and 30 with Figure 3, A–G for the remaining genes). Surprisingly, although the synchrony of meiosis as measured by the nuclear division cycle is diminished by the lack of a starvation step (Averbeck et al., 2005; Supplemental Figure S1), cellular asynchrony does not appear to be reflected in the molecular analyses of RNA accumulation, as the peaks are equally sharp in the starved and unstarved cultures (compare Figures 1B and 2, rows 2, 4, 5, 22, and 30 with Figure 3, A–G).
Mutations in RNA surveillance factors differentially affect meiotic transcript accumulation
In our earlier study, we showed that in mitotically growing mmi1-ts mutants shifted to the nonpermissive temperature, crs1 RNA is cleaved and polyadenylated at the same sites as in meiotic cells, in addition to accumulating to substantially higher levels (McPheeters et al., 2009). In light of the high frequency of genes in our survey that, like crs1, are transcribed in vegetative and early meiotic cells (yet fail to accumulate RNA), we examined the contribution of this specialized surveillance pathway. As a prelude to seeking new mmi1-sensitive transcripts, we tested whether the mRNAs identified in the original study (Harigaya et al., 2006) acquire poly(A) tails only at the nonpermissive temperature in mmi1-ts mutants (Figure 4A). All 12 transcripts examined accumulate dramatically increased levels of poly(A)+ RNA under restrictive conditions in the mmi1-ts3 and -ts6 mutant strains, but not in a wild-type strain subjected to the same temperature shift, similar to crs1 (McPheeters et al., 2009; top panel). Thus, we proceeded to use enhanced polyadenylation in the mutants as a diagnostic tool to identify candidate mmi1 targets (Figure 4B).
FIGURE 4:
Effects of mutating factors implicated in meiotic and general nuclear RNA turnover on accumulation of meiotic transcripts. (A) Analysis of poly(A)+ RNA accumulation for mmi1 targets identified in the original study (Harigaya et al., 2006). The two temperature-sensitive mutants (ts3 and ts6) or an isogenic wild-type control (WT) were grown to early log phase at the permissive temperature (26ºC) and then shifted to the nonpermissive temperature (36ºC) for 3 h prior to RNA extraction. Polyadenylation assays were performed as in Figure 2. One of the 13 genes up-regulated in mmi1-ts mutants, SPCP20C8.03 (Harigaya et al., 2006), is not included as we were unable to map a polyadenylation site, consistent with the recent reclassification of the locus as a pseudogene. (B) Analysis of poly(A)+ RNA accumulation for a subset of the meiotic genes (see text under “Mutations in RNA surveillance factors…“) in the mmi1-ts, rrp6Δ, and cid14Δ strains. The mmi1 strains were processed as in A, while the deletion mutants were grown to early log phase at 30ºC (the standard growth temperature for fission yeast) before harvesting RNA. To assess the impact of mutations, we assigned a numerical value from 0 (undetectable) to 5 to the signal intensity in each lane; these values are shown at the right.
In addition to rec8, one of the eight previously identified mmi1-sensitive transcripts (Harigaya et al., 2006), two other early meiotic genes in our survey accumulate higher levels of polyadenylated RNA at the restrictive temperature in the mmi1-ts mutants (meu13 and ght3, Figure 4B, rows 1 and 3), whereas fbp1 does not (Figure 4B, row 4). Due to undetectable signals in both wild type and mmi1-ts strains, Figure 4B does not include data for the late genes, nor for 10 of the 26 middle genes. Among the 16 remaining middle genes, five, in addition to the known mmi1 target mei4 (row 5), show no detectable polyadenylated RNA in wild-type cells and distinct bands in both mmi1-ts mutants (Figure 4B, rows 7, 9, 11, 12, and 23). Two other middle genes show visible bands in the wild-type lane that are clearly enhanced in both mmi1-ts mutants (Figure 4B, rows 8 and 24), and the remaining middle genes are not candidates for destruction by the mmi1 pathway as bands of approximately equal intensity are present in all three lanes. Interestingly, the genes with mapped DSRs cluster near the top of the mRNA accumulation/Pol II occupancy ranking (Table 2, columns 6–8). Neither vegetative transcript shows increased poly(A)+ RNA in the mmi1-ts mutants after the temperature shift (Figure 4B, rows 33 and 34), consistent with specificity of this pathway for meiotic transcripts.
As our previous study demonstrated that the exosome is also critical for preventing crs1 expression in proliferating cells (McPheeters et al., 2009), we tested the effect of deleting the rrp6 gene encoding a nucleus-specific 3′ to 5′ ribonuclease on accumulation and 3′ processing of the meiotic transcripts in our survey. Notably, the four transcripts that showed the most dramatic increases in the levels of poly(A)+ RNA in the mmi1-ts mutants are also most sensitive to rrp6Δ (Figure 4B, rows 1–3, 5). Of the other mmi1-sensitive transcripts, SPAPB17E12.09 shows a band of comparable intensity in the rrp6Δ strain (Figure 4B, row 7), whereas for meu21, mug131, and meu6, the band present in rrp6Δ is less prominent than in the mmi1-ts strains (Figure 4B, rows 8, 9, and 12). Thus, among meiotic genes, there is a strong correlation between sensitivity to mmi1-ts mutants and rrp6Δ, but the magnitude of the effect is not uniform. Of the two control genes, only act1 is sensitive to rrp6Δ (Figure 4B, row 33).
Finally, we tested the effect of deleting cid14, which encodes the sole fission yeast ortholog of Trf4 and Trf5, the noncanonical poly(A) polymerases found in the TRAMP (Trf4/5, Air2, and Mtr4 polyadenylation) complex, which marks multiple classes of RNA for exosome-mediated turnover in budding yeast (LaCava et al., 2005) but did not affect accumulation of polyadenylated crs1 RNA (McPheeters et al., 2009). Intriguingly, meu4, a poly(A)+ transcript proposed to function as a noncoding RNA (Watanabe et al., 2001), shows a band in cid14Δ but not in any of the other strains examined (Figure 4B, row 13), and pronounced bands are also observed in the cid14Δ strain for fbp1, SPAC1610.02c, meu21, SPAC10F6.15, mug142, and mde7 (Figure 4B, rows 4, 6, 8, 18, 24, and 26). The absence of either Cid14p or Rrp6p leads to increased act1 mRNA levels (Figure 4B, row 33), consistent with roles for the TRAMP complex and nuclear exosome in turnover of some RNA polymerase II transcripts in fission yeast (Wang et al., 2008).
Splicing and polyadenylation of meiotic mRNAs are not obligatorily coupled
In our earlier study, we discovered that polyadenylation and splicing of crs1 primary transcripts are mechanistically coupled, which was unexpected in that this mode of RNA processing was believed to reflect exon definition and, therefore, to be restricted to metazoans (McPheeters et al., 2009). To test whether coupling of polyadenylation and splicing is a general feature of meiotic gene regulation, we assayed splicing of meu6 and atg8, the two intron-containing transcripts in our survey that had not been examined previously for meiosis-specific splicing. RT-PCR analysis of RNA from a meiotic time course with primers corresponding to the 5′ and 3′ exons of meu6 and atg8 revealed that it is mature mRNA, not unspliced or partially spliced RNA, that accumulates in vegetative cells and throughout meiosis (Figure 5). In contrast, crs1 RNA was barely detectable in proliferating cells, and the few transcripts present were neither spliced nor polyadenylated (McPheeters et al., 2009). Thus, although both meu6 and atg8 transcripts show meiosis-specific polyadenylation (Figure 2, rows 12 and 30), they are constitutively spliced. The lack of obligatory coupling between splicing and polyadenylation is consistent with the large number of genes in our survey that lack introns yet display meiosis-specific polyadenylation.
FIGURE 5:
Analyses of meu6 and atg8 splicing over a meiotic time course. Splicing was assayed by RT-PCR using primers complementary to the terminal exons (both genes contain two introns; Table 1). Signal-recognition particle (SRP) RNA was used as an internal loading control as in our previous study (McPheeters et al., 2009).
DISCUSSION
In this study, we analyzed nascent RNA synthesis, accumulation, and processing, as well as the effects of trans-acting RNA surveillance mutants, to assess the relative contributions of DNA-level and RNA-level regulation during the transition from vegetative growth to meiosis in fission yeast. As discussed in all four sections below, our results highlight the importance of events once believed to occur post-transcriptionally but that are now known to be initiated while transcript synthesis is still ongoing (reviewed in Perales and Bentley, 2009).
Biological implications: A regulatory cascade initiated by transcription of meiotic genes during vegetative growth
In addition to relief of mmi1-mediated destruction of meiotic transcripts, the pathway through which wild-type fission yeast cells initiate meiotic development in response to nitrogen starvation involves up-regulation of multiple genes by the ste11-encoded transcription factor (e.g., Li and McLeod, 1996; Mata and Bähler, 2006; Helmlinger et al., 2008). In most of the experiments presented here, this pathway is bypassed and accumulation of unphosphorylated Mei2p, which is necessary and sufficient to trigger meiosis (Iino and Yamamoto, 1985; Watanabe et al., 1997), is achieved solely through heat inactivation of the pat1–114 mutant kinase (Figure 6, top left). Mei2p sequesters Mmi1p in a nuclear “dot” that also contains the noncoding meiRNA (Harigaya et al., 2006), leading to stabilization of a number of transcripts including mei4 (Figure 6, center). This mRNA is translated to produce the Mei4p transcription factor (Figure 6, center), which in turn regulates a large number of genes expressed in mid-meiosis (Figure 6, bottom). Taking all of these findings into account, we propose that the mmi1 pathway, which directly blocks 3′ processing and promotes nuclear turnover of specific transcripts and affects others indirectly (see next section), sits at the apex of a regulatory cascade that operates alongside the well-known, starvation-induced transcriptional cascade initiated by ste11. As the direct mmi1 target spo5 (Yamanaka et al., 2010) encodes an RNA-binding protein that forms Mei2 dotlike foci (Kasama et al., 2006), it may encode a component of the regulatory apparatus.
FIGURE 6:
Model for a meiotic gene regulatory cascade initiated by inactivation of a pathway that promotes turnover of meiotic transcripts produced in vegetatively growing cells. During proliferation (top), Mei2p is maintained in the inactive phosphorylated state by the Pat1p kinase (Iino and Yamamoto, 1985), and Mmi1p is available to target certain transcripts for destruction by the nuclear exosome, which contains Rrp6p. Upon entry into meiosis (below dotted line), Mmi1p is sequestered in a nuclear dot via association with Mei2p and the noncoding meiRNA (Harigaya et al., 2006), thereby allowing its substrate RNAs to accumulate. The only middle meiotic transcript known to be a direct target of the mmi1 pathway is mei4, which encodes a transcription factor that in turn regulates a large number of target genes (bottom). Some of these transcripts, highlighted in bold in both halves of the figure, also display enhanced accumulation in one or more surveillance mutants (Figure 4B). For most of the mei4 target genes, there is no discernible peak in transcription (Figure 1A; middle right); these genes include mei4 itself, which is subject to autoregulation (Abe and Shimoda, 2000). Two genes containing the Mei4p recognition element (see text under “Not all targets of the Mei4p transcription factor…“) show a spike in transcription at 3 h, whereas seven show a peak at 5 h (bottom left). Only two genes fit none of the regulatory paradigms defined to date (bottom middle).
Both stages of the proposed regulatory cascade impact the chromosome transactions that provide familiar meiotic landmarks. Two direct mmi1 targets, rec8 and meu13, encode proteins with roles in meiotic recombination (Davis et al., 2008), whereas mei4-regulated transcripts include spo6, encoding a protein kinase regulatory subunit required for spindle formation (Nakamura et al., 2000). Although our previous proposal that meiosis-specific splicing controls the abundance of rec8, meu13, and spo6 (Averbeck et al., 2005) cannot be ruled out, the current data suggest that activation of 3′ RNA processing occupies a higher position in the regulatory hierarchy. Indeed, two transcripts that exhibit meiosis-specific polyadenylation (Figure 2, rows 12 and 30) are constitutively spliced (Figure 5). Downstream events in meiotic gene expression may also be part of the post-transcriptional cascade, as the mei4-regulated genes in our survey include several that encode predicted RNA binding proteins (Table 1 and Figure 1, rows 23, 26, and 28).
The mmi1 pathway is likely to function in tandem with general nuclear RNA surveillance
In light of the unusual features of crs1 regulation (McPheeters et al., 2009), a central goal of our study was to assess the prevalence of this control mechanism. Whereas five genes in our survey were originally identified as mmi1 pathway substrates (Harigaya et al., 2006), a more recent investigation revealed that only rec8 and mei4 are direct targets (Yamanaka et al., 2010), together with crs1 (McPheeters et al., 2009). A fourth gene in our survey, meu13, is similar to crs1, rec8, and mei4 in showing dramatically increased RNA accumulation in the rrp6Δ mutant but insensitivity to cid14Δ (McPheeters et al., 2009), consistent with a shared regulatory mechanism culminating in elimination from mitotically growing cells by the exosome. Whereas the kinetics of transcription, RNA accumulation, and 3′ processing profiles for meu13 and rec8 are similar to those of crs1 (McPheeters et al., 2009), the longer lag prior to RNA accumulation for mei4 (Figure 1, rows 1, 2, and 5) may be related to the unusual location of the mei4 DSR, which is located near the middle of the ORF, in contrast to the 3′ location of the four other mapped DSRs (Harigaya et al., 2006; McPheeters et al., 2009).
A physical feature that the meu13, rec8, and mei4 genes share with crs1 is the presence of two closely spaced polyadenylation signals (Figure 2), which may facilitate targeting of nascent transcripts for destruction. Indeed, our proposal that bypass of meiosis-specific polyadenylation signals in proliferating cells promotes turnover of newly synthesized transcripts (McPheeters et al., 2009) has been bolstered by two recently published studies. First, in mammalian cell extracts, a functional but unused poly(A) signal triggers degradation of 3′ extended nascent transcripts (Kazerouninia et al., 2010). Second, in Drosophila embryos, suppression of polyadenylation serves as a regulatory mechanism to limit production of Notch mRNA (Shepherd et al., 2009). At first glance, our data and interpretation appear to contradict the recent proposal that polyadenylation of mmi1 target transcripts precedes delivery to the exosome (Yamanaka et al., 2010). To reconcile the two models, we propose that they operate in tandem, with the general surveillance mechanism originally identified in the context of small nucleolar RNA (snoRNA) metabolism (Lemay et al., 2010) mediating destruction of mmi1 substrates that escape the meiotic transcript-specific mechanism. This interpretation is consistent with the finding that the general pathway, which requires recognition by Pab2p, a nuclear poly(A) binding protein, affects many meiotic transcripts that are not targets of the mmi1 pathway (St-André et al., 2010).
In the initial report, the mmi1 pathway was proposed to protect mitotic cells from the deleterious effects of aberrantly produced meiotic gene products (Harigaya et al., 2006). Our discovery, however, that an mmi1 target gene is transcribed in vegetative and early meiotic cells but fails to accumulate RNA raised a different possibility, that certain meiotic genes are maintained in an active configuration to ensure rapid availability of their products (McPheeters et al., 2009). The results reported here indicate that transcription of meiotic genes in growing cells is widespread and not limited to direct targets of the mmi1 pathway. Mechanistically, this surprising phenomenon may be related to the transcription of “silent” heterochromatin into RNAs that are rapidly destroyed by the exosome (Bühler et al., 2007; Zofall et al., 2009). Although we believe that mmi1-mediated regulation has a role beyond preventing untimely meiotic gene expression, the general (pab2-mediated) surveillance pathway may well function in a “fail-safe” capacity to eliminate inappropriately expressed meiotic transcripts in fission yeast including, but not limited to, those that escape mmi1-mediated destruction (St-André et al., 2010).
Two early meiotic genes in our survey do not fit the crs1 regulatory paradigm discussed earlier in this article. Both ght3 and fbp1 transcripts, which encode products involved in sugar metabolism, persist into late meiosis following peak polyadenylation at 4–5 h (Figure 2, rows 3 and 4). In addition, the two widely separated 3′ processing sites in fbp1 are not activated concurrently, and use of the proximal signal is exclusive to the late stages of meiosis (9 and 7 h with and without nitrogen starvation, respectively; Figure 2, row 4 and Figure 3C), in contrast to the previously identified, less tightly regulated examples of differential polyadenylation in S. pombe (Girard et al., 1993; Jang et al., 1996; Munoz et al., 2002). Truncation of the 3′ UTR might alter stability or translation of fbp1 RNA, analogous to the widespread shortening of oncogene 3′ UTRs observed in cancerous mammalian cells (Mayr and Bartel, 2009) or the widespread cytoplasmic polyadenylation that orchestrates late meiotic events in metazoans (reviewed in Richter, 2000; Wickens et al., 2000).
Not all targets of the Mei4p transcription factor show meiotic peaks in RNA synthesis
Importantly, our study is the first to provide direct evidence for increased transcription of meiotic genes during differentiation. Interestingly, however, the three that show a spike 3 h after induction (Figure 1, rows 6, 12, and 13) lack the recognition element for the DSC1/MBF (DNA synthesis control-like/Mlu1 box binding) transcription factor known to regulate other early meiotic genes, including the mmi1 substrates meu13 and rec8 (Sugiyama et al., 1994; Ding and Smith, 1998; Cunliffe et al., 2004). Instead, two of these genes contain perfect copies of the motif recognized by Mei4p in their upstream DNA (Horie et al., 1998; Mata et al., 2007), as do all seven genes that show a spike in RNA synthesis 5 h after initiation of meiosis (Figure 6, lower left). One of these, spo6, contains introns (Table 1, row 22), and appears to share the transcription-coupled splicing regulatory mechanism proposed for rem1 (Moldon et al., 2008). Despite containing presumptive Mei4p recognition elements, however, meu6 and atg8 are spliced constitutively (Figure 5) and thus are not subject to this form of regulation.
Our survey included 18 other meiotic genes that contain upstream Mei4p recognition motifs and are bona fide targets of this transcription factor based on their failure to show increased RNA accumulation during meiosis in a mei4Δ strain (Mata et al., 2007). Further investigation will be required to determine whether these genes, which include members of the early, middle, and late classes (Mata et al., 2002), share a regulatory mechanism with the genes that show a mid-meiotic transcriptional spike as all of them, together with mei4 itself, show maximal transcription in vegetative and early meiotic cells but little or no RNA accumulation until 4–5 h after meiotic induction (Figure 1, rows 3, 4, 7–11, 14–19, 23–25, 30, and 32). In the case of mei4, this pattern may result from the superimposition of mmi1-mediated targeting for destruction and transcriptional auto-regulation (Abe and Shimoda, 2000). Although none of the other mei4-regulated transcripts appears to be directly targeted by the mmi1 pathway (Harigaya et al., 2006; Yamanaka et al., 2010), several show increased accumulation when surveillance factors are inactivated (Figure 4, rows 3, 4, 7–11, 14, 18, 23, and 25), presumably as a secondary consequence of increased Mei4p levels in these cells.
CONCLUSION
Counter to the previous proposal that the successive waves of mRNA accumulation observed in microarray studies represent the unfolding of a transcriptional program (Mata et al., 2002), less than a third of the genes in our survey display a peak in RNA synthesis during meiosis, and transcription of all but one is readily detectable in proliferating cells. Despite a feature common to all meiotic genes analyzed in fission yeast, that the temporal profile of RNA accumulation parallels polyadenylation (Figures 1B and 2), not transcription (Figure 1A), multiple mechanisms, in many cases acting combinatorially and/or sequentially, can be discerned (Figure 6). In light of the prominent role of changes in 3′ processing of primary transcripts, it is tempting to speculate that regulated nuclear polyadenylation may have evolved into the cytoplasmic polyadenylation-dependent mechanism that stimulates translation of meiotic mRNAs during the mitosis–meiosis decision in Caenorhabditis elegans (Suh et al., 2006). Indeed, one of the middle meiotic fission yeast genes (mpf1) is a member of the Pumilio family, which functions in translational control during nematode meiotic development (Wickens et al., 2001), thus potentially representing the first step toward a dominant role for post-transcriptional regulatory mechanisms based in the cytoplasm rather than the nucleus.
MATERIALS AND METHODS
Fission yeast strains and manipulations
For the experiments shown in Figures 1, 2, and 5 and in Supplemental Figures S1–S4, ectopic meiosis was induced by shifting the F90 (h− pat1–114 leu1–32 ura4-D18) strain from the permissive to the nonpermissive temperature (23.5ºC → 34ºC), as in earlier studies (Averbeck et al., 2005; McPheeters et al., 2009). For the experiment shown in Figure 3, a 2-h nitrogen starvation step was imposed prior to the temperature shift (Mata et al., 2002). For the temperature-sensitive mmi1-ts3 and -ts6 mutants (Figure 4), the temperature was shifted from 26ºC to 36ºC as previously described (McPheeters et al., 2009).
TRO analysis
The TRO assay protocol was as described in McPheeters et al. (2009). The probes immobilized on the filters hybridized with the labeled TRO products were PCR fragments corresponding to coding sequences near the 3′ end of each gene. The average length was 515 nt (range 379–770 nt), with variations as needed to optimize primer annealing for amplification from genomic DNA. Although these probes do not provide strand-specific information, genomic tiling microarray analyses indicated that the corresponding segments produce little or no antisense RNA during either vegetative growth or meiosis (Dutrow et al., 2008; Wilhelm et al., 2008). Disregarding differences in probe length and base composition, the signals obtained in any given assay correspond to the rate of transcription over that region of an individual gene.
RNA analyses
The accumulation of total RNA for each gene (Figure 1B) was determined by RT-PCR using the same primers as those used to generate probes for the TRO assays. Polyadenylated RNAs (Figure 2) were analyzed as described previously (McPheeters et al., 2009). qRT-PCR assays were carried out as described previously (McPheeters et al., 2009) except that, in the present studies, fold changes rather than absolute levels of RNA were measured.
Oligonucleotides
The sequences of primers used for plasmid construction, mutagenesis, and RT-PCR assays are available upon request.
Supplementary Material
Acknowledgments
We thank David S. McPheeters for performing the experiments shown in Figures 1A and 5 and Supplemental Figures S1–S3 as well as some of the assays shown in Figures 2 and 4. We are grateful to our colleagues Jeff Coller, Jonathan Karn, Hua Lou, Tim Nilsen, and Helen Salz for helpful discussions and critical reading of the manuscript. This research was funded by National Institutes of Health grants R01-GM38070 and R01-GM073217 (awarded to J.A.W.) and R01-GM064682 (awarded to David McPheeters).
Abbreviations used:
- DSR
determinants of selective removal
- ORF
open reading frame
- qRT-PCR
real-time PCR
- rNTP
ribonucleoside triphosphate
- snoRNA
small nucleolar RNA
- SRP
signal-recognition particle
- TRO
transcription run-on.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-05-0448) on December 9, 2010.
REFERENCES
- Abe H, Shimoda C. Autoregulated expression of Schizosaccharomyces pombe meiosis-specific transcription factor Mei4p and a genome-wide search for its target genes. Genet. 2000;154:1497–1508. doi: 10.1093/genetics/154.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Àlvarez B, Moreno S. Fission yeast Tor2 promotes cell growth and represses cell differentiation. J Cell Sci. 2006;119:4475–4485. doi: 10.1242/jcs.03241. [DOI] [PubMed] [Google Scholar]
- Averbeck N, Sunder S, Sample N, Wise JA, Leatherwood J. Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol Cell. 2005;18:491–498. doi: 10.1016/j.molcel.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Bähler J, Schuchert P, Grimm C, Kohli J. Synchronized meiosis and recombination in fission yeast: observations with pat1–114 diploid cells. Curr Genet. 1991;19:445–451. doi: 10.1007/BF00312735. [DOI] [PubMed] [Google Scholar]
- Bühler M, Haas W, Gygi SP, Moazed D. RNA-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- Chalmel F, et al. The conserved transcriptome in human and rodent male gametogenesis. Proc Natl Acad Sci USA. 2007;104:8346–8351. doi: 10.1073/pnas.0701883104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe L, White S, McInerny CJ. DSC1-MCB regulation of meiotic transcription in Schizosaccharomyces pombe. Mol Genet Genomics. 2004;271:60–71. doi: 10.1007/s00438-003-0956-6. [DOI] [PubMed] [Google Scholar]
- Davis L, Rozalen AE, Moreno S, Smith GR, Martin-Castellanos C. Rec25 and Rec27, novel linear-element components, link cohesin to meiotic DNA breakage and recombination. Curr Biol. 2008;18:849–854. doi: 10.1016/j.cub.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, Smith GR. Global control of meiotic recombination genes by Schizosaccharomyces pombe rec16 (rep1) Mol Gen Genet. 1998;258:663–670. doi: 10.1007/s004380050780. [DOI] [PubMed] [Google Scholar]
- Dutrow N, Nix DA, Holt D, Milash B, Dalley B, Westbroek E, Parnell TJ, Cairns BR. Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping. Nat Genet. 2008;40:977–986. doi: 10.1038/ng.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard JP, Feliu J, Caizergues-Ferrer M, Lapeyre B. Study of multiple fibrillarin mRNAs reveals that 3′ end formation in Schizosaccharomyces pombe is sensitive to cold shock. Nucleic Acids Res. 1993;21:1881–1887. doi: 10.1093/nar/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K, Birse CE, Proudfoot NJ. Nascent transcription from the nmt1 and nmt2 genes of Schizosaccharomyces pombe overlaps neighbouring genes. EMBO J. 1998;17:3066–3077. doi: 10.1093/emboj/17.11.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- Helmlinger D, Marguerat S, Villen J, Gygi SP, Bähler J, Winston F. The S. pombe SAGA complex controls the switch from proliferation to sexual differentiation through the opposing roles of its subunits Gcn5 and Spt8. Genes Dev. 2008;22:3184–3195. doi: 10.1101/gad.1719908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- Horie S, Watanabe Y, Tanaka K, Nishiwaki S, Fujioka H, Abe H, Yamamoto M, Shimoda C. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol Cell Biol. 1998;18:2118–2129. doi: 10.1128/mcb.18.4.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino Y, Yamamoto M. Negative control for the initiation of meiosis in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1985;82:2447–2451. doi: 10.1073/pnas.82.8.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang YK, Jin YH, Myung K, Seong RH, Hong SH, Park SD. Differential expression of the rhp51+ gene, a recA and RAD51 homolog from the fission yeast Schizosaccharomyces pombe. Gene. 1996;169:125–130. doi: 10.1016/0378-1119(96)83099-x. [DOI] [PubMed] [Google Scholar]
- Kasama T, Shigehisa A, Hirata A, Saito TT, Tougan T, Okuzaki D, Nojima H. Spo5/Mug12, a putative meiosis-specific RNA-binding protein, is essential for meiotic progression and forms Mei2 dot-like nuclear foci. Eukaryot Cell. 2006;5(8):1301–1313. doi: 10.1128/EC.00099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazerouninia A, Ngo B, Martinson HG. Poly(A) signal-dependent degradation of unprocessed nascent transcripts accompanies poly(A) signal-dependent transcriptional pausing in vitro. RNA. 2010;16:197–210. doi: 10.1261/rna.1622010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida M, Nagai T, Nakaseko Y, Shimoda C. Meiosis-dependent mRNA splicing of the fission yeast Schizosaccharomyces pombe mes1+ gene. Curr Genet. 1994;25:497–503. doi: 10.1007/BF00351668. [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bähler J. A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell. 2007;26:145–155.. doi: 10.1016/j.molcel.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay JF, D’Amours A, Lemieux C, Lackner DH, St.-Sauver VG, Bähler J, Bachand F. The nuclear poly(A)-binding protein interacts with the exosome to promote synthesis of noncoding small nucleolar RNAs. Mol Cell. 2010;15:34–45. doi: 10.1016/j.molcel.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Li P, McLeod M. Molecular mimicry in development: identification of ste11+ as a substrate and mei3+ as a pseudosubstrate inhibitor of ran1+ kinase. Cell. 1996;87:869–880. doi: 10.1016/s0092-8674(00)81994-7. [DOI] [PubMed] [Google Scholar]
- Malapeira J, Moldón A, Hidalgo E, Smith GR, Nurse P, Ayté J. A meiosis-specific cyclin regulated by splicing is required for proper progression through meiosis. Mol Cell Biol. 2005;25:6330–6337. doi: 10.1128/MCB.25.15.6330-6337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Bähler J. Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proc Natl Acad Sci USA. 2006);103:15517–15522. doi: 10.1073/pnas.0603403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bähler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- Mata J, Wilbrey A, Bähler J. Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol. 2007;8:R217. doi: 10.1186/gb-2007-8-10-r217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters DS, Cremona N, Sunder S, Chen HM, Averbeck N, Leatherwood J, Wise JA. A complex gene regulatory mechanism that operates at the nexus of multiple RNA processing decisions. Nat Struct Mol Biol. 2009;16:255–264. doi: 10.1038/nsmb.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldón A, Malapeira J, Gabrielli N, Gogol M, Gómez-Escoda B, Ivanova T, Seidel C, Ayté J. Promoter-driven splicing regulation in fission yeast. Nature. 2008;455:997–1000. doi: 10.1038/nature07325. [DOI] [PubMed] [Google Scholar]
- Munoz MJ, Daga RR, Garzon A, Thode G, Jimenez J. Poly(A) site choice during mRNA 3′-end formation in the Schizosaccharomyces pombe wos2 gene. Mol Genet Genomics. 2002;267:792–796. doi: 10.1007/s00438-002-0710-5. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Kishida M, Shimoda C. The Schizosaccharomyces pombe spo6+ gene encoding a nuclear protein with sequence similarity to budding yeast Dbf4 is required for meiotic second division and sporulation. Genes Cells. 2000;5:463–479. doi: 10.1046/j.1365-2443.2000.00343.x. [DOI] [PubMed] [Google Scholar]
- Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, et al. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Richter JD. Translational control in vertebrate development. In: Translational Control of Gene Expression, ed. In: Hershey JWB, Mathews MB, Sonenberg N, editors. New York: Cold Spring Harbor Laboratory Press; 2000. pp. 785–806. [Google Scholar]
- Shepherd AK, Singh R, Wesley CS. Notch mRNA expression in Drosophila embryos is negatively regulated at the level of 3′ processing. PLoS ONE. 2009;4(11) doi: 10.1371/journal.pone.0008063. doi:10.1371/journal.pone.0008063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoseki M, Shimoda C. The 5′ terminal region of the Schizosaccharomyces pombe mes1 mRNA is crucial for its meiosis-specific splicing. Mol Genet Genomics. 2001;265:673–682. doi: 10.1007/s004380100462. [DOI] [PubMed] [Google Scholar]
- St-André O, Lemieux C, Perreault A, Lackner DH, Bähler J, Bachand F. Negative regulation of meiotic gene expression by the nuclear poly(A)-binding protein in fission yeast. J Biol Chem. 2010;285:27859–27868. doi: 10.1074/jbc.M110.150748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama A, Tanaka K, Okazaki K, Nojima H, Okayama H. A zinc finger protein controls the onset of premeiotic DNA synthesis of fission yeast in a Mei2-independent cascade. EMBO J. 1994;13:1881–1887. doi: 10.1002/j.1460-2075.1994.tb06457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Jedamzik B, Eckmann CR, Wickens M, Kimble J. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc Natl Acad Sci USA. 2006;103:15108–15112. doi: 10.1073/pnas.0607050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Stevenson AL, Kearsey SE, Watt S, Bähler J. Global role for polyadenylation-assisted nuclear RNA degradation in posttranscriptional gene silencing. Mol Cell Biol. 2008;28:656–665. doi: 10.1128/MCB.01531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Miyashita K, Saito TT, Yoneki T, Kakihara Y, Nabeshima K, Kishi YA, Shimoda C, Nojima H. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 2001;29:2327–2337. doi: 10.1093/nar/29.11.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Shinozaki-Yabana S, Chikashige Y, Hiraoka Y, Yamamoto M. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature. 1997;386:187–190. doi: 10.1038/386187a0. [DOI] [PubMed] [Google Scholar]
- Wellmer F, Alves-Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM. Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet. 2006;2:e117. doi: 10.1371/journal.pgen.0020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M, Bernstein D, Crittenden S, Luitjens C, Kimble J. PUF proteins and 3′UTR regulation in the Caenorhabditis elegans germ line. Cold Spring Harb Symp Quant Biol. 2001;66:337–343. doi: 10.1101/sqb.2001.66.337. [DOI] [PubMed] [Google Scholar]
- Wickens M, Goodwin EB, Kimble J, Strickland S, Hentze M. In: Translational Control of Gene Expression. ed. In: Hershey JWB, Mathews MB, Sonenberg N, editors. New York:: Cold Spring Harbor Laboratory Press; 2000. pp. 295–370. Translational control in developmental decisions. [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bähler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- Wood V, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J. 2010;29:2173–2181. doi: 10.1038/emboj.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Fischer T, Zhang K, Zhou M, Cui B, Veenstra TW, Grewal SIS. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature. 2009;461:419–422. doi: 10.1038/nature08321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.