We show that SUMO-specific protease SENP6 can cleave mixed SUMO-1 and SUMO-2/3 chains. Depletion of SENP6 results in accumulation of SUMO-2/3 and SUMO-1 conjugates in promyelocytic leukemia (PML) nuclear bodies. Inactivation of SENP6 results in its accumulation at the SUMO-2/3-rich core of PML nuclear bodies. Biochemical analysis indicates that SUMO-modified PML is a SENP6 substrate.
Abstract
Promyelocytic leukemia protein (PML) is the core component of PML-nuclear bodies (PML NBs). The small ubiquitin-like modifier (SUMO) system (and, in particular, SUMOylation of PML) is a critical component in the formation and regulation of PML NBs. SUMO protease SENP6 has been shown previously to be specific for SUMO-2/3–modified substrates and shows preference for SUMO polymers. Here, we further investigate the substrate specificity of SENP6 and show that it is also capable of cleaving mixed chains of SUMO-1 and SUMO-2/3. Depletion of SENP6 results in accumulation of endogenous SUMO-2/3 and SUMO-1 conjugates, and immunofluorescence analysis shows accumulation of SUMO and PML in an increased number of PML NBs. Although SENP6 depletion drastically increases the size of PML NBs, the organizational structure of the body is not affected. Mutation of the catalytic cysteine of SENP6 results in its accumulation in PML NBs, and biochemical analysis indicates that SUMO-modified PML is a substrate of SENP6.
INTRODUCTION
The small ubiquitin-like modifier (SUMO) system is responsible for the modification of a large pool of cellular proteins. Modification by the near identical SUMO-2 and -3 and the distinct family member SUMO-1 achieves a diverse range of effects from regulating subcellular localization to transcription factor activity, protein stability, and cell stress responses. SUMO modification is achieved through an enzymatic pathway consisting of an E1 activating enzyme (SAE-2/1), an E2 conjugating enzyme (Ubc9), and a number of E3 ligases. Ubc9 is capable of directly modifying substrates through interaction with a SUMO conjugation motif (ΨKXD/E, where Ψ is a large hydrophobic amino acid and X is any amino acid); however, E3 ligases appear to add specificity and increase the efficiency of the conjugation reaction. SUMO-2 and -3 both possess an N-terminal SUMO conjugation motif (VKTE) that allows their polymerization to form SUMO chains (Tatham et al., 2001; Bylebyl et al., 2003; Matic et al., 2008). SUMO-1 and SUMO-2/3 are also conjugated to a distinct range of substrates and are therefore likely to have different roles, although knockout of SUMO-1 can be compensated for by SUMO-2/3 (Evdokimov et al., 2008; Zhang et al., 2008a), and any SUMO paralogue can rescue the effects of knockout of all three SUMO members in zebrafish (Yuan et al., 2010). Although SUMO modification of particular lysine residues may prevent other post-translational modifications (Desterro et al., 1998), SUMO generally acts by altering binding interfaces on substrate proteins. Recently a single SUMO interaction motif (SIM) was identified: It consists of a short hydrophobic core (V/L/I-X-V/L/I-V/L/I or V/L/I-V/L/I-X-V/L/I), often flanked by several acidic residues. This hydrophobic core forms a β-strand that can insert in either orientation into a hydrophobic groove between the second β-strand and the α-helix in SUMO (Song et al., 2004; Hannich et al., 2005; Song et al., 2005; Hecker et al., 2006). Recently it has been shown that increasing the acidity of the region flanking the hydrophobic core of the SIM by phosphorylation increases the affinity of the SIM for SUMO (Hecker et al., 2006; Stehmeier and Muller, 2009).
The balance between SUMO conjugation and deconjugation is maintained by a family of six SUMO proteases (SENP1—3 and 5–7) that are also responsible for the maturation of the immature SUMO translation product. The six proteases possess distinct substrate specificities due not only to differential preference for SUMO family members but also to differing subcellular localizations. SENP1 and SENP2 have been shown to localize to the nuclear pore complex and nucleoplasm, as well as to temporarily shuttle to the cytoplasm (Bailey and O’Hare, 2002; Hang and Dasso, 2002; Zhang et al., 2002; Itahana et al., 2006; Li et al., 2008). In contrast, the two related SENPs (SENP3 and SENP5) localize to the nucleolus (Di Bacco et al., 2006; Gong and Yeh, 2006; Haindl et al., 2008; Kuo et al., 2008; Yun et al., 2008) although SENP5 also has been reported to be involved in mitochondrial morphology (Zunino et al., 2007, 2009). Both SENP6 and 7 have been shown to localize to the nucleoplasm (Choi et al., 2006; Mukhopadhyay et al., 2006; Shen et al., 2009). All SUMO proteases except SENP1 (which shows little discrimination) appear to show a substrate preference for SUMO-2/3 deconjugation over that of SUMO-1 (Di Bacco and Gill, 2006; Gong and Yeh, 2006; Mukhopadhyay et al., 2006; Reverter and Lima, 2006; Shen et al., 2006, 2009). SENP6 and SENP7 appear to differ, however, from other proteases in their preference for SUMO-2/3 chain depolymerization over that of SUMO-2/3 deconjugation (Mukhopadhyay et al., 2006; Lima and Reverter, 2008; Shen et al., 2009) and in this respect are more homologous to one of the two yeast SUMO proteases (Ulp2) that depolymerize SUMO chains (Li and Hochstrasser, 2000; Bylebyl et al., 2003; Schwartz et al., 2007). The recent determination of specific roles for SUMO-2/3 polymers in vivo indicates that this form of Ubl modification is distinct from the signals provided by SUMO-1 or SUMO-2/3 mono-modification and that control of SUMO-2/3 polymers must be tightly regulated (Lallemand-Breitenbach et al., 2008; Tatham et al., 2008; Zhang et al., 2008b; Skilton et al., 2009; Lin et al., 2010; Sekiyama et al., 2010). One of the first identified roles for polySUMO modification in higher eukaryotes was the arsenic-induced ubiquitination and degradation of poly-SUMOylated promyelocytic leukemia (PML) by the ubiquitin E3-ligase RNF4 (Lallemand-Breitenbach et al., 2008; Tatham et al., 2008).
PML is a member of the tripartite motif (TRIM) family of proteins and, as such, contains the characteristic RBCC motif consisting of Really Interesting New Gene (RING), B-Box (one or two), and a coiled coil domain. PML is the defining component of PML nuclear bodies (PML NBs) of which 1–20 are found in the nucleus depending on cell type/development and cellular stresses (Salomoni and Pandolfi, 2002; Bernardi and Pandolfi, 2007; Reineke and Kao, 2009). Diverse roles have been suggested for PML NBs, including involvement in the DNA damage response, apoptosis induction, angiogenesis, telomere maintenance, cell proliferation, and senescence. PML NBs are also important in a number of disease states, including viral infection and cancer (Salomoni and Pandolfi, 2002; Bernardi and Pandolfi, 2007). PML was one of the first identified targets of the SUMO pathway; indeed, an original name for SUMO was PML Interacting Clone (PIC)-1 (Boddy et al., 1996). PML has subsequently been identified as having three SUMO modification sites (Kamitani et al., 1998a; Duprez et al., 1999; Lallemand-Breitenbach et al., 2001), and these sites, along with the coiled-coil oligomerization domain, are required for the formation of mature interphase PML NBs (Kamitani et al., 1998a, 1998b; Duprez et al., 1999; Ishov et al., 1999; Zhong et al., 2000; Jensen et al., 2001; Ayaydin and Dasso, 2004; Fu et al., 2005).
It has been shown previously that the preferred substrates for SENP6 are polySUMO-2/3 chains, and depletion of SENP6 results in overexpressed SUMO accumulating in PML NBs (Mukhopadhyay et al., 2006; Lima and Reverter, 2008). Here, we probe the substrate specificity of SENP6 and demonstrate that it can cleave SUMO-1 and SUMO-2 mixed dimers suggesting that SENP6 can remove SUMO-1 “caps” from SUMO-2/3 polymers in vivo. We show that, consistent with this substrate specificity, depletion of SENP6 results in the accumulation of both endogenous SUMO-2/3 and SUMO-1 conjugates that are localized to an increased number of PML NBs. Whereas SENP6 depletion increases the size of PML NBs, high-resolution imaging reveals that the overall structure of the body is unaffected. Mutation of the catalytic cysteine of SENP6 results in its accumulation in PML NBs, and biochemical analysis indicates that SUMO-modified PML is a substrate of SENP6.
RESULTS
SENP6 can cleave mixed SUMO-1–SUMO-2 dimers
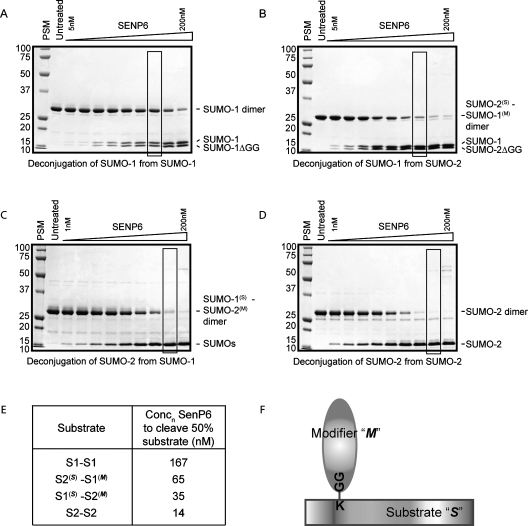
SENP6 has been defined previously as a SUMO-2/3 specific protease with a preference for depolymerization of SUMO-2/3 chains (Mukhopadhyay et al., 2006; Lima and Reverter, 2008). It remains unclear, however, whether this preference requires the presence of a single SUMO-2/3 moiety in the polymer or whether SENP6 can only cleave chains solely consisting of SUMO-2/3. Defining this preference is particularly important as SUMO-1 (which does not readily form polymers in vivo or in vitro) can “cap” SUMO-2/3 chains, terminating their growth and potentially altering their susceptibility to depolymerization by the SUMO specific proteases. To address whether SENP6 requires SUMO-2/3 moieties in both the “substrate” and “modifier” positions (see Figure 1F for schematic explanation) to depolymerize SUMO chains, we used SUMO-1, SUMO-2, and mixed SUMO dimers as substrates. A fixed concentration (20 μM) of each substrate was incubated with a range of concentrations of SENP6, and the extent of cleavage was determined by SDS–PAGE and Coomassie Blue staining (Figure 1, A–D). An estimate of the efficiency of substrate cleavage was calculated from the concentration of SENP6 required to cleave 50% of the substrate in 1 h (Figure 1E). For comparison, the cleavage of substrate by 100 nM SENP6 is indicated by the boxed region in Figure 1, A–D.
FIGURE 1:
SENP6 exhibits a preference for substrates containing SUMO-2/3. The ability of the catalytic domain to cleave SUMO dimers consisting of SUMO-1-SUMO-1 (A), SUMO-2(S)-SUMO-1(M) (B), SUMO-1(S)-SUMO-2(M) (C), and SUMO-2-SUMO-2 (D) was assessed by incubating 20 μM substrate with SENP6 (A and B – 0, 5, 10, 20, 40, 60, 80, 100, 150, 200 nM; C and D – 0, 1, 2, 4, 8, 12, 25, 50, 100, 200 nM) for 1 h at 37ºC. For ease of comparison, boxed regions indicate a reaction carried out with 100 nM protease. Substrate cleavage/product appearance was determined by SDS–PAGE and Coomassie Blue staining (molecular weight markers and position of SUMO dimer/monomers are indicated) and could then be used to assess the concentration of SENP6 required to cut 50% substrate in this time frame (E). A schematic is included for ease of identification of substrate (S) and modifier (M) in mixed SUMO dimers (F).
As expected, SENP6 shows little activity against SUMO-1 dimers as demonstrated by the relatively low level of substrate cleavage by 100 nM SENP6 after 1 h (Figure 1A). Comparatively, SENP6 shows ∼10-fold more activity against SUMO-2 dimers, and 100 nM SENP6 cleaves all SUMO-2 dimer substrate within the same timescale (Figure 1, B and E).
To assess the requirement for SUMO-2/3 in either the “substrate” (S) or “modifier” (M) position (as demonstrated in Figure 1F), mixed SUMO dimers were conjugated consisting of SUMO-1 and SUMO-2 in either the substrate or modifier position and used as substrates for SENP6. Compared to the activity of SENP6 against SUMO-1 dimers (Figure 1A), the replacement of SUMO-1 with SUMO-2 in the substrate position (SUMO-2(S)-SUMO-1(M)) resulted in an approximately threefold increase in the ability of SENP6 to cleave the dimer (Figure 1, C and E). Replacement of SUMO-1 with SUMO-2 in the modifier position (SUMO-1(S)-SUMO-2(M)) resulted in a greater increase in substrate cleavage (an almost fivefold increase) compared with SUMO-1 in the substrate position (Figure 1, D and E). The single replacements of SUMO-1 with SUMO-2, however, did not completely recover the activity of SENP6 seen against SUMO-2 dimers and were more than fourfold and twofold less for SUMO-2(S)-SUMO-1(M) and SUMO-1(S)-SUMO-2(M), respectively. Thus, these data confirm the specificity of SENP6 for SUMO-2/3–modified proteins, in particular for polymeric SUMO-2/3 chains (Mukhopadhyay et al., 2006; Lima and Reverter, 2008) but also indicate that SENP6 can cleave mixed chains of SUMO-1 and SUMO-2. The ability of SENP6 to cleave SUMO-2 modified with SUMO-1 may be important in vivo as one of the postulated roles of SUMO-1 is the capping of SUMO-2/3 polymers, preventing their further elongation and possibly protecting them from specific chain depolymerizing enzymes such as SENP6 and SENP7. The ability (albeit approximately fourfold reduced) of SENP6 to cleave SUMO-2/3 chains capped with SUMO-1 suggests that SENP6 could cleave these capped substrates in vivo.
SENP6 controls endogenous levels of SUMO conjugates and is required for cell viability
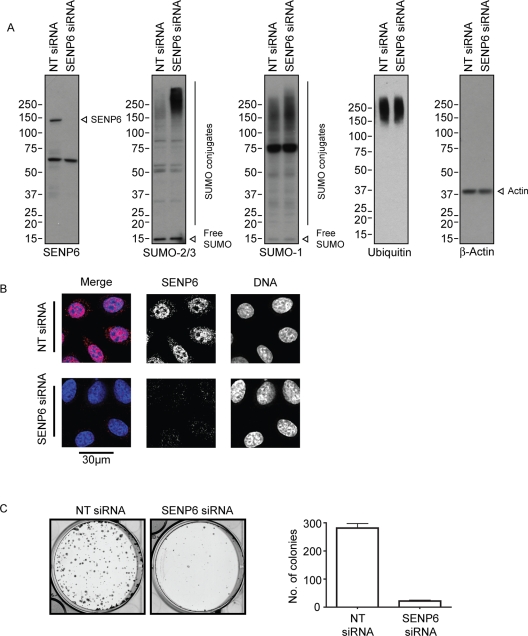
Depletion of SENP6 has been shown previously to affect the subcellular distribution of overexpressed SUMO-2/3 with little observed effect on the distribution of endogenous SUMO conjugates analyzed by immunoblotting or immunofluorescence (Mukhopadhyay et al., 2006). However, upon SENP6 depletion with a pool of four SENP6-targeting small interfering RNAs (siRNAs; Figure 2A), we observed a substantial increase in endogenous high-molecular-weight SUMO-2/3 conjugates and a more modest increase in high-molecular-weight SUMO-1 conjugates (Figure 2A). This shift in the profile of endogenous SUMO-1 conjugates is consistent with the findings in Figure 1 and reflects the predominant role of SENP6 as a SUMO-2/3 or chain depolymerizing enzyme. In this situation the modest increase in SUMO-1 conjugates may be due to an increased number of SUMO-1 “caps” on SUMO-2/3 chains reflecting the ability of SENP6 to cleave these substrates. By contrast, depletion of SENP6 had little discernible effect on the total ubiquitin conjugate profile (Figure 2A). Depletion of SENP6 using individual siRNA duplexes confirmed that the level of depletion correlated with the effects seen on the profile of SUMO conjugates (Supplemental Figure 1A). Analysis of SENP6 depletion by immunofluorescence confirmed effective depletion of the protease as well as its previously described localization to the nucleoplasm (Figure 2B; Mukhopadhyay et al., 2006). To determine the requirement for SENP6 in HeLa cells, clonogenic survival assays were performed following SENP6 depletion. These assays show a drastic reduction in long-term cell survival in cells treated with SENP6 siRNA compared to those treated with nontargeting (NT) siRNA (Figure 2C). Depletion of SENP6 with individual siRNA duplexes also resulted in a loss of cell viability that was directly related to the extent of knockdown (Supplemental Figure 1B). The loss of cell viability following SENP6 depletion may represent the inability of SENP6-depleted cells to transit mitosis as noted in Mukhopadhyay et al. (2010) and Hattersley et al. (unpublished data).
FIGURE 2:
SENP6 controls endogenous levels of SUMO conjugates and is required for cell viability. (A) HeLa cells were treated with either NT or SENP6 siRNA for 48 h before harvesting in SDS-loading buffer. Samples were analyzed by SDS–PAGE and immunoblotting with the indicated antibodies (individual proteins are indicated). (B) HeLa cells were treated with either NT or SENP6 siRNA for 48 h before staining with SENP6 antibodies. Scale bar represents 30 μm. (C) HeLa cells were treated with siRNA as with (A) before trypsinization and reseeding at low cell density and grown to allow colonies to form before fixation and staining with Giemsa to identify number of surviving cells.
SENP6 depletion results in an increased recruitment of PML and SUMO to PML NBs
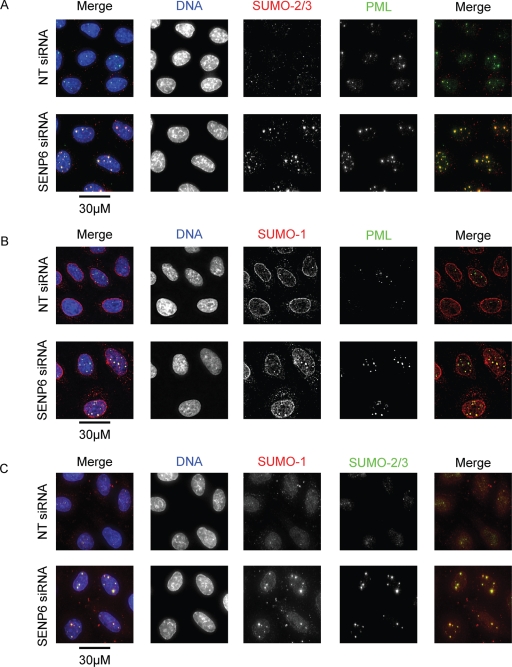
Although SENP6 has been shown to regulate SUMO and PML NBs in cells overexpressing SUMO-2/3 (Mukhopadhyay et al., 2006), it was important to establish that SENP6 depletion affected the localization of endogenous SUMO and PML. Costaining of SUMO-2/3 and PML in cells treated with either NT or SENP6 siRNA shows clear localization of the two proteins in PML NBs. Following SENP6 depletion, the recruitment of SUMO-2/3 and PML to PML NBs and the total number of bodies is clearly increased, although the actual number of bodies varies from cell to cell (Figure 3, A–C). The distribution of SUMO-1 was also investigated in SENP6-depleted cells and showed a similarly increased recruitment to PML NBs. There was no obvious effect, however, on the level of SUMO-1 staining to be found at the nuclear rim (conjugated to RanGAP1; Figure 3, B and C). Depletion of SENP6 with individual siRNA duplexes also resulted in an accumulation of SUMO-2/3 and PML in PML NBs (Supplemental Figure 1C). The change in distribution of both SUMO-1 and SUMO-2/3 following SENP6 siRNA treatment correlates with the effects seen on both SUMO paralogue groups in Figure 2A, indicating that SENP6 regulates the subcellular distribution of all SUMO paralogues in vivo.
FIGURE 3:
SENP6 depletion results in an increased recruitment of PML and SUMO-to-PML NBs. HeLa cells were treated with either NT or SENP6 siRNA for 48 h before immunostaining with (A) PML and SUMO-2/3, (B) PML and SUMO-1, or (C) SUMO-1 and SUMO-2/3 antibodies. Images are presented as projections of Z-slices. Scale bars represent 30 μm.
SENP6 depletion results in an increased number of PML NBs
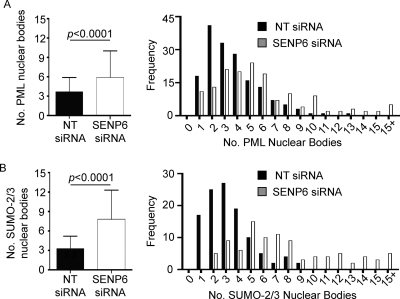
As noted in Figure 3, the number of PML NBs in cells treated with either NT or SENP6 siRNA is variable. To assess the impact of SENP6 depletion on either PML or SUMO-2/3 stained nuclear foci we carried out a quantitative analysis of NBs in ∼100 cells. This analysis shows that, in NT siRNA-treated cells, the number of PML or SUMO-2/3 foci generally varied from between 1 and 6 per nucleus, with a mean of 3–4 (Figure 4, A and B). Following SENP6 depletion, the mean number of PML/SUMO-2/3 bodies per cell increased to between 6 (in the case of PML) and 8 (in the case of SUMO-2/3) bodies per cell (Figure 4, A and B) with a far greater proportion of cells with ≥10 PML/SUMO-2/3 foci (2% and 0% for PML and SUMO-2/3 bodies, respectively, in NT siRNA-treated cells and 16% and 28% for PML and SUMO-2/3 bodies, respectively, in SENP6 siRNA-treated cells). The considerable variance in the number of PML bodies has been linked to the stage of the cell cycle; however, we note no increase of PML body number within a specific phase of the cell cycle (G1, S, or G2). Rather we observe an increase in all phases (data not shown) indicating that during interphase, SENP6 does not regulate PML body number in a cell cycle–specific manner. Interestingly, although the number of PML and SUMO-2/3 nuclear foci is similar in cells treated with NT siRNA, there is a larger increase in the number of SUMO-2/3 foci than in the number of PML NBs in cells depleted of SENP6. This finding suggests that some SUMO-2/3 foci that appear in response to SENP6 depletion are not PML related and that SENP6 may also control SUMO recruitment to other nuclear structures.
FIGURE 4:
SENP6 depletion results in an increased number of PML NBs. HeLa cells were treated with either NT or SENP6 siRNA for 48 h before immunostaining with PML or SUMO-2/3 antibodies. The number of PML (A) or SUMO-2/3 (B) NBs per cell was assessed and plotted as either mean +/− SD number of bodies or as frequency histograms of body numbers per cell. Cells treated with NT siRNA have a mean value of 3.7 PML NBs and 3.3 SUMO NBs per cell with SDs of 2.3 and 1.9, respectively. SENP6-depleted cells have a mean of 5.9 PML NBs and 7.8 SUMO NBs per cell with SDs of 4.1 and 4.5, respectively. Cells with > 15 NBs/cell were classed as 15+ for this purpose (n > 90 cells per staining/siRNA condition).
FIGURE 5:
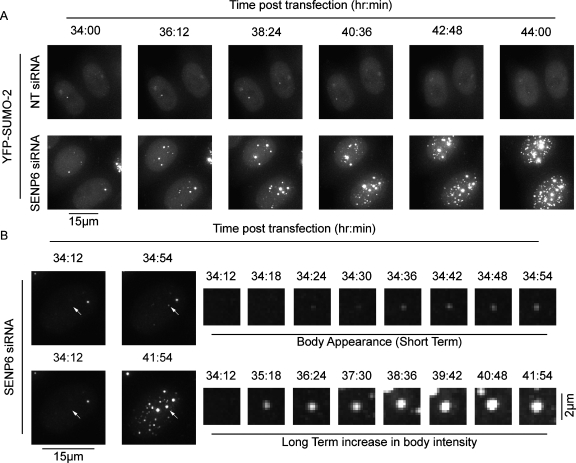
SENP6 depletion results in the de novo formation of NBs. HeLa cells stably expressing YFP-SUMO-2 were treated with NT (top panels) or SENP6 (bottom panels) siRNA for 24 h before live cell imaging (A). (B) Appearance (top panels) and the increase in intensity (bottom panels) of one NB. Images are presented as projections of individual Z-slices.
SENP6 depletion results in de novo formation of PML NBs
SENP6 depletion clearly results in an increased number of PML NBs. Increases in PML NB number previously have been shown to occur in S phase, with contacts between PML NBs and chromatin resulting in PML NB fission as DNA is replicated (Dellaire et al., 2006). Our analysis indicates, however, that the increase in PML NBs following SENP6 depletion is not specific to a particular phase of the cell cycle (data not shown). To further investigate the manner in which PML NBs increase in number, we used live cell imaging of cells stably expressing yellow fluorescent protein (YFP)–SUMO-2 (Figure 5, A and B). Following SENP6 depletion in these conditions we did not observe fission events, but rather the increase in bodies appeared to occur by de novo formation. YFP-SUMO-2 foci formed without links or associations with other bodies (Figure 5, A and B, Supplemental Movies 1–3). Large numbers of new foci began to appear approximately 30–40 h posttransfection, which subsequently increased in intensity over a period of several hours (Figure 5, A and B, Supplemental Movies 1–3). SUMO modification of PML is required for the formation of mature interphase PML NBs (Kamitani et al., 1998a, 1998b; Duprez et al., 1999; Ishov et al., 1999; Zhong et al., 2000; Jensen et al., 2001; Ayaydin and Dasso, 2004; Fu et al., 2005), and we hypothesize that a shift in the dynamics of the conjugation/deconjugation balance due to depletion of SENP6 affects the SUMO modification status of PML NB components, driving the de novo formation of new PML NBs. To test the effects of SENP6 depletion on the mobility of PML, we used HeLa cells stably expressing YFP-PML-III (Geoffroy et al., 2010) to carry out fluorescence recovery after photobleaching (FRAP). Depletion of SENP6, however, did not significantly alter the rate of fluorescence recovery of PML into PML NBs (Supplemental Figure 2B). This finding suggests that SENP6 depletion, rather that affecting the rate of PML import into PML NBs, regulates the export of PML from the bodies and that this regulation is responsible for the increased amount of PML in PML NBs following SENP6 depletion.
FIGURE 6:
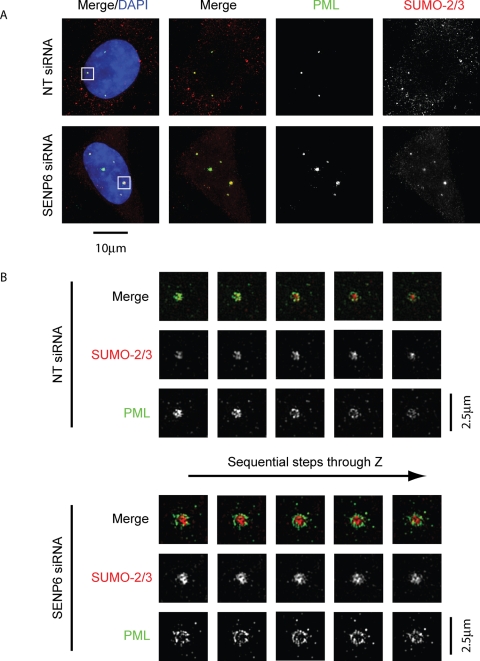
SENP6 depletion increases PML NB size but does not affect PML NB structure. HeLa cells were treated with either NT or SENP6 siRNA for 48 h before immunostaining with PML or SUMO-2/3 antibodies. (A) Conventional deconvolution microscopy was performed on samples and presented as projected images. Scale bar represents 10 μm. (B) To achieve higher resolution of the indicated PML NBs in (A), cells were analyzed by structured illumination and are images presented as individual Z-slices. Scale bars represent 2.5 μm.
SENP6 depletion increases PML NB size but does not affect PML NB structure
SENP6 depletion results in an increased recruitment of PML NB components to PML NBs in addition to increasing the number of PML NBs. A number of electron microscopic and immunofluorescence studies have determined that PML NBs are spherical structures and that PML itself forms a shell around the periphery of the body (Koken et al., 1994; Weis et al., 1994; Boisvert et al., 2000; Lang et al., 2010). Recently 4Pi microscopy revealed that PML localized to the shell of the NBs, whereas SUMO-2/3 was found at the inner/core region of the bodies (Lang et al., 2010).
To investigate the effect of SENP6 depletion on PML NB structure and the distribution of proteins therein, we used three-dimensional (3D) structured illumination microscopy (Schermelleh et al., 2008) to visualize PML NBs. Costaining of endogenous PML and SUMO-2/3 and visualization by conventional deconvolution microscopy show colocalization of SUMO-2/3 and PML to PML NBs (Figure 6A). Structured illumination of these samples shows that, as noted in Lang et al. (2010), PML forms a shell arrangement around the inner mass of SUMO-2/3 (Figure 6B). Following SENP6 depletion, the PML NB clearly shows a similar arrangement of PML and SUMO-2/3 in which the whole structure is enlarged. SUMO-2/3 and PML occupy almost completely distinct regions of the body although there are some regions of colocalization. Furthermore, the distribution of SUMO-2/3 is not uniform within the PML NB and shows regions of varying intensity, indicating distinct regions within the body (Figure 6B). These data indicate that, although SENP6 appears to regulate the recruitment of proteins to, as well as the total number of PML NBs, SENP6 depletion does not compromise the overall structure of PML NBs. Rather the bodies are able to increase in size to accommodate the extra protein component while maintaining organization and structure. 3D structured illumination was also used to determine the comparative localization of SUMO-1 with PML as well as SUMO-1 and SUMO-2/3. In slight contrast to the findings of Lang et al. (2010), we found that the SUMO-1 signal could appear as partially colocalizing with the PML-shell region but was also found in the inner domain of the body, colocalizing with SUMO-2/3 (Supplemental Figures 1 and 2). Costaining of PML with SUMO-1 and SUMO-1 with SUMO-2/3, following SENP6 depletion, showed similar patterns of localization and changes in size as that previously observed for PML and SUMO-2/3 (Supplemental Figures 3 and 4).
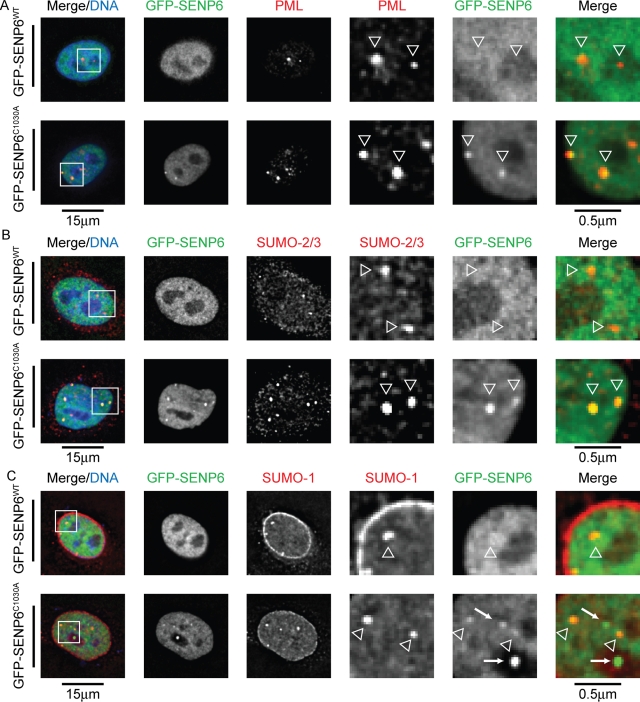
Catalytically inactive SENP6 accumulates in PML NBs
Exogenous SENP6 previously has been shown to localize to the nucleoplasm due to the presence of a number of nuclear localization signal (NLS) motifs (Mukhopadhyay et al., 2006). Overexpression of green fluorescent protein (GFP)–tagged SENP6 confirms this localization, however, expressed GFP–SENP6 containing a cysteine-to-alanine mutation of the catalytic cysteine (C1030A) was detected in nuclear foci (Figure 7, A–C). As SUMO is a component of PML NBs, cells expressing GFP–SENP6WT/C1030A were costained with PML, SUMO-1, or SUMO-2/3. This experiment revealed that SENP6C1030A colocalized with all three proteins in PML NBs (Figure 7, A–C, arrowheads). Occasionally, this colocalization can also be seen in cells overexpressing wild-type SENP6 and weakly when staining for endogenous SENP6 (data not shown). These data suggest that catalytically inactive SENP6 accumulates in PML NBs. Under normal circumstances, SENP6 would normally transit to these structures, perform its SUMO protease role, and dissociate. The inactivating mutation of the catalytic cysteine to alanine prevents catalysis and traps SENP6 bound to its substrates in PML bodies. Whereas a number of GFP–SENP6C1030A foci colocalize with PML NBs, other nuclear SENP6C1030A foci can be seen that do not localize with either SUMO or PML (Figure 7C, arrows). The function of these accumulations of SENP6 is unclear and requires further study. Analysis of GFP–SENP6WT and GFP–SENP6C1030A overexpression by immunoblotting indicates that overexpression of GFP–SENP6WT only and not GFP–SENP6C1030A causes a decrease in SUMO-2/3 (and to a lesser extent SUMO-1) conjugates (Supplemental Figure 5).
FIGURE 7:
Catalytically inactive SENP6 accumulates in PML bodies. (A–C) HeLa cells were transfected with either GFP-SENP6WT/C1030A for 36 h before fixation with paraformaldehyde, permeabilization with Triton X-100, and staining with (A) PML, (B) SUMO-2/3, or (C) SUMO-1 antibodies. Enlargements of selected regions are shown, and PML/SUMO NBs are indicated with open arrowheads. SENP6 nuclear foci are indicated by solid arrows (C). Scale bars represent 15 μm and 0.5 μm (enlarged sections).
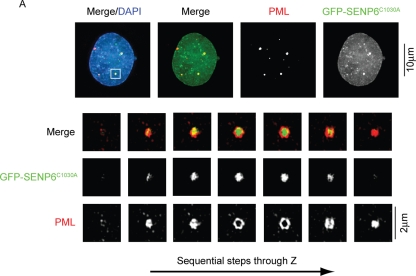
Catalytically inactive SENP6 localizes to the core domain of PML NBs
As Figure 7 shows, a catalytically inactive mutant of SENP6C1030A localizes to PML NBs due to its accumulation at the sites of its substrates. To establish whether SENP6 localized to the PML-rich shell region of the NB or to the SUMO-2/3–rich core domain, we used structured illumination of PML NBs in cells expressing GFP–SENP6C1030A and costained with PML. This analysis indicates that, in the majority of cases (57%), the catalytically inactive protease localizes to the SUMO-2/3–rich core of the PML NBs with the PML shell encircling the GFP–SENP6 signal (Figure 8). In some instances, however, GFP–SENP6C1030A did not localize to the core domain but formed a ring structure (17%; Supplemental Figure 6A) or localized throughout the body in both the core and outer domain (26%; Supplemental Figure 6B). It should be noted, however, that, in the cases of GFP–SENP6C1030A localization throughout the NB, PML also showed localization throughout the body suggesting that the overall NB structure was perturbed.
FIGURE 8:
GFP-SENP6C1030A localizes to the core domain of PML NBs. HeLa cells were transfected with GFP-SENP6C1030A 36 h before immunostaining with PML antibodies. Conventional deconvolution microscopy was performed on samples and presented as projected images. Scale bar represents 10 μm (top panels). To achieve higher resolution of the indicated PML NBs, cells were analyzed by structured illumination and are images presented as individual Z-slices. SENP6 was found to localize to the core domain in 57% of cases. Scale bars represent 2 μm.
Poly-SUMO–modified PML is a SENP6 substrate
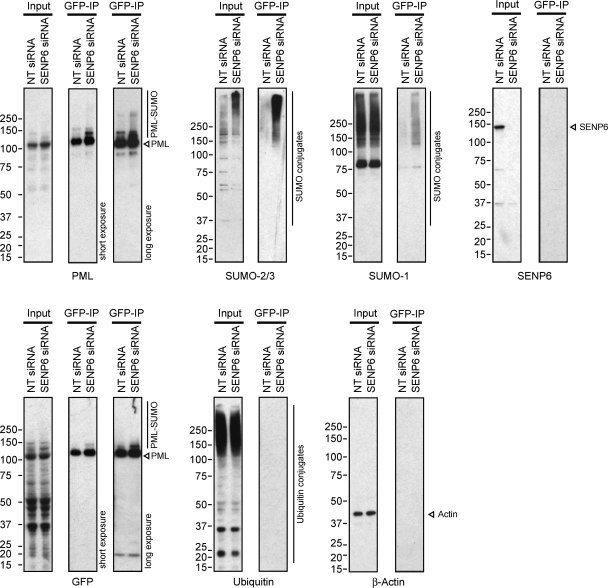
The ability of SENP6 to regulate the size, number, and recruitment of proteins to PML NBs suggests that SENP6 may directly regulate the SUMO conjugation status of PML itself, a known SUMO substrate whose modification is required for formation of mature interphase PML NBs. To determine whether SENP6 regulates the SUMO modification status of PML, we used cells stably expressing YFP-PML to perform GFP immunoprecipitation (GFP-IP) to isolate PML (Rothbauer et al., 2008). Figure 9 shows that, following GFP-IP to isolate PML and PML conjugates, high-molecular-weight PML, SUMO-2/3, and SUMO-1 species are increased in SENP6-depleted cells, indicating that SENP6 regulates the SUMO modification of PML. In contrast, the depletion of SENP6 does not alter the level of ubiquitin found following GFP-IP of PML. Semiquantitative immunoblotting analysis indicates that depletion of SENP6 does not change the total amount of PML, but does result in an accumulation of SUMO-modified PML species (Supplemental Figure 7). As the poly-SUMO–dependent, ubiquitin E3 ligase RNF4 has been shown to specifically bind and ubiquitinate poly-SUMO–modified PML (Tatham et al., 2008), it could be expected that increased SUMO chain modification of PML in response to SENP6 depletion could also result in the ubiquitination and subsequent degradation of PML, triggered by RNF4 binding. A SENP6 depletion-dependent increase in PML SUMO modification, however, does not appear to trigger the degradation of PML but rather to increase the polymeric SUMO modification of PML as well as to increase the number and size of PML NBs.
FIGURE 9:
Poly-SUMO–modified PML is a SENP6 substrate. HeLa cells stably expressing YFP-SUMO-2 were treated with either NT or SENP6 siRNA before harvesting and nuclear fractionation. Lysed nuclei were then used for GFP-IP, and input and elution samples were analyzed by SDS–PAGE and immunoblotting with antibodies as indicated.
DISCUSSION
The SUMO system has long been recognized as a regulatory component of PML NBs. Here, we further define the substrate specificity of the SUMO chain-editing protease SENP6, show that it is recruited to PML NBs, and demonstrate that SUMO-modified PML is a SENP6 substrate in vivo. As a consequence, SENP6 regulates the recruitment of endogenous PML and SUMO to PML NBs, controlling the size of PML NBs and the number of PML NBs per cell.
SENP6 previously has been characterized as having specificity for SUMO-2/3 conjugates, in particular SUMO-2/3 polymeric chains (Mukhopadhyay et al., 2006; Lima and Reverter, 2008). Although SUMO-2/3 chains are clearly the preferred substrate, SENP6 is also capable of cleaving mixed chains of SUMO-1 and SUMO-2/3. Therefore, although SUMO-1 may be able to act as a SUMO chain terminator in vivo, SUMO-1–capped polymers would not be resistant to SENP6, one of the two characterized chain-remodeling enzymes. Whether SENP7 shares this ability to cleave mixed SUMO conjugates remains to be determined.
Consistent with the in vitro characterization of SENP6, depletion of SENP6 from HeLa cells causes the accumulation of high-molecular-weight SUMO-2/3 conjugates as well as SUMO-1 conjugates. Although accumulation of SUMO-1 on the ends of SUMO-2/3 chains may account for the increase in high-molecular-weight SUMO-1 conjugates, they could also arise due to the increase in SUMO-2/3 chain length on substrates that are also modified by SUMO-1 at another lysine residue. These effects are evident on a particular substrate, PML, which shows increased SUMO-1– and SUMO-2/3–associated species following SENP6 depletion. It is not clear in this case, however, whether SENP6 regulates SUMO-1 caps on SUMO-2/3 chains or whether SUMO-1 is found on one of the three alternate SUMO conjugation sites on PML. Converse to SENP6 depletion, the overexpression of wild-type, GFP-tagged SENP6 (but not catalytically inactive SENP6) results in the loss of high-molecular-weight SUMO conjugates. Although Smt3 polymers are not required for yeast viability (Bylebyl et al., 2003), the loss of the yeast chain editing SUMO protease Ulp2 results in a severe phenotype (Li and Hochstrasser, 2000). Similarly, in human cells, loss of SENP6 causes a substantial decrease in long-term cell viability (Figure 2) and errors in mitotic progression (Mukhopadhyay et al., 2010) and Hattersley et al. (unpublished data).
PML NBs often contact chromatin and are surrounded by transcriptionally active regions of DNA (Boisvert et al., 2000; Eskiw et al., 2004; Xie and Pombo, 2006). Numerous studies using electron microscopy as well as high-resolution fluorescence microscopy and electron spectroscopic imaging have shown that, within PML NBs, PML itself appears to form the shell of an approximately spherical structure (Koken et al., 1994; Weis et al., 1994; Boisvert et al., 2000; Eskiw et al., 2003; Xie and Pombo, 2006; Lang et al., 2010). This outer domain also has been shown to include other PML NB components, such as SUMO-1 and SP100, whereas the inner, more electron-dense core contains proteins such as SUMO-2/3 (Lang et al., 2010). The localization of SUMO-1 and SUMO-2/3 and the requirement of PML’s SUMO modification clearly make the SUMO system a critical component in PML NB function. Therefore, perturbation of proteins involved in SUMO conjugation (such as Ubc9 and the E3 ligase RanBP2) and the proteases involved in deconjugation lead to alterations in PML NB number and size (Bailey and O’Hare, 2002, 2004; Best et al., 2002; Nacerddine et al., 2005; Saitoh et al., 2006; Han et al., 2010). Furthermore, the depletion of SENP7 was also shown recently to increase PML NB number (Shen et al., 2009). Finally, in a system overexpressing SUMO-3, the depletion of SENP6 results in a dramatic increase in PML NBs (Mukhopadhyay et al., 2006).
The catalytically inactive C1030A mutation of SENP6 allows it to accumulate in PML NBs and other unidentified nuclear foci. This observation suggests that SENP6C1030A is recruited to SUMO conjugate–rich PML NBs but, as it is unable to hydrolyze the SUMO polymers, it is trapped bound to its substrates. In contrast, GFP-SENP6WT does not accumulate in PML NBs as it is likely to rapidly dissociate after substrates have been hydrolyzed. Consistently, in the majority of cases GFP-SENP6C1030A localizes to the SUMO-2/3–rich core domain. In some cases, however, GFP-SENP6C1030A is found on the periphery of the structure or throughout the apparently deformed NBs. These observations may be a consequence of the time taken by SENP6 to accumulate and transit to the center of the body or may be affected by the balance of SUMO-1 and SUMO-2/3 conjugates within the body. Consistent with the localization of its substrates, SENP6 depletion results in an accumulation of SUMO as well as PML in PML NBs. PML NBs, however, are able to increase in size to accommodate accumulated proteins while maintaining their structural organization, characterized by PML and the partially colocalizing SUMO-1 forming a shell around the SUMO-2/3– and SUMO-1–rich core domain.
While SENP6 depletion increases the size of PML NBs as well as their protein recruitment, it also affects the total number of PML NBs. The number of PML NBs has been shown previously to be specifically increased in S-phase by fission, where the chromatin contacts of the body pull it apart as DNA is replicated (Dellaire et al., 2006). In the case of SENP6 depletion, however, the increase in body number is not specific to a particular phase of the cell cycle (increases in body number are observed at all phases of the cell cycle). Furthermore, the increase in PML NB number does not appear to occur by fission events but by the de novo formation of new bodies. As seen in Figure 2, the depletion of SENP6 clearly shifts the balance of the SUMO system to conjugation and thus causes the redistribution of SUMO and PML from the nucleoplasm to more abundant and larger PML NBs. This effect is also seen following heat stress, proteasome inhibition, and short-term arsenic treatment (Muller et al., 1998; Sternsdorf et al., 1999; Saitoh and Hinchey, 2000; Bailey and O’Hare, 2005; Lallemand-Breitenbach et al., 2008; Schimmel et al., 2008; Tatham et al., 2008; Matafora et al., 2009), all of which also increase PML NB number. We hypothesize that, by altering the balance of the SUMO system, SENP6 also alters the dynamics of PML NB formation. The increase in SUMO conjugates (in particular PML itself) results in the accumulation of PML–SUMO conjugates that are able to spontaneously form new PML NBs, although existing bodies are capable of increasing in size to accommodate their extra protein content. Interestingly, SENP6 depletion does not affect the rate of import of YFP–PML into PML NBs (Supplemental Figure 2), suggesting that the depletion of SENP6 results in the retention of PML in PML NBs (decreased export rate) and that this retention results in the increased size of PML NBs. De novo formation of PML NBs from previously nucleoplasmic PML and SUMO also argues that SENP6 does not solely regulate SUMO conjugates in PML NBs and that, in terms of PML NBs, its role in the nucleoplasm is equally important. Furthermore, in contrast to arsenic treatment, which causes the hyperSUMO modification of PML, resulting in it being targeted by the ubiquitin E3 ligase RNF4 and subsequently degraded (Lallemand-Breitenbach et al., 2008; Tatham et al., 2008), SENP6 treatment results only in the accumulation of SUMO-modified PML and PML NBs. This lack of an RNF4 response suggests that the RNF4 system may be overwhelmed by the overall increase in SUMO polymers in PML NBs and throughout the nucleus. These results indicate that SENP6 is an important regulator of SUMO conjugation and the dynamics of PML NB formation.
METHODS
Recombinant protein expression and SUMO conjugation and deconjugation reactions
SUMO-1 (residues 1–97), SUMO-1ΔGG (residues 1–95), SUMO-2 (residues 1–93), SUMO-2ΔGG (residues 1–91), and SUMO-1 mutant (D15V) were cloned into BamHI and HindIII cleaved pHisTEV. SENP6 catalytic domain (residues 635–1112) was cloned into BamHI and HindIII cleaved expression vector pLou3 (a gift from Jim Naismith, University of St. Andrews). All clones were verified by automated DNA sequence analysis and shown to be identical with those previously reported for SENP6, SUMO-1, and SUMO-2 (GeneBank accession nos. Q9GZR1, AAH53528, and NP_008867, respectively). SENP6 mutants (C1030A) were generated using PCR-based mutagenesis and verified by DNA sequence analysis (Dundee University DNA Sequencing Unit).
Recombinant SENP6 was expressed as a 6HisMBP-tagged fusion protein in Rosetta (DE3; Novagen Merck, Nottingham, UK) by induction with 0.1 mM isopropyl-β-D-thio-galactoside (IPTG) at 20°C for 12 h. 6HisMBP-tagged SENP6 was purified by Ni-NTA affinity chromatography. The 6HisMBP tag was cleaved from the fusion protein by 6His-tagged Tobacco Etch Virus (TEV) protease, and SENP6 was further purified by Ni-NTA affinity chromatography and gel filtration (Superdex200; Ge Healthcare, Buckinghamshire, UK). Protein purity was evaluated by SDS–PAGE and Coomassie Blue staining. Concentrations of the purified proteins were determined by the Bradford method.
All in vitro isopeptidase assays were performed in a buffer containing 50 mM Tris-HCl (pH 7.5), 2 mM MgCl2, 150 mM NaCl, and 5 mM β-mercaptoethanol. To make SUMO-1(S)ΔGG–SUMO-1(M) and SUMO-1(S) – SUMO-2(M), SUMO-1(D15V) was generated by PCR-based mutagenesis. The isopeptide-linked SUMO dimers were made by conjugation in vitro and purified by gel filtration (Superdex75; Amersham-Pharmacia). Isopeptidase assays were performed in 50-μl reactions containing substrates at a concentration of 20 μM and a range of SENP6 concentrations (1 nM to 200 μM) for 1 h at 37ºC. Reactions were terminated by adding 2× protein sample buffer. Samples were fractionated by SDS–PAGE (10%) followed by Coomassie Blue staining. Protein quantification was performed using a charge-coupled device (CCD) camera system (LAS1000 Plus system; Fujifilm, Tokyo, Japan) after separation by SDS–PAGE (10% gels).
Cell culture, siRNA and DNA transfection, and clonogenic survival assays
HeLa cells were maintained at 37°C in DMEM supplemented with 10% fetal calf serum (FCS), 5% CO2, and penicillin and streptomycin at 100 U/ml. Cells stably expressing YFP-PML-III or YFP-SUMO-2 were maintained at 37°C in DMEM supplemented with 10% FCS, 5% CO2, penicillin and streptomycin at 100 U/ml, and blasticidin at 1 μg/ml or G418 at 100 μg/ml, respectively (Geoffroy et al., 2010).
Cells were transfected with 2 nM SENP6 siRNA (Dharmacon onTarget+, as either individual or as a smartpool of four duplexes as indicated. Antisense strands SENP6; 1 – GAAAGUGACCCUCGUUAUAUU, 2 – GGACAAAUCUGCUCAGUGUUU, 3 – GAGCUUUGAUCAUGAAGAAUU, 4 – GUAGAGGACAGUUGUAUUUUU) with Lipofectamine siRNAmax (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions. Control transfections were performed with a NT duplex at the same concentration. Cells were cultured for 48 h after transfection before analysis. Clonogenic survival assays were performed by siRNA transfection for 48 h as mentioned earlier in text before trypsinization and reseeding 1 × 103 cells per well (six-well plate) and allowing colonies to form over 10–14 days before fixation and staining with Giemsa/methanol. SENP6 was cloned into EGFP-C1 (BglII-ApaI cleaved), and the catalytic mutant was prepared by mutating the catalytic cysteine (C1030) to alanine. Empty vector (GFP), GFP-SENP6WT, or GFP-SENP6C1030A (0.2 μg) was transfected using GeneJuice, (Novagen Merck, Nottingham, UK), according to the manufacturer’s instructions for 36 h before harvesting.
Immunofluorescence, live-cell imaging, and FRAP
HeLa cells were grown on coverslips prior to treatment, fixed for 10 min in 4% paraformaldehyde/phosphate-buffered saline (PFA/PBS) at 37°C and permeabilized in 0.2% Triton X-100/PBS. Cells were then blocked in 5% bovine serum albumin (BSA), 0.1% Tween, and PBS for 30 min before washing in 1% BSA, 0.1% Tween, and PBS (PBS–T/BSA). Cells were stained with primary antibody diluted in PBS–T/BSA for 1 h at room temperature before washing in PBS–T/BSA and incubating with secondary antibodies. Cells were then stained with 4’,6-diamidino-2-phenylindole (DAPI) at 0.1 μg/ml and mounted for imaging with VECTASHIELD mounting medium. Images were collected using a DeltaVision DV3 widefield microscope and processed using Softworx (both Applied Precision, Issaquah, WA). Images are presented as maximal intensity projections or single Z-slices as specified. PML NB number analysis was performed using GraphPad prism software (GraphPad Software, Inc., La Jolla, CA).
Live-cell imaging was performed by seeding HeLa cells stably expressing YFP–SUMO-2 into LabTek (Bloomington, IN) dishes and treated as indicated. Prior to imaging, cells were washed and incubated for 30 min in Liebovitz CO2 independent medium and imaged on a DeltaVision Spectris widefield microscope fitted with a 37°C environment chamber (Solent Scientific, Segensworth, United Kingdom). Images were processed using SoftWorx.
FRAP was performed by treating YFP-PML– or YFP-SUMO-2–expressing HeLa cells as indicated before bleaching individual PML/SUMO bodies with a 488-nm argon laser at 100% intensity and performing conventional live-cell imaging as mentioned earlier in text to monitor fluorescence recovery. Recovery curves were calculated by first standardizing intensity values as percentages of the prebleach NB intensity before calculating the percentage of fluorescence recovery values using the formula rI = (I –postB) / (100 – postB) × 100 (in which rI is the percentage of fluorescence recovery, I is the fluorescence intensity, and postB is the postbleach intensity. Fluorescence recovery data were analyzed using GraphPad Prism 4.0c to calculate T1/2 and mobile/immobile fractions.
Structured illumination
The protocol applied was based on that described (Schermelleh et al., 2008). Images were acquired using a UPlanSApochromat 100× 1.4NA, oil immersion objective lens (Olympus, Center Valley, PA) and back-illuminated Cascade II 512 × 512 electron-multiplying charge-coupled device (EMCCD) camera (Photometrics, Tucson, AZ) on the OMX version 2 system (Applied Precision) equipped with 405-, 488-, and 593-nm solid-state lasers. Samples were illuminated by a coherent scrambled laser light source that had passed through a diffraction grating to generate the structured illumination by interference of light orders in the image plane to create a 3D sinusoidal pattern, with lateral stripes approximately 0.2 μm apart. The pattern was shifted laterally through five phases and through three angular rotations of 60’ for each Z-section, separated by 0.125 μm. Exposure times were typically between 200 and 500 ms, and the power of each laser was adjusted to achieve optimal intensities of between 2,000 and 4,000 counts in a raw image of 16-bit dynamic range, at the lowest possible laser power to minimize photo bleaching. Each frame acquisition was separated by a 300-ms pause. Raw images were processed and reconstructed to reveal structures with greater resolution (Gustafsson, 2008). The channels were then aligned in x, y, and rotationally using predetermined shifts as measured using a target lens and the Softworx alignment tool (Applied Precision).
GFP-IP
Cells were transfected as described before washing twice in PBS/100 mM iodoacetamide and harvested by scraping. Cells were collected by centrifugation at 430 × g, resuspended in ice-cold Buffer A (10 mM HEPES pH7.9, 1.5 mM MgCl, and 10 mM KCl) with 100 mM iodoacetamide, and incubated at 4°C for 15 min to swell cells. Nuclei were then prepared from cells lysed by passage through a 26-gauge needle, collected by centrifugation at 2,000 × g for 5 min, and washed twice in Buffer A with 10 mM iodoacetamide. Nuclei were resuspended and lysed in 2–5 volumes RIPA buffer (50 mM Tris pH6.8, 150 mM NaCl, 1% Nonidet P-40 [NP-40], 0.5% deoxycholate) and 10 mM iodoacetamide and sonicated to shear DNA. Cellular debris was cleared by centrifugation at 17,000 × g and supernatant was precleared with sepharose beads before incubation with GFP beads (ChromoTek, Planegg-Martinsried, Germany) for 1–3 h at 4°C. Beads were washed twice in RIPA buffer before elution with SDS-loading buffer and analysis by SDS–PAGE and immunoblotting.
Antibodies
Antigen affinity-purified sheep anti-SUMO-1 and -SUMO-2/3 were as described (Tatham et al., 2008) and antigen affinity-purified sheep anti-SENP6 was prepared in house using glutathione-S-transferase (GST)–SENP6 catalytic domain (residues 635–1112) as antigen. Anti-PML (5e10, immunofluorescence) was a gift from R. Van Driel (Stuurman et al., 1992), anti-PML (83, immunoblotting) was a gift from J. Seeler, and anti-SUMO-2 monoclonal antibody was a gift from M. Matunis. Anti-β-actin was obtained from Oncogene (Cambridge, MA), anti-GFP from Roche (Basel, Switzerland), anti-HA from Berkeley Antibody Company (BAbCO, Richmond, CA), and anti-ubiquitin conjugates from Enzo Life Sciences (Exeter, UK). Secondary antibodies for fluorescence microscopy were obtained from Jackson Immunochemicals (West Grove, PA) and Invitrogen (San Diego, CA). Quantitation of PML immunoblots was performed using Image Gauge v4.21 software (Fuji Photofilm, Bedford, UK).
Supplementary Material
Acknowledgments
We thank members of the Hay lab for help and advice. We also thank M. Posch and E. King for support with structured illumination studies. Use of the OMX microscope was supported by the Scottish University Life Sciences Alliance. This work was funded by Cancer Research UK. N.H. was funded by a Biotechnology and Biological Sciences Research Council (BBSRC) studentship.
Abbreviations used:
- SUMO
small ubiquitin-like modifier
- SENP
sentrin (SUMO) protease
- PML
promyelocytic leukemia protein
- PML NB
PML nuclear body
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-06-0504) on December 9, 2010.
REFERENCES
- Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell. 2004;15:5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D, O’Hare P. Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease. J Gen Virol. 2002;83:2951–2964. doi: 10.1099/0022-1317-83-12-2951. [DOI] [PubMed] [Google Scholar]
- Bailey D, O’Hare P. Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J Biol Chem. 2004;279:692–703. doi: 10.1074/jbc.M306195200. [DOI] [PubMed] [Google Scholar]
- Bailey D, O’Hare P. Comparison of the SUMO1 and ubiquitin conjugation pathways during the inhibition of proteasome activity with evidence of SUMO1 recycling. Biochem J. 2005;392:271–281. doi: 10.1042/BJ20050873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Best JL, Ganiatsas S, Agarwal S, Changou A, Salomoni P, Shirihai O, Meluh PB, Pandolfi PP, Zon LI. SUMO-1 protease-1 regulates gene transcription through PML. Mol Cell. 2002;10:843–855. doi: 10.1016/s1097-2765(02)00699-8. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- Boisvert FM, Hendzel MJ, Bazett-Jones DP. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J Cell Biol. 2000;148:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 2003;278:44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- Choi SJ, Chung SS, Rho EJ, Lee HW, Lee MH, Choi HS, Seol JH, Baek SH, Bang OS, Chung CH. Negative modulation of RXRalpha transcriptional activity by small ubiquitin-related modifier (SUMO) modification and its reversal by SUMO-specific protease SUSP1. J Biol Chem. 2006;281:30669–30677. doi: 10.1074/jbc.M604033200. [DOI] [PubMed] [Google Scholar]
- Dellaire G, Ching RW, Dehghani H, Ren Y, Bazett-Jones DP. The number of PML nuclear bodies increases in early S phase by a fission mechanism. J Cell Sci. 2006;119:1026–1033. doi: 10.1242/jcs.02816. [DOI] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- Di Bacco A, Gill G. SUMO-specific proteases and the cell cycle. An essential role for SENP5 in cell proliferation. Cell Cycle. 2006;5:2310–2313. doi: 10.4161/cc.5.20.3367. [DOI] [PubMed] [Google Scholar]
- Di Bacco A, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G. The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol. 2006;26:4489–4498. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprez E, Saurin AJ, Desterro JM, Lallemand-Breitenbach V, Howe K, Boddy MN, Solomon E, de Thé H, Hay RT, Freemont PS. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J Cell Sci. 1999;112 (Pt 3):381–393. doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- Eskiw CH, Dellaire G, Bazett-Jones DP. Chromatin contributes to structural integrity of promyelocytic leukemia bodies through a SUMO-1-independent mechanism. J Biol Chem. 2004;279:9577–9585. doi: 10.1074/jbc.M312580200. [DOI] [PubMed] [Google Scholar]
- Eskiw CH, Dellaire G, Mymryk JS, Bazett-Jones DP. Size, position and dynamic behavior of PML nuclear bodies following cell stress as a paradigm for supramolecular trafficking and assembly. J Cell Sci. 2003;116:4455–4466. doi: 10.1242/jcs.00758. [DOI] [PubMed] [Google Scholar]
- Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- Fu C, et al. Stabilization of PML nuclear localization by conjugation and oligomerization of SUMO-3. Oncogene. 2005;24:5401–5413. doi: 10.1038/sj.onc.1208714. [DOI] [PubMed] [Google Scholar]
- Geoffroy M-C, Jeffray EG, Walker KJ, Hay RT. Arsenic-induced SUMO-dependent recruitment of RNF4 into PML nuclear bodies. Mol Biol Cell. 2010;21:4227–4239. doi: 10.1091/mbc.E10-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L, Yeh ET. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem. 2006;281:15869–15877. doi: 10.1074/jbc.M511658200. [DOI] [PubMed] [Google Scholar]
- Gustafsson MG. Super-resolution light microscopy goes live. Nat Methods. 2008;5:385–387. doi: 10.1038/nmeth0508-385. [DOI] [PubMed] [Google Scholar]
- Haindl M, Harasim T, Eick D, Muller S. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 2008;9:273–279. doi: 10.1038/embor.2008.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, et al. SENP3-mediated de-conjugation of SUMO2/3 from promyelocytic leukemia is correlated with accelerated cell proliferation under mild oxidative stress. J Biol Chem. 2010;285:12906–12915. doi: 10.1074/jbc.M109.071431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang J, Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem. 2002;277:19961–19966. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, Hochstrasser M. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss, 3rd JF, Maul GG. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana Y, Yeh ET, Zhang Y. Nucleocytoplasmic shuttling modulates activity and ubiquitination-dependent turnover of SUMO-specific protease 2. Mol Cell Biol. 2006;26:4675–4689. doi: 10.1128/MCB.01830-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20:7223–7233. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Nguyen HP, Wada H, Fukuda-Kamitani T, Yeh ET. Identification of three major sentrinization sites in PML. J Biol Chem. 1998a;273:26675–26682. doi: 10.1074/jbc.273.41.26675. [DOI] [PubMed] [Google Scholar]
- Kamitani T, Nguyen HP, Kito K, Fukuda-Kamitani T, Yeh ET. Covalent modification of PML by the sentrin family of ubiquitin-like proteins. J Biol Chem. 1998b;273:3117–3120. doi: 10.1074/jbc.273.6.3117. [DOI] [PubMed] [Google Scholar]
- Koken MH, et al. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo ML, den Besten W, Thomas MC, Sherr CJ. Arf-induced turnover of the nucleolar nucleophosmin-associated SUMO-2/3 protease Senp3. Cell Cycle. 2008;7:3378–3387. doi: 10.4161/cc.7.21.6930. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de Thé H. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, et al. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J Exp Med. 2001;193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M, et al. Three-dimensional organization of promyelocytic leukemia nuclear bodies. J Cell Sci. 2010;123:392–400. doi: 10.1242/jcs.053496. [DOI] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol. 2000;20:2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, et al. SENP1 mediates TNF-induced desumoylation and cytoplasmic translocation of HIPK1 to enhance ASK1-dependent apoptosis. Cell Death Differ. 2008;15:739–750. doi: 10.1038/sj.cdd.4402303. [DOI] [PubMed] [Google Scholar]
- Lima CD, Reverter D. Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. J Biol Chem. 2008;283:32045–32055. doi: 10.1074/jbc.M805655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FM, Lai YJ, Shen HJ, Cheng YH, Wang TF. Yeast axial-element protein, Red1, binds SUMO chains to promote meiotic interhomologue recombination and chromosome synapsis. EMBO J. 2010;29:586–596. doi: 10.1038/emboj.2009.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matafora V, D’Amato A, Mori S, Blasi F, Bachi A. Proteomic analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol Cell Proteomics. 2009;8:2243–2255. doi: 10.1074/mcp.M900079-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal AC. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Arnaoutov A, Dasso M. The SUMO protease SENP6 is essential for inner kinetochore assembly. J Cell Biol. 2010;188:681–692. doi: 10.1083/jcb.200909008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Ayaydin F, Kolli N, Tan SH, Anan T, Kametaka A, Azuma Y, Wilkinson KD, Dasso M. SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J Cell Biol. 2006;174:939–949. doi: 10.1083/jcb.200510103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Reineke EL, Kao HY. Targeting promyelocytic leukemia protein: a means to regulating PML nuclear bodies. Int J Biol Sci. 2009;5:366–376. doi: 10.7150/ijbs.5.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat Struct Mol Biol. 2006;13:1060–1068. doi: 10.1038/nsmb1168. [DOI] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- Saitoh N, Uchimura Y, Tachibana T, Sugahara S, Saitoh H, Nakao M. In situ SUMOylation analysis reveals a modulatory role of RanBP2 in the nuclear rim and PML bodies. Exp Cell Res. 2006;312:1418–1430. doi: 10.1016/j.yexcr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108:165–170. doi: 10.1016/s0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- Schermelleh L, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel J, Larsen KM, Matic I, van Hagen M, Cox J, Mann M, Andersen JS, Vertegaal AC. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol Cell Proteomics. 2008;7:2107–2122. doi: 10.1074/mcp.M800025-MCP200. [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Felberbaum R, Hochstrasser M. The Ulp2 SUMO protease is required for cell division following termination of the DNA damage checkpoint. Mol Cell Biol. 2007;27:6948–6961. doi: 10.1128/MCB.00774-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiyama N, Arita K, Ikeda Y, Hashiguchi K, Ariyoshi M, Tochio H, Saitoh H, Shirakawa M. Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45. Proteins. 2010;78:1491–1502. doi: 10.1002/prot.22667. [DOI] [PubMed] [Google Scholar]
- Shen LN, Dong C, Liu H, Naismith JH, Hay RT. The structure of SENP1-SUMO-2 complex suggests a structural basis for discrimination between SUMO paralogues during processing. Biochem J. 2006;397:279–288. doi: 10.1042/BJ20052030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen LN, Geoffroy MC, Jaffray EG, Hay RT. Characterization of SENP7, a SUMO-2/3-specific isopeptidase. Biochem J. 2009;421:223–230. doi: 10.1042/BJ20090246. [DOI] [PubMed] [Google Scholar]
- Skilton A, Ho JC, Mercer B, Outwin E, Watts FZ. SUMO chain formation is required for response to replication arrest in S. pombe. PLoS One. 2009;4:e6750. doi: 10.1371/journal.pone.0006750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- Stehmeier P, Muller S. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol Cell. 2009;33:400–409. doi: 10.1016/j.molcel.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Sternsdorf T, Puccetti E, Jensen K, Hoelzer D, Will H, Ottmann OG, Ruthardt M. PIC-1/SUMO-1-modified PML-retinoic acid receptor alpha mediates arsenic trioxide-induced apoptosis in acute promyelocytic leukemia. Mol Cell Biol. 1999;19:5170–5178. doi: 10.1128/mcb.19.7.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci. 1992;101((Pt 4)):773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Xie SQ, Pombo A. Distribution of different phosphorylated forms of RNA polymerase II in relation to Cajal and PML bodies in human cells: an ultrastructural study. Histochem Cell Biol. 2006;125:21–31. doi: 10.1007/s00418-005-0064-2. [DOI] [PubMed] [Google Scholar]
- Yuan H, Zhou J, Deng M, Liu X, Le Bras M, de Thé H, Chen SJ, Chen Z, Liu TX, Zhu J. Small ubiquitin-related modifier paralogs are indispensable but functionally redundant during early development of zebrafish. Cell Res. 2010;20:185–196. doi: 10.1038/cr.2009.101. [DOI] [PubMed] [Google Scholar]
- Yun C, Wang Y, Mukhopadhyay D, Backlund P, Kolli N, Yergey A, Wilkinson KD, Dasso M. Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J Cell Biol. 2008;183:589–595. doi: 10.1083/jcb.200807185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FP, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Janne OA. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol. 2008a;28:5381–5390. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol Cell Biol. 2002;22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD, Goeres J, Zhang H, Yen TJ, Porter AC, Matunis MJ. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell. 2008b;29:729–741. doi: 10.1016/j.molcel.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
- Zunino R, Braschi E, Xu L, McBride HM. Translocation of SenP5 from the nucleoli to the mitochondria modulates DRP1-dependent fission during mitosis. J Biol Chem. 2009;284:17783–17795. doi: 10.1074/jbc.M901902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunino R, Schauss A, Rippstein P, Andrade-Navarro M, McBride HM. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J Cell Sci. 2007;120:1178–1188. doi: 10.1242/jcs.03418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.