Abstract
The extremely halophilic archaeon Halobacterium sp. NRC-1 can grow phototrophically by means of light-driven proton pumping by bacteriorhodopsin in the purple membrane. Here, we show by genetic analysis of the wild type, and insertion and double-frame shift mutants of Bat that this transcriptional regulator coordinates synthesis of a structural protein and a chromophore for purple membrane biogenesis in response to both light and oxygen. Analysis of the complete Halobacterium sp. NRC-1 genome sequence showed that the regulatory site, upstream activator sequence (UAS), the putative binding site for Bat upstream of the bacterio-opsin gene (bop), is also present upstream to the other Bat-regulated genes. The transcription regulator Bat contains a photoresponsive cGMP-binding (GAF) domain, and a bacterial AraC type helix–turn–helix DNA binding motif. We also provide evidence for involvement of the PAS/PAC domain of Bat in redox-sensing activity by genetic analysis of a purple membrane overproducer. Five additional Bat-like putative regulatory genes were found, which together are likely to be responsible for orchestrating the complex response of this archaeon to light and oxygen. Similarities of the bop-like UAS and transcription factors in diverse organisms, including a plant and a γ-proteobacterium, suggest an ancient origin for this regulon capable of coordinating light and oxygen responses in the three major branches of the evolutionary tree of life. Finally, sensitivity of four of five regulon genes to DNA supercoiling is demonstrated and correlated to presence of alternating purine–pyrimidine sequences (RY boxes) near the regulated promoters.
Halobacterium species produce a specialized region in the cell membrane, named purple membrane, which consists of a two-dimensional crystalline lattice of a single chromoprotein, bacteriorhodopsin (BR). BR contains a 1:1 complex between a protein component bacterio-opsin and a chromophore retinal, and carries out light-driven proton pumping across the membrane (1, 2). BR is highly induced in late exponential to stationary phase of growth in response to high light intensity and low oxygen tension (3, 4). Proton pumping by BR is used to drive ATP synthesis and can sustain a period of phototrophic growth (1, 2). The mechanism of proton translocation across the membrane by BR has been the subject of extensive study by biophysical and mutational approaches (5, 6). Recently, retinal proteins similar to BR have been discovered in some fungi and uncultivated marine planktonic bacteria, indicating a much wider distribution in nature than originally appreciated (7, 8).
In Halobacterium sp. NRC-1, the proteins for biogenesis of the purple membrane are coded by the bop gene, specifying bacterio-opsin, and several nearby genes, e.g., brp, bat, blp, and crtB1, thought to be involved in synthesis of the chromophore, genetic regulation, or biogenesis of the membrane (Fig. 1a) (9, 10). Early studies showed that insertions into bacterio-opsin gene activator (bat) or bacterio-opsin related protein (brp), which are organized as an operon, block the transcription of bop. The bat gene restored regulation of bop expression by oxygen in cis–trans tests and encoded a 74-kDa predicted protein (Bat) with a PAS/PAC domain thought to be responsive to the redox status of cells (11, 12). Regulation of the bop gene was also studied by extensive mutagenesis of the 53-bp minimal promoter, which identified three cis-acting elements, a TATA box, for transcription factor and polymerase recruitment, an RY box, a DNA supercoiling sensitivity site, and an upstream activator sequence (UAS), the likely site for regulation by Bat (13, 14).
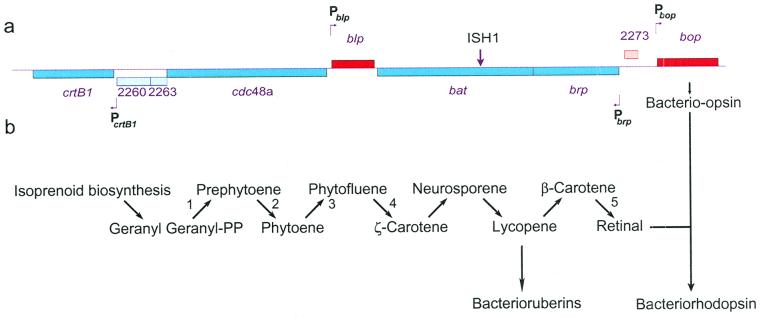
Figure 1.
Map of the purple membrane regulon and carotenoid biosynthesis pathway. (a) A map for the chromosomal locus coding for bop, brp, bat, blp, and crtB1 genes. The site for ISH1 insertion in bat in strain SD20 is indicated. The four mapped promoters, Pbop, Pbrp, Pblp, and PcrtB1, are indicated with bent arrows either above or below the map. (b) The pathway for carotenoid biosynthesis. Steps 1 and 2 in bacteria are catalyzed by the crtB1 gene product: phytoene synthase; steps 3 and 4 in plants are catalyzed by phytoene desaturase (Pds); and step 5 in Halobacterium sp. is catalyzed by Brp.
In this report, we have used a combination of genomic and genetic approaches to more fully characterize the purple membrane regulon. Evidence is provided for coordinate regulation of bacterio-opsin and retinal chromophore synthesis, and we also discuss mechanisms for sensing oxygen and light, by the pleiotropic regulator, Bat.
Materials and Methods
Halobacterium Strains and Culturing.
Halobacterium NRC-1 is the wild-type strain. Other strains used include Halobacterium S9, a purple membrane (Pum) constitutive strain, and SD20, a Pum− strain derivative of S9 with an ISH1 insertion in the bat gene (Fig. 1a) (12). Culturing of Halobacterium strains was done at 37°C in CM (complete medium) containing trace metals as described (15). Culturing for studying effects of treatment with novobiocin (Sigma) was conducted according to Yang et al. (12). Briefly, novobiocin was added to cultures, grown to an OD600 of 0.2 (early exponential phase), to a final concentration of 0.05 μg/ml; the cultures were allowed to grow further, in the presence of the drug, to stationary phase (OD600 > 1.8), at which point the cells were harvested for RNA preparations.
Identification of UAS.
The UAS consensus sequence (5′-ACCcnactagTTnGG-3′) (13, 14) was used in FINDPATTERNS analysis (16) (with 1–4 mismatches) of the whole Halobacterium NRC-1 genome sequence. The criteria used for subsequent screening of hits were the following: (i) the UAS should be upstream to a TATA box consensus sequence (5′-rtyTT/aTa-3′) (13, 14), (ii) the TATA box is at least 25–30 bp upstream to the first codon of the coding sequence, (iii) the spacing between the UAS and TATA box is not greater than 7 bp, and (iv) the putative UAS-containing promoter does not overlap a neighboring gene. However, considering the presence of multiple TATA box-binding proteins and transcription factor B (TFB) homologs in Halobacterium NRC-1, the alternate TATA box consensus sequence, and lack of a TFB responsive element (in the bop promoter, refs. 13 and 14), the screening process was also relaxed to allow for UAS-TATA box-coding sequence organizations (10, 17). It was considered imperative that the search process identified at least the bop and blp promoters, which are known to contain a UAS.
Primer Extension.
For RNA preparation, cells were grown as 10-ml (miniprep) or 100-ml (midiprep) cultures in CM supplemented with trace metals, to the desired cell density. Total RNA was prepared from 1.5-ml cultures by using the RNeasy mini-kit (Qiagen, Chatsworth, CA). Larger amounts of RNA were prepared from 10-, 50-, or 100-ml cultures (depending on cell density) by using the RNeasy midi-kit. Message levels were estimated by primer extension analysis performed with 5 pM (bop) or 100 fM (blp, brp, and crtB1) of 32P end-labeled oligonucleotide, on 10 μg (bop, blp, and crtB1) or 20 μg (brp) of RNA. The experiments were done in duplicate or triplicate, and the message levels were quantified by Phosphoimager analysis. Primer extension analysis of the pyrimidine nucleotide–disulphide oxidoreductase homolog message performed with 100 fM of primer on 10 μg of RNA was used as a control in all experiments.
Sequence Analysis of bat.
The bat gene was amplified from 1 μg of total genomic DNA prepared from Halobacterium NRC-1 and S9 by PCR using oligonucleotides flanking the coding sequence. The bat gene PCR products were sequenced on both strands by using the fluorescent dye-terminator chemistry (Amersham Pharmacia) on an ABI373A sequencer (Perkin–Elmer). Sequences were analyzed by using the Genetics Computer Group (Madison, WI) software package (16) running on a SGI O2 workstation (Silicon Graphics, Mountain View, CA).
Identification of GAF Domains.
A GAF domain profile was generated by using the program PROFILEMAKE (16) and a seed alignment available from the PFAM web site (18). This profile was then used to search the complete proteomes of Halobacterium NRC-1, Archaeoglobus fulgidus, Methanococcus jannaschii, Methanobacterium thermoautotrophicum, Aeropyrum pernix, and Pyrococcus abyssi. GAF domains in candidate proteins identified by this method were confirmed by profile search at the PFAM web site.
Results
Identification of Purple Membrane Regulon Genes.
To define the genes other than bop belonging to the purple membrane regulon (e.g., those controlled by Bat), we searched the genome of Halobacterium NRC-1 for ORFs containing properly positioned bop-like UAS sequences, which would suggest coordinate regulation with bop (10). Sequences most similar to the bop gene UAS were identified upstream to the three nearby genes, blp, brp, and crtB1 (17) (Fig. 1a). Interestingly, the predicted function of the crtB1 gene product, a phytoene synthase homolog, is to catalyze the first unique step of retinal and carotenoid biosynthesis, condensation of two molecules of geranyl–geranyl pyrophosphate yielding the C-40 isoprenoid compound phytoene (19), whereas the brp gene product likely catalyzes the final step in the formation of retinal, oxidative cleavage of β-carotene (20) (Fig. 1b). Although the function for the blp gene product is unknown, it is highly homologous to a conserved hypothetical protein in the green nonsulfur bacterium Chloroflexus aurantiacus. More importantly, the blp homolog in the bacterium is clustered with genes involved in bacteriochlorophyll/porphyrin biosynthesis, carotenoid biosynthesis, and photosynthetic electron transfer (21). Alignment of the 5′ regions of the four genes (Fig. 2) indicates a high degree of similarity within the UASs and the presence of nearby TATA boxes. Also, bop-like RY boxes with alternating purine–pyrimidine sequences were identified near two regulon promoters (brp: −12-ACGTGTGTAT−3; crtB1: +28-CGCACACGCAC-+38).
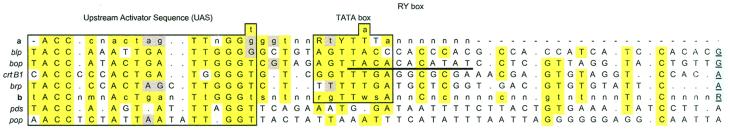
Figure 2.
Alignments for Halobacterium NRC-1 promoters (blp, bop, brp, and crtB1) with the bop promoter consensus derived by saturation mutagenesis (a) (13, 14), the consensus representing nucleotides conserved within the four promoters (b), the tomato pds gene, and the γ-proteobacterium pop gene promoters. Nucleotides conserved in at least three Halobacterium NRC-1 promoters are shaded in yellow, other identities to a are shaded in gray, and gaps are indicated with a dot. The transcription start site (+1) is shaded in blue. The bop promoter RY box is underlined; the sixth and tenth nucleotides within this element were identified to be the sites for supercoiling sensitivity (14).
A search for bop-like UAS sequences in GenBank led to the identification of several possible homologous regulatory sites, including one upstream to the recently discovered bop-like proteo-opsin gene (pop) in a marine γ-proteobacterium (8), and another within the transcribed noncoding 5′-leader region of the tomato phytoene desaturase (pds) gene (22) (Fig. 2).
Regulation of Purple Membrane Regulon by Bat.
To evaluate expression profiles of genes in the purple membrane regulon (bop, brp, bat, blp, and crtB1), we used primer extension analysis with RNA isolated at various stages of growth in batch culture (Fig. 3). All but one gene, brp, in the wild-type strain, NRC-1, were induced at late-exponential and stationary phases, a time when BR is induced. In strain S9, a purple membrane overproducer, transcript levels were constitutively high, and, in strain SD20, a bat∷ISH1 derivative of S9, transcript levels were undetectable. Relatively low levels of brp message were detected in the early exponential phase of both NRC-1 and SD20 cultures. This alternate profile for brp can be explained by the presence of an alternate TATA box downstream to the bop-like UAS (23) (Fig. 2). Because brp and bat are organized as an operon, with transcription being initiated at the promoter upstream to brp, both of these gene products are likely to be produced constitutively at low levels by means of an auto-repression mechanism (Fig. 3) (3). These observations indicate that Bat is a transcriptional regulator for all five genes in the purple membrane regulon.
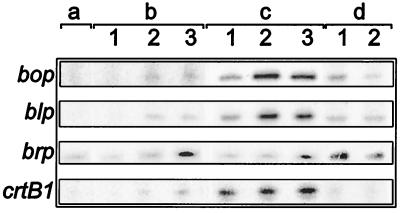
Figure 3.
Expression profiles for bop, blp, brp, and crtB1. Message levels for the four bop regulon genes in stationary phase cultures of the bat∷ISH1 strain, SD20 (a); early (1), mid- to late-exponential (2), and stationary phase (3) cultures of the wild-type strain NRC-1 (b); and early (1), mid- to late-exponential (2), and stationary phase (3) cultures of the purple membrane overproducing strain, S9 (c). (d) Sensitivity of transcription to changes in DNA supercoiling was tested by quantifying message levels from cultures grown with (1) and without (2) novobiocin.
In addition to Pbop, transcription from two other purple membrane regulon promoters, namely Pbrp and PcrtB1, showed sensitivity to novobiocin, a DNA-gyrase inhibitor (Fig. 3d) albeit at a lower magnitude (3). Addition of the drug to early exponential phase cultures resulted in >25–30% reduction in brp and crtB1 transcript levels. The blp transcript level was insensitive to novobiocin.
Bat Domain Structure.
The expression profiling implicated Bat as the purple membrane regulator. We therefore subjected Bat to computational analysis by using the profile search algorithms PFAM (18), HTHSCAN, and MOTIFS (16), which identified multiple domains. Interestingly, we found a putative cGMP-binding domain, GAF, which has been shown to be responsive to changes in light intensity (24, 25). A bacterial AraC type helix–turn–helix (HTH) motif was also identified, indicating that Bat binds directly to DNA (Fig. 4a). This type of HTH motif is common in Halobacterium sp. NRC-1, with a total of 138 HTH-containing genes identified in the genome (S.P.K. and S.D., unpublished observations). Previously, one halophage HTH protein was shown to bind DNA in band-shift assays (26). Finally, the redox-sensing PAS/PAC region was also present in Bat (3, 11).
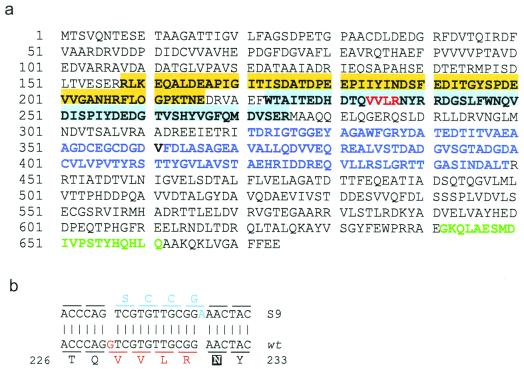
Figure 4.
Bat, the transcription regulator of the purple membrane regulon. (a) Positions and amino acids within the redox-responsive PAS (shaded orange)/PAC (shaded blue) domain, the photoresponsive GAF domain (purple font), and the HTH DNA binding motif (green font) are shown. The amino acids altered within the PAC domain in S9 Bat are indicated with a red font. (b) The double-frameshift mutation in the S9 bat gene. Deletion of a “G” (red font) and an insertion of an “A” (blue font) four codons apart has resulted in an altered amino acid sequence (shown below and above the aligned DNA sequences). The amino acid corresponding to the N85C mutation in the E. coli Aer protein is shaded in black (27).
Mutations in the S9 Bat PAC Domain.
Considering the ability of NRC-1 Bat to complement the deregulation of purple membrane synthesis in S9, we targeted the bat gene from both strains for further analysis (11). The bat gene was amplified from total genomic DNA from strains NRC-1 and S9, sequenced, and compared to elucidate the mechanism underlying the constitutive purple membrane overproducing phenotype of Halobacterium strain S9. We confirmed the role of the Bat PAS/PAC domain in redox-sensing by localizing double-frameshift mutations in the S9 bat gene, which altered the amino acid sequence near the N terminus of the PAC domain from 233-VVLR-236 to 233-SCCG-236. A similar mutation (N85C) has recently been reported in a homologous region of the Escherichia coli Aer protein responsible for regulating aerotaxis, resulting in a constant signal-on phenotype (27) (Fig. 4b).
Multiple GAF-Domain Linked Regulators.
Because multiple photosignaling pathways are known to exist in Halobacterium sp. (28), we searched NRC-1 genome sequence for more GAF-domain containing proteins. In addition to bat, five other genes coding proteins with putative GAF domains were identified. Two of these genes, boa1 and boa4, had been identified as homologs of bat by BLAST analysis (10). The three others identified include phoR, coding a putative regulatory protein, and two coding conserved hypothetical proteins, CHP2037 and CHP2334. The GAF domains were linked to redox-sensing PAS/PAC or PAS domains for the boa4, phoR, CHP2037, and CHP2334 gene products. Furthermore, three genes may code for histidine kinases (phoR, CHP2037, CHP2334), whereas the remaining two may code for DNA binding proteins (boa1, boa4) (Fig. 5). Similar computational analysis identified GAF domains in three other predicted archaeal signal transduction proteins, two in Methanobacterium thermoautotrophicum and one in Archaeoglobus fulgidus (Fig. 5).
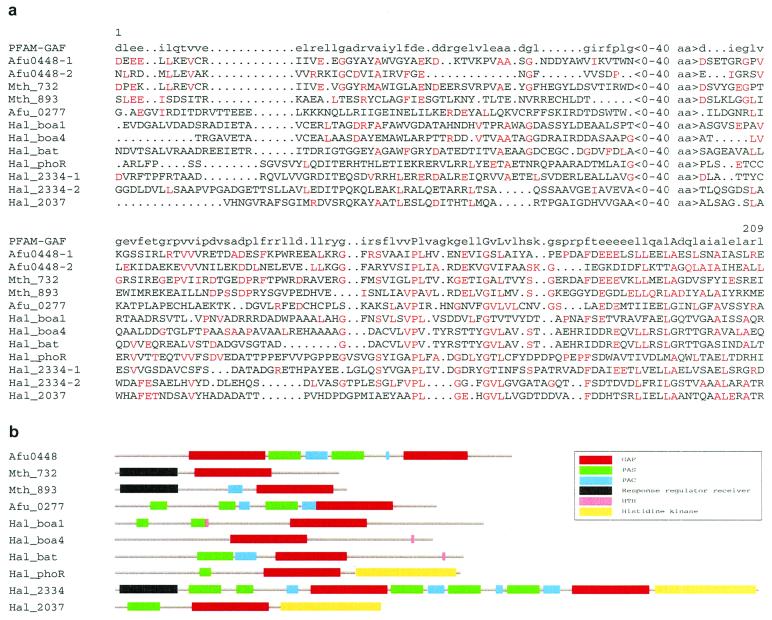
Figure 5.
GAF and other domains in predicted archaeal proteins. (a) Multiple sequence alignment of archaeal GAF domains. The amino acids with identities to the signature are indicated in a red font. Alignments for both GAF domains identified in Archaeoglobus fulgidus Afu0448 and Halobacterium CHP2334 (Hal_2334) are included in the alignment. (b) Domain organization in the archaeal putative cytoplasmic signal-transducing proteins. Regions containing GAF domains are shown as red boxes, PAS domains as green boxes, PAC domains as blue boxes, HTH domains as pink boxes, and histidine kinase domains as yellow boxes.
Discussion
The genes responsible for purple membrane synthesis in Halobacterium sp. constitute a regulon, probably to ensure equimolar synthesis of the structural protein, bacterio-opsin, and the chromophore, retinal, which together constitute BR. We have demonstrated a direct correlation between the presence of a bop-like UAS, the putative binding site for Bat, upstream to all genes critical in synthesis of BR and the requirement of a functional bat gene for expression and regulation of these genes. We therefore conclude that Bat, a pleiotropic transcription regulator, regulates purple membrane synthesis in Halobacterium sp. in response to both light and oxygen by directly binding a bop-like UAS in the regulon promoters.
Presence of a bop-like RY box, near the novobiocin-sensitive brp and crtB1 promoters, suggests that this regulon is also tuned to respond to changes in DNA supercoiling. DNA supercoiling-mediated regulation of bop has been shown to rely on presence of two critical nucleotides within the RY box, an alternating purine–pyrimidine sequence in the bop promoter. It has been speculated that the RY box attenuates bop transcription in response to changes in DNA-supercoiling by modulating either open complex formation or binding of TATA boxbinding protein to the TATA box (14). The brp and crtB1 promoters may respond to changes in DNA supercoiling by a similar phenomenon.
Bat is probably capable of responding to both light and oxygen because of presence of a photoresponsive GAF domain, a redox-responsive PAS/PAC domain, and a HTH DNA-binding motif. In bacteria and eukaryotes, the GAF domain is conserved in two classes of proteins, phytochromes and cGMP-specific phosphodiesterases. Phytochromes, which can exist in two photoconvertible forms, Pr (generally inactive) and Pfr (active), induce cGMP-mediated cascades to control the expression of light-regulated genes. This mechanism of light signal transduction is hypothesized to be an evolutionary link between phototransducing proteins from bacteria and eukaryotes (25). In an alternate mechanism, the Pfr form of phytochrome B (PfrB), a GAF domain-containing protein, translocates into the nucleus to directly interact with the G box-bound PIF3, a transcription factor (29). A similar mechanism of light-regulation may be operative in Halobacterium sp. NRC-1, a prokaryote; however, here the GAF domain and the HTH DNA-binding motif are fused in Bat probably because of lack of a nuclear translocation step.
The association of GAF and PAS/PAC domains with a DNA-binding motif in Bat classifies it as a member of a superfamily of cytoplasmic sensory transduction proteins responsible for translating changes in light intensity and/or redox-potential into transcription signals (22). In other organisms, the transcription signals can be mediated by means of different transducer domains like serine–threonine protein kinases [e.g., NPH1, which is responsible for mediating phototropism in Arabidopsis thaliana (30)], histidine kinases [e.g., NodV, a nodulation gene in Bradyrhizobium japonicum (31)], or helix–loop–helix domains [e.g., PIF3, in A. thaliana (29)].
Interestingly, the tomato pds gene region containing the putative bop-like UAS could restore enhanced and regulated expression in promoter-GUS-fusion reporter-gene analysis (22). Because plants induce the genes for carotenoid synthesis, like Halobacterium, under photo-oxidative stress, it suggests the intriguing possibility that a similar mechanism is operating in both systems. No data are available on the regulation of the marine γ-proteobacterium pop gene because this organism has not yet been cultured in the laboratory (8).
We have reported on the characterization of a regulon for coordinate synthesis of bacterio-opsin protein and retinal chromophore in the purple membrane of the extremely halophilic archaeon, Halobacterium sp. NRC-1. The transcription regulator Bat contains features that resemble both bacterial and eukaryotic signal transduction systems. The bop-like UAS, which Bat likely acts at to regulate light- and oxygen-responsive gene expression, may be conserved in highly diverse organisms, such as some plants and bacteria. The occurrence of a family of regulators similar to Bat suggests that a complex regulatory network exists in Halobacterium for responding to frequent and sudden changes in light and oxygen availability in its extraordinarily dynamic environment. Physiological capabilities of this extremely halophilic archaeon include facultatively anaerobic and phototrophic growth, phototaxis, gas vesicle-mediated flotation, circadian rhythm, and photorepair of DNA. Given the complete genome sequence (10) and the ability to apply postgenomic methodologies to Halobacterium (15, 32), we are in an ideal position to further decipher the regulation of multigene systems in an archaeon.
Acknowledgments
This work was supported by research grants from the National Science Foundation (MCB-9604443, MCB-0095700, and MCB-9812330, to S.D.) (MCB-9900497 and MCB-0000-4124, to L.H.).
Abbreviations
- BR
bacteriorhodopsin
- bop
bacterio-opsin gene
- bat
bacterio-opsin gene activator
- brp
bacterio-opsin related protein
- UAS
upstream activator sequence
- CM
complete medium
- pop
proteo-opsin gene
- pds
phytoene desaturase
- HTH
helix–turn–helix
References
- 1.Oesterhelt D, Stoeckenius W. Proc Natl Acad Sci USA. 1973;70:2853–2857. doi: 10.1073/pnas.70.10.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sumper M, Reitmeier H, Oesterhelt D. Angew Chem Int Ed Engl. 1976;16:187–194. doi: 10.1002/anie.197601871. [DOI] [PubMed] [Google Scholar]
- 3.Yang C-F, DasSarma S. J Bacteriol. 1990;172:4118–4121. doi: 10.1128/jb.172.7.4118-4121.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shand F R, Betlach M C. J Bacteriol. 1991;173:4692–4699. doi: 10.1128/jb.173.15.4692-4699.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luecke H, Richter H T, Lanyi J K. Science. 1998;280:1934–1937. doi: 10.1126/science.280.5371.1934. [DOI] [PubMed] [Google Scholar]
- 6.Krebs M P, Khorana H G. J Bacteriol. 1993;175:1555–1560. doi: 10.1128/jb.175.6.1555-1560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spudich J L, Yang C S, Jung K H, Spudich E N. Annu Rev Cell Dev Biol. 2000;16:365–392. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 8.Beja O, Aravind L, Koonin E V, Suzuki M T, Hadd A, Nguyen L P, Jovanovich S B, Gates C M, Feldman R A, Spudich J L, et al. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 9.Leong D, Pfeifer F, Boyer H, Betlach M C. J Bacteriol. 1988;170:4903–4909. doi: 10.1128/jb.170.10.4903-4909.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng W V, Kennedy S P, Mahairas G G, Berquist B, Pan M, Shukla H D, Lasky S R, Baliga N S, Thorsson V, Sbrogna J, et al. Proc Natl Acad Sci USA. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. . (First Published October 3, 2000; 10.1073/pnas.190337797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gropp F, Betlach M C. Proc Natl Acad Sci USA. 1994;91:5475–5479. doi: 10.1073/pnas.91.12.5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang C-F, Kim J-M, Molinari E, DasSarma S. J Bacteriol. 1996;178:840–845. doi: 10.1128/jb.178.3.840-845.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baliga N S, DasSarma S. J Bacteriol. 1999;181:2513–2518. doi: 10.1128/jb.181.8.2513-2518.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baliga N S, DasSarma S. Mol Microbiol. 2000;36:1175–1183. doi: 10.1046/j.1365-2958.2000.01915.x. [DOI] [PubMed] [Google Scholar]
- 15.DasSarma S, Fleischmann E M, Rodriguez-Valera F. Archaea: A Laboratory Manual-Halophiles. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. [Google Scholar]
- 16.Genetics Computer Group. wisconsin package. Madison, WI: Genetics Computer Group; 1999. , version 10.1. [Google Scholar]
- 17.Gropp F, Gropp R, Betlach M C. Mol Microbiol. 1995;16:357–364. doi: 10.1111/j.1365-2958.1995.tb02307.x. [DOI] [PubMed] [Google Scholar]
- 18.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armstrong G A. Annu Rev Microbiol. 1997;52:629–659. doi: 10.1146/annurev.micro.51.1.629. [DOI] [PubMed] [Google Scholar]
- 20.Peck, R. F., Echavarri-Erasun, C., Johnson, E. A., Ng, W. V., Kennedy, S. P., Hood, L., DasSarma, S. & Krebs, M. P. (November 22, 2000) J. Biol. Chem., 10.1074/jbc.M009492200. [DOI] [PubMed]
- 21.Xiong J, Fischer W M, Inoue K, Nakahara M, Bauer C E. Science. 2000;289:1724–1730. doi: 10.1126/science.289.5485.1724. [DOI] [PubMed] [Google Scholar]
- 22.Corona V, Aracri B, Kosturkova G, Bartley G E, Pitto L, Giorgetti L, Scolnik P A, Giuliano G. Plant J. 1996;9:505–512. doi: 10.1046/j.1365-313x.1996.09040505.x. [DOI] [PubMed] [Google Scholar]
- 23.Baliga N S, Goo Y A, Ng W V, Hood L, Daniels C J, DasSarma S. Mol Microbiol. 2000;36:1184–1185. doi: 10.1046/j.1365-2958.2000.01916.x. [DOI] [PubMed] [Google Scholar]
- 24.Quail P H. BioEssays. 1997;19:571–579. doi: 10.1002/bies.950190708. [DOI] [PubMed] [Google Scholar]
- 25.Aravind L, Ponting C P. Trends Biochem Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 26.Ken R, Hackett N R. J Bacteriol. 1991;173:955–960. doi: 10.1128/jb.173.3.955-960.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Repik A, Rebbapragada A, Johnson M S, Haznedar J O, Zhulin I B, Taylor B L. Mol Microbiol. 2000;36:806–816. doi: 10.1046/j.1365-2958.2000.01910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoff W D, Jung K H, Spudich J L. Annu Rev Biophys Biomol Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Garcia J F, Huq E, Quail P H. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- 30.Christie J M, Reymond P, Powell G K, Bernasconi P, Raibekas A A, Liscum E, Briggs W R. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 31.Loh J, Garcia M, Stacey G. J Bacteriol. 1997;179:3013–3020. doi: 10.1128/jb.179.9.3013-3020.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peck R F, DasSarma S, Krebs M P. Mol Microbiol. 2000;35:667–676. doi: 10.1046/j.1365-2958.2000.01739.x. [DOI] [PubMed] [Google Scholar]