Abstract
Ralstonia solanacearum, which causes bacterial wilt of diverse plants, produces copious extracellular polysaccharide (EPS), a major virulence factor. The function of EPS in wilt disease is uncertain. Leading hypotheses are that EPS physically obstructs plant water transport, or that EPS cloaks the bacterium from host plant recognition and subsequent defense. Tomato plants infected with R. solanacearum race 3 biovar 2 strain UW551 and tropical strain GMI1000 upregulated genes in both the ethylene (ET) and salicylic acid (SA) defense signal transduction pathways. The horizontally wilt-resistant tomato line Hawaii7996 activated expression of these defense genes faster and to a greater degree in response to R. solanacearum infection than did susceptible cultivar Bonny Best. However, EPS played different roles in resistant and susceptible host responses to R. solanacearum. In susceptible plants the wild-type and eps− mutant strains induced generally similar defense responses. But in resistant Hawaii7996 tomato plants, the wild-type pathogens induced significantly greater defense responses than the eps− mutants, suggesting that the resistant host recognizes R. solanacearum EPS. Consistent with this idea, purified EPS triggered significant SA pathway defense gene expression in resistant, but not in susceptible, tomato plants. In addition, the eps− mutant triggered noticeably less production of defense-associated reactive oxygen species in resistant tomato stems and leaves, despite attaining similar cell densities in planta. Collectively, these data suggest that bacterial wilt-resistant plants can specifically recognize EPS from R. solanacearum.
Introduction
Plants resist many potential pathogens with low-amplitude innate immunity defenses that are triggered by recognition of microbe-associated molecular patterns (MAMPs) such as bacterial flagellin [1], [2]. R-gene mediated plant disease resistance typically involves much higher amplitude defense responses launched in response to pathogen effectors (avirulence factors) that the pathogen needs for full virulence and that the resistant plant has evolved to recognize [1]. The triggers and mechanisms of horizontal plant disease resistance are poorly understood, although this type of resistance is often stable and is widely deployed in agriculture [3].
As for many other plant diseases, resistance breeding is the best control for bacterial wilt (BW), a serious vascular disease caused by the soilborne bacterium Ralstonia solanacearum [4]. There is no single-gene resistance to BW in tomato, an economically important natural host of R. solanacearum. The most widely used resistance source is Hawaii7996 (H7996), a breeding line that carries at least five QTLs that together confer resistance to most pathogen strains via unknown mechanism(s) [5], [6], [7]. However, this horizontally-resistant line is not immune to the pathogen, and latent infections occur frequently (3). The defense signaling pathways triggered by BW disease development in tomato are not known, and these have direct implications for understanding and selecting BW-resistant germplasm. Thus, one aim of this study was to describe the kinetics of defense responses in susceptible and resistant tomato plants infected by two biologically distinct strains of R. solanacearum.
Extracellular polysaccharide (EPS) is a major virulence factor of R. solanacearum [8]. Site-directed mutants unable to synthesize EPS I, a heterogenous polymer of N-acetylated monosaccharides, are nearly avirulent and do not colonize plant xylem vessels as well as wild-type [9], [10]. R. solanacearum is a genetically diverse species complex, but the EPS structure is sufficiently well-conserved that an anti-EPS antibody can recognize all members of the group [11], [12]. EPS synthesis is regulated by the PhcA quorum sensing system such that it is produced abundantly at high cell densities in culture or when the bacterium grows in the confines of host plant xylem vessels [13]. However, it is not known how EPS contributes to BW disease development. It has been suggested that EPS directly causes wilting by physically blocking water flow in the densely-colonized xylem vessels of infected hosts [14]. It has also been hypothesized that the pathogen needs EPS to form biofilms on vessel surfaces during disease development; that EPS helps R. solanacearum survive desiccation or antibiosis in soil during periods away from host plants; and finally that EPS protects R. solanacearum from plant antimicrobial defenses by cloaking bacterial surface features that could be recognized by hosts [9], [14], [15], [16].
To test the latter hypothesis, we measured expression of defense genes and production of defensive reactive oxygen species (ROS) in susceptible and resistant tomato plants infected by wild-type and eps− mutants of two R. solanacearum strains. We found that eps− bacteria triggered similar defense signal pathway expression in a BW-susceptible tomato, undermining the cloaking hypothesis. Unexpectedly, BW-resistant H7996 plants expressed reduced defenses against the eps− strain, but they did activate the salicylic acid defense pathway in response to cell-free purified EPS. These results suggest that BW-resistant tomato plants recognize EPS, an abundantly-expressed and indispensible virulence factor of R. solanacearum.
Results
Temperate R. solanacearum strain UW551 breaks the BW resistance of H7996 tomato
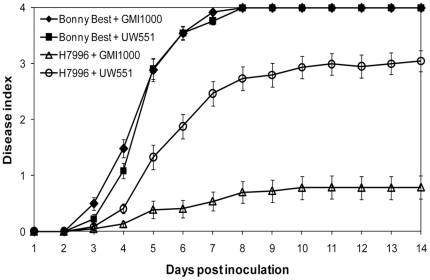
R. solanacearum strains GMI1000 and UW551 were both highly virulent on susceptible tomato cv. Bonny Best (Figure 1). All inoculated plants were dead by 8 dpi and the strains had indistinguishable disease progress curves. In contrast, tomato breeding line H7996, a widely-used source of BW disease resistance, was quite resistant to tropical strain GMI1000; only 12% of the plants were dead by 14 dpi (Figure 1). However, H7996 was susceptible to R. solanacearum UW551, a typical sequevar 1 (Race 3 biovar 2) strain that causes losses in temperate zones and tropical highlands [17]. UW551 killed about 80% of H7996 plants within 14 dpi. The virulence of strains GMI1000 and UW551 was significantly different (P<0.001) on the resistant tomato plants.
Figure 1. Virulence of Ralstonia solanacearum strains GMI1000 and UW551 on resistant and susceptible tomato plants.
Unwounded susceptible (cv. Bonny Best) and horizontally resistant (H7996) tomato plants were soil-soak inoculated to a concentration of ∼1×108 CFU/g soil and incubated at 28°C. Plants were rated daily on a 0 to 4 disease index scale where 0 = healthy and 4 = 100% wilted. Each point represents the mean disease index (± SE) for four independent experiments, each containing 16 plants per treatment.
Tomato plants responded to R. solanacearum infection by upregulating marker genes for the salicylic acid (SA) and ethylene (ET) defense pathways
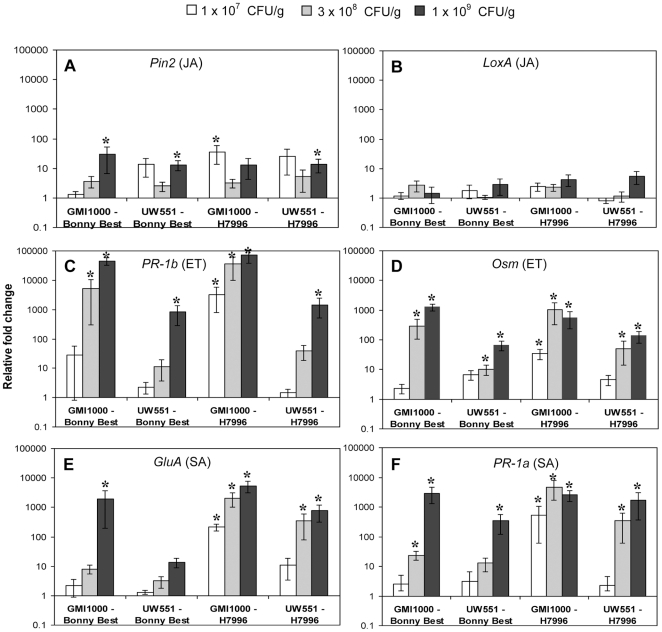
Quantitative RT-PCR gene expression analysis in susceptible and resistant tomato plants infected with R. solanacearum revealed little or no activation of the jasmonic acid (JA) pathway marker genes Pin-2 and LoxA [18], [19]. However, both PR-1b and Osm, which are ET-induced [20], [21], [22], and GluA and PR-1a, which are regulated by the SA pathway [20], [22], [23], were expressed at significantly higher levels in plants with pathogen cell densities ≥3×108 CFU/g, relative to water-inoculated controls (Figure 2).
Figure 2. Expression of tomato defense genes following soil-soak inoculation with Ralstonia solanacearum strains GMI1000 or UW551 in susceptible cultivar Bonny Best or horizontally resistant line H7996.
Genes represent activation of the jasmonic acid (JA) pathway (A: Pin2, B: LoxA), the ethylene (ET) pathway (C: PR-1b, D: Osm), and the salicylic acid (SA) pathway (E: GluA, F: PR-1a). Gene expression was measured by qRT-PCR in response to three pathogen cell densities: 1×107 CFU/g stem (symptomless plants, white bars), 3×108 CFU/g (symptomless or first wilting signs, grey bars), and 1×109 CFU/g (early disease corresponding to DI = 1, black bars). Asterisks above bars indicate significant differences (P>0.05) in gene expression between mock and R. solanacearum inoculated tomatoes. P-values reflecting differences between cell densities (CFU), tomato cultivars and strains are shown in Table S2. Bars show normalized mean fold induction relative to mock-inoculated control plants (± SE). N = 6 to 12 plants for each cell density and strain, >3 independent experiments.
Resistant tomato plants activated the SA and ET defense pathways more rapidly than a susceptible cultivar
BW-resistant H7996 responded to large populations of both R. solanacearum strains by increasing expression of genes in the ET and SA signaling pathways by two to three orders of magnitude (Figure 2). Defense genes in H7996 were noticeably induced even at lower pathogen cell densities (1×107 CFU of GMI1000/gm stem and 3×108 CFU of UW551/gm stem). In contrast, susceptible cv. Bonny Best had no detectable defense response to 1×107 CFU/gm. This result is consistent with the general observation that disease-resistant plants have faster and stronger defense responses [24].
Large populations of strain GMI1000 triggered strong defense pathway gene expression in both susceptible and resistant tomato plants
R. solanacearum GMI1000, a broad host range tropical strain originally isolated from tomato, readily infected susceptible cv. Bonny Best. Resistant H7996 was less frequently infected and disease developed more slowly in this line, as is characteristic of horizontal resistance. However, when either Bonny Best or H7996 plants contained 1×109 CFU of GMI1000/g stem, populations typical of full-blown wilt disease, their expression of PR-1b and Osm (ET pathway) and GluA and PR-1a (SA pathway) was two to four orders of magnitude larger than in plants at an early stage of colonization, containing just 1×107 CFU/gm (Figure 2). This result suggests that GMI1000 induces similar defense responses in both susceptible and resistant tomato, but that the timing of response is different in the two hosts.
Strain UW551 was able to avoid or calm defense responses in a susceptible tomato cultivar, but did activate defense gene transcription in BW resistant line H7996
We observed a strikingly different pattern of tomato responses to strain UW551, a temperate strain with a relatively narrow host range limited to potato, tomato, and some related species. At high bacterial cell densities in resistant H7996, UW551 elicited PR-1a, Osm and GluA expression levels similar to those induced by GMI1000. Only PR-1b expression was two orders of magnitude lower after infection with UW551 compared to GMI1000 (Figure 2). However, in susceptible Bonny Best UW551 had remarkably little effect on defense gene expression, which was two to three orders of magnitude lower than that elicited by GMI1000. Induction of GluA, representative of SA pathway activation, was especially weak (Figure 2E).
Bacterial EPS plays different roles in susceptible and resistant tomato host responses
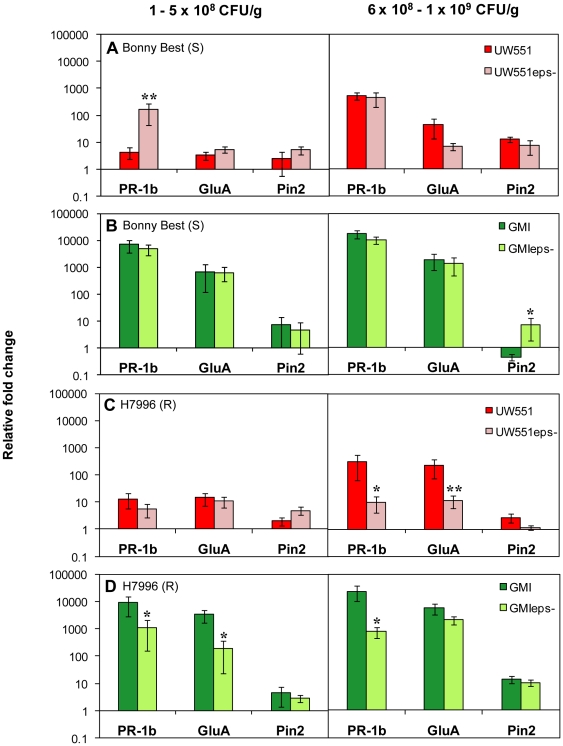
As documented for other R. solanacearum strains, an EPS-deficient mutant of UW551, UW551ΔepsB, was dramatically reduced in virulence on both susceptible and resistant tomato plants (P<0.001) and rarely killed the host (Figure S1). To test the hypothesis that EPS cloaks R. solanacearum from recognition by its plant host, we measured tomato defense gene expression following infection by wild-type and ΔepsB strains of the pathogen. The defense-associated PR-1b gene of susceptible Bonny Best was upregulated 40-fold (P = 0.001) in response to an eps − mutant of R. solanacearum UW551 compared to PR-1b expression triggered by wild-type bacteria. This finding is consistent with the cloaking hypothesis. This effect was observed up to a pathogen cell density of about 5×108 CFU/g stem (Figure 3A, left). However, at higher cell densities (>5×108 CFU/gm stem) led to increased defense gene expression (Fig. 3A, right). However, infection with wild-type and EPS-deficient strain GMI1000 elicited defense gene expression at comparable magnitudes in susceptible Bonny Best (Figure 3B).
Figure 3. Expression of tomato defense genes following petiole inoculation with Ralstonia solanacearum wild-type strains and extracellular polysaccharide-deficient ΔepsB mutants.
Gene expression was measured in A: BW-susceptible (S) cv. Bonny Best infected with UW551 or UW551ΔepsB; B: BW-susceptible cv. Bonny Best infected with GMI1000 or GMI1000ΔepsB; and C: horizontally resistant (R) line H7996 infected with UW551 or UW551ΔepsB; D: horizontally resistant line H7996 infected with GMI1000 or GMI1000ΔepsB. Plants were inoculated through the cut petiole of the first true leaf. Genes represent activation of ET pathway (PR-1b), SA pathway (GluA), and JA pathway (Pin2). Gene expression was measured in response to two pathogen cell densities in tomato stem tissue: 1 to 5×108 CFU/g stem and 6×108 to1×109 CFU/g. Asterisks above bars indicate significant differences in gene expression between wild-type strain and ΔepsB mutant (* = P>0.05, ** = P = 0.001). Bars show normalized mean fold induction relative to mock-inoculated control plants (± SE). UW551: N = 8 to 15 plants per treatment, with 4 independent experiments; GMI1000: N = 6 to 11 plants per treatment, with 3 independent experiments.
Surprisingly, the opposite was true in BW resistant H7996 tomato plants. At cell densities below 5×108 CFU/gm stem, the host responded to infections with wild-type and eps− R. solanacearum strain UW551 by slightly upregulating tomato defense genes (Figure 3C, left). But when the pathogen exceeded 5×108 CFU/gm stem, the wild-type strain elicited 30-fold higher PR-1b (P = 0.03) and 20-fold higher GluA expression (P = 0.001) than did the ΔepsB mutant (Figure 3C, right). Similarly, wild-type strain GMI1000 triggered a significantly stronger response than the ΔepsB mutant at all pathogen concentration tested. Even at 1×105 CFU/g stem PR-1b expression was 8-fold higher (P = 0.049) and GluA expression showed an 18-fold increase (P = 0.015) after infection with the wild-type strain compared to the EPS-deficient strain. At cell densities above 5×108 CFU/gm stem, the effect of EPS on defense gene expression became even more apparent since the wild-type strain elicited 30-fold higher PR-1b expression (P = 0.04) than GMI1000ΔepsB. Collectively, these results suggested that the resistant tomato can recognize the EPS produced by R. solanacearum.
Resistant plants recognized cell-free EPS
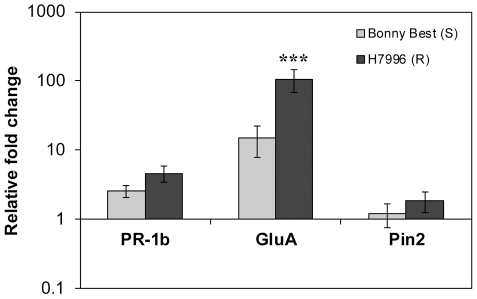
Plant defense expression levels could be affected by the effectors and enzymes secreted by live bacteria. To more directly test the hypothesis that pathogen EPS triggers defense responses in wilt-resistant plants, we measured tomato transcriptional response to a biologically relevant amount (20 µg) [9] of extensively purified EPS from UW551. Purified EPS activated the SA pathway (GluA) in H7996 to a significantly greater degree (7-fold, P = 0.00003) than in Bonny Best. This indicates that the resistant host perceives and responds to R. solancearum EPS. Interestingly, although live cells of the wild-type pathogen triggered much higher PR-1b expression in H7996 than UW551ΔepsB did, cell-free EPS alone did not significantly increase PR-1b expression, suggesting that EPS activates only a subset of defense-associated responses (Figure 4). Alternatively, full-spectrum signal transduction may require interaction of EPS with specific tissues in ways that occur during natural infection but not when EPS is introduced directly into the stem.
Figure 4. Expression of tomato defense genes in response to purified Ralstonia solanacearum extracellular polysaccharide.
Gene expression was measured in bacterial wilt-susceptible cv. Bonny Best (S) and horizontally resistant line H7996 (R) by qRT-PCR 24 h after injection of 20 µg purified EPS through the cut petiole of the first true leaf directly into the vascular system. Genes represent activation of the ET pathway (PR-1b), the SA pathway (GluA), and the JA pathway (Pin2). Asterisks above bars indicate significant differences in gene expression between BW susceptible Bonny Best and horizontally resistant H7996 (*** = P>0.0001). Bars show normalized mean fold induction relative to mock-inoculated control plants (± SE). N = 40 plants per treatment, in four independent experiments.
EPS triggered a strong oxidative burst in resistant plants
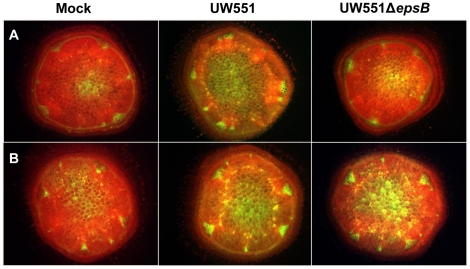
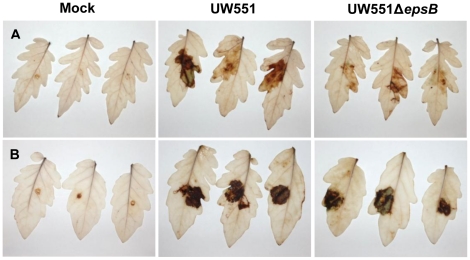
To determine if the defense-associated gene expression patterns we observed in response to wild-type and EPS-deficient R. solanacearum cells correlated with biochemical indicators of active plant defenses, we used the fluorescent dye dihydrorhodamine123 to assess tomato stem levels of ROS, a common element of plant antimicrobial defenses [1]. This qualitative dye revealed that infection by wild-type R. solanacearum UW551 triggered a strong oxidative burst in the vascular bundles of both resistant and susceptible tomato plants (Figure 5). In contrast, H7996 plants infected with 104 to 105 CFU/g of UW551ΔepsB accumulated noticeably less ROS than did H7996 stems carrying similar populations of wild-type R. solanacearum (Figure 5A). No such response was observed in cv. Bonny Best, where stems containing 104 to 105 CFU/g of UW551ΔepsB had ROS levels indistinguishable from those in stems infected by the wild-type strain (Figure 5B). These differences in ROS accumulation triggered by wild-type and EPS-deficient bacteria were also seen in tomato leaves, indicating that this phenomenon is not unique to stem tissue (Figure 6).
Figure 5. Accumulation of reactive oxygen species (ROS) in tomato stem tissue.
ROS were determined in A: horizontally BW-resistant tomato H7996 and B: susceptible cv. Bonny Best 48 h after infection with Ralstonia solanacearum wild-type strain UW551 or UW551ΔepsB or water (mock-inoculated control). At 48 h post-inoculation, stem cross-sections containing 105 CFU/g bacteria were stained with 50 µM dihydrorhodamine 123 (DHR 123) and fluorescence microscopy was used to visualize the green fluorescence of rhodamine 123 generated by oxidizing DHR 123 by ROS. Three independent experiments each contained eight plants per treatment; representative results are shown.
Figure 6. Reactive oxygen species (ROS) accumulation in tomato leaves.
ROS were determined in A: horizontally BW-resistant line H7996 and B: BW-susceptible cv. Bonny Best 48 h after infusion with 1×109 CFU/ml R. solanacearum strain UW551 or EPS I mutant UW551ΔepsB. ROS appears as a brown precipitate after leaves were stained with the in situ endogenous peroxidase-dependent histochemical stain 3,3′-diaminobenzidine (DAB). The experiment was repeated five times; representative results are shown.
Discussion
Host resistance is the optimal strategy for controlling BW disease, but the specific triggers and mechanisms responsible for horizontal wilt resistance in tomato are not known. We found that the tomato ET and SA signaling pathways are activated during BW disease resistance. This is consistent with the finding that VIGS-mediated disruption of the JA, ET, SA and MAPK pathways increased colonization of stem bases and/or mid-stems in H7996 by R. solanacearum strain Pss4 [25], [26]. In addition, overexpressing the ET pathway decreased wilting symptoms in susceptible L390 tomato [25]. These results suggested that the JA, ET and SA defense signaling pathways interact synergistically in the resistance of tomato against BW. However, under our experimental conditions, JA pathway marker genes were not substantially upregulated in response to either R. solanacearum strain in resistant or in susceptible tomato plants.
Arabidopsis has been used as model plant for the study of R. solanacearum-host interactions, but it is not a natural host of R. solanacearum and artifactual inoculation methods are required to generate symptoms [27]. Our results and those of others [28] suggest that this model plant may react differently to R. solanacearum than the natural host tomato. Disease development and proliferation of GMI1000 in Arabidopsis was not SA-dependent, but inactivation of ET-related signaling pathways resulted in decreased symptom development in susceptible plants, indicating that ET-regulated defenses reduce disease severity [29]. In contrast, resistance of Arabidopsis ecotype Nd-1, which unlike tomato carries a single vertical resistance gene (RSS1), was partially dependent on SA [30], but appeared to be independent of ET signaling [29]. Further, the JA signaling pathway may suppress Arabidopsis defense against R. solanacearum, since JA-insensitive Arabidopsis plants displayed milder disease symptoms [31]. Overall, the responses of Arabidopsis plants to R. solanacearum appear to differ significantly from those of tomato.
We describe here the kinetics of tomato defense gene expression against two biologically distinct R. solanacearum strains in BW-resistant and susceptible hosts. Our results are consistent with the general observation that major differences between resistant and susceptible responses are quantitative and/or kinetic, and not necessarily caused by the expression of different sets of genes [32], [33]. H7996 resistance was characterized by a faster response kinetic; the ET and SA pathways were activated at much lower threshold pathogen cell densities in H9776 xylem than in susceptible Bonny Best. The ultimate magnitude of the plant response to high pathogen cell densities was comparable between susceptible and resistant plants, but it differed strikingly between the two strains. In particular, susceptible Bonny Best launched stronger defenses against strain GMI1000 than against strain UW551.
Genetic differences between the strains could explain why these two pathogen strains trigger different responses from the plant host. The GMI1000 genome contains about 1000 coding sequences not present in UW551, while about 500 genes are unique to UW551 [34]; most of these genes encode hypothetical proteins. The products of any of those strain-specific genes, or differential regulation of common genes, could lead to differential recognition of the pathogen and might explain how temperate strain UW551 calms or evades host recognition. One likely explanation is that strain UW551 deploys Type 3-secreted (Hrp) effectors that specifically suppress defense responses in Bonny Best. R. solanacearum strains do produce Type 3-secreted effectors that reduce plant innate immunity [14], and indeed we found that expression of genes in the ET and SA signaling pathways was reduced in tomato plants infected by a hrp mutant (A. Milling and J. M. Jacobs, unpublished results). Further, a recent in planta microarray analysis in our lab revealed expression trend differences between UW551 and GMI1000. Of 31 orthologous genes encoding Type 3-secreted effectors or HrpB-dependent secretion system structural components, 25 were upregulated to a significantly greater degree in UW551 than in GMI1000 (Jacobs et al., in preparation). This is an intriguing topic for future study.
Interestingly, although strain GMI1000 triggered stronger expression of the ET and SA pathway genes than UW551, these strains induced indistinguishable rapid disease progress in susceptible tomato plants. In contrast, the resistance of H7996 to GMI1000 may result from more rapid induction of the ET and SA pathways. It seems likely that the differential expression of defense signaling pathways we observed in Bonny Best and H7996 is accompanied by expression of diverse additional plant genes that confer specific aspects of wilt susceptibility or tolerance.
R. solanacearum's nitrogen- and carbohydrate-rich EPS is metabolically expensive and its production is tightly regulated by a complex network. Nonetheless, it is abundantly produced at high cell densities and inside host plants [9], [13] and it is critical for bacterial wilt virulence [16]. Why? It has been hypothesized that EPS protects R. solanacearum from plant antimicrobial defenses by cloaking bacterial surface features from host recognition. It would seem advantageous for hosts to recognize an abundantly expressed extracellular molecule required for virulence. However, bacterial EPS is generally not perceived by eukaryotes as a MAMP, but rather enables bacteria to evade immunity [35]. We did observe that susceptible plants upregulated the ET pathway to a greater degree in response to an eps− mutant of one R. solanacearum strain, temperate R3bv2 strain UW551, at least at lower pathogen cell densities. Overall, however, the susceptible cultivar responded similarly to wild-type and eps− strains, which does not support the cloaking hypothesis.
It has been suggested that the EPS of many bacterial plant pathogens, including R. solanacearum, non-specifically suppresses MAMP-triggered immunity via sequestration of apoplastic calcium ions, which play a role in defense signaling [36]. However, we found that R. solanacearum EPS does not suppress plant defenses, but rather plays a more specific role in inducing plant defenses. Our experiments with eps− mutants and with purified EPS from two different R. solanacearum strains demonstrate that EPS can specifically elicit defense gene expression and ROS production in at least one resistant tomato genotype. The SA defense signaling pathway in H7996 appears especially responsive to EPS-induced signaling. The susceptible cultivar generally responded similarly to wild-type and EPS-deficient strains, which is also inconsistent with the calcium hypothesis.
EPS is a virulence factor for many plant pathogenic bacteria, and the chemical structures of these polysaccharides vary among species, suggesting diversifying selection pressure [15]. Some experiments have suggested that certain plants can recognize EPS from specific bacteria. Potato cultivars have membrane-bound receptors that recognize EPS from Clavibacter michiganensis pv. sepedonicus and induce defense responses [37]. EPS extracts from Pseudomonas syringae pv. ciccaronei and P. savastonoi pv. nerii caused necrotic lesions when infiltrated into tobacco leaves, induced H2O2 release from tobacco cells in culture medium, and decreased cytosolic ascorbate peroxidase (APX), one of the main enzymes for ROS scavenging in plant cells; EPS extracts from the related plant pathogen P. caryophylli had no such effects [38]. EPS from an incompatible isolate of Xanthomonas campestris pv. vesicatoria elicited phytoalexin production in pepper leaves [39]. Moreover, the EPS produced by bacteria present in the mammalian gut can increase certain host immune responses [40], [41].
Recognition of R. solanacearum EPS, either specifically or as a MAMP, could give BW-resistant H7996 tomato plants a crucial advantage by triggering faster defense responses. To evade host recognition, plant pathogenic bacteria are known to vary MAMP structure both within and across species [27], [42]. Our result suggests that polymorphisms also exist on the host side for elicitor perception. Bacterial wilt resistance in H7996 is polygenic and complex, so EPS-triggered defenses can explain only part of its resistance. Nonetheless, if H7996 proves unique among tomato lines in its ability to perceive R. solanacearum EPS, this may explain why it has consistently ranked as the most wilt resistant tomato line in multiple comparative field trials [5], [43], [44].
Identifying the presumptive EPS receptor could elucidate the mechanism by which H7996 recognizes EPS. However, it is unclear which and how many additional R. solanacearum elicitors or MAMPs are detected by the tomato host. Insights acquired through expression profiling of single genes as presented here are necessarily incomplete. A more comprehensive microarray analysis could monitor global transcriptional responses to R. solanacearum in susceptible and resistant tomato hosts to generate a broader understanding of BW resistance and identify targets for marker-assisted breeding of wilt-resistant plants.
Materials and Methods
Bacterial strains and growth conditions
R. solanacearum strains used in these experiments were tropical strain GMI1000 (phylotype I, sequvar 18, biovar 3) [45], [46] and temperate strain UW551 (phylotype II, sequevar 1, biovar 2, historically known as Race 3) [17], [34]. To facilitate enumeration of R. solanacearum in the natural plant microbial background, all inoculations were performed with rifampicin-resistant R. solanacearum strains [47]. We confirmed that the rif-resistant strains had wild-type virulence and elicited plant gene expression comparable to the wild-type strain. R. solanacearum was grown on CPG solid medium [48] at 28°C for 48 h. If required, antibiotics were added at final concentrations of 25 mg/l kanamycin and 25 mg/l rifampicin. Medium components were from Difco Laboratories (Detroit, MI). All other chemicals and antibiotics were from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Hanover Park, IL).
Construction of an eps− mutant
An approximately 2,000-bp internal DNA fragment of the epsB gene of R. solanacearum strain UW551 (RRSL_1061) was amplified by PCR using primers UW551EPSBi-F: 5′-GACGAATTCGTCAGCTTCTTGGGCTTCAC and UW551EPSBi-R: 5′-GACTCTAGACTCAAGACGCTGGAGATGCT. To delete epsB, this amplicon was digested with EcoR1 and XbaI and ligated into the suicide vector pVIK112 [49] to create pEPSBi. This construct was moved into UW551 by conjugation with selection for kanamycin resistance. Correct deletion of epsB was verified by PCR and by comparing colony morphologies of wild-type and mutant strains. Extraction of cell-free EPS confirmed that UW551ΔepsB produced less than 5% of the EPS made by wild-type UW551. The corresponding mutant was constructed in strain GMI1000 by moving the ΔepsB construct into the GMI1000 genome by natural transformation. Correct deletion of the gene was verified by PCR and the eps− phenotype was verified by colony morphology.
Plant inoculations and tissue collection
We compared the virulence of R. solanacearum strains GMI1000 and UW551 in susceptible cultivar Bonny Best and horizontally resistant line Hawaii7996 (H7996) by means of a naturalistic soil soak inoculation [50]. Briefly, unwounded 19 to 21 day old plants were inoculated by pouring a bacterial suspension onto the soil to a final density of approximately 1×108 CFU/g soil, followed by incubation at 28°C. Control plants were mock-inoculated with sterile water. Symptoms were scored daily by a rater blind to treatment identity on a 0-to-4 disease index, where 0 indicates no disease, 1 indicates 1 to 25% of leaves wilted, 2 indicates 25 to 50% of leaves wilted, 3 indicates 51 to 75% of leaves wilted, and 4 indicates 76 to 100% of leaves wilted. Each experiment contained 16 plants per treatment, and experiments were repeated at least three times. To measure plant gene expression, Bonny Best tissue was sampled 4 to 7 dpi, while H7996 samples were collected 7 to 14 dpi due to slower disease development in this resistant host.
We measured disease progress and host defense responses to wild-type strains UW551 and GMI1000 and to UW551ΔepsB and GMI1000ΔepsB in the two tomato cultivars by directly inoculating 21-day old plants with either R. solanacearum wild-type (2×103 cells) or ΔepsB (2×105 cells) through the cut petiole of the first true leaf. Higher inoculum levels were necessary for the ΔepsB strains because of their reduced colonization rate. Control plants were inoculated with sterile water. Samples were taken 3 to 4 dpi from plant stems containing 1×108 to 1×109 CFU/g R. solanacearum.
RNA extraction
Samples (100 mg) from randomly selected individual tomato plants were taken from mid-stem just above the cotyledon. One sub-sample was ground in sterile water, and dilution plated in triplicate to determine pathogen population size in the plant. Colonies were counted after 48 h incubation at 28°C. Another sub-sample was immersed in RNAlater (Ambion Inc., Austin, TX) for 24 h at 4°C to preserve RNA integrity before storage at −80°C. Ultimately, RNA was extracted from tomato samples that contained the target bacterial cell densities of about 1×107 CFU/g (symptomless plants) and 1×108 or 1×109 CFU/g (disease index 1, early disease) using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA) including DNaseI treatment according to the manufacturer's instructions. RNA quantity and quality were assessed with micro-spectrophotometry (NanoDrop Technologies Inc., Wilmington, DE).
Gene expression analysis using quantitative real-time PCR
To measure plant mRNA levels, 1 µg of total RNA from each sample was reverse transcribed into cDNA using Superscript III reverse transcriptase First-Strand Synthesis SuperMix (Invitrogen, Carlsbad, CA) containing oligo (dT) and random hexamer primers according to the manufacturer's instructions. Quantitative RT-PCR primers for selected tomato defense genes and three constitutively expressed normalization genes (Table S1) were designed using Biology Workbench software from the relevant GenBank (NCBI) tomato mRNA sequences. Quantitative RT-PCR amplifications were performed in duplicate 25 µl reactions using PowerSYBR Green Mastermix (Applied Biosystems, Warrington, UK) and consisted of 1X Mastermix, 400 nM forward and reverse primer, and 50 ng template cDNA. Reactions were run on an ABI PRISM 7300 Real-Time PCR System (Applied Biosystems, Foster, CA). Reaction parameters were: 10 min polymerase activation, followed by 40 cycles of 95°C for 15 s and 57°C for 1 min. Gene expression was quantified separately for each cDNA sample. Controls were cDNA samples lacking reverse transcriptase to check for DNA contamination and no-template reactions. Reaction efficiencies were between 94 and 105% for all primers, calculated by generating a standard curve and plotting the threshold cycle (CT) against the logarithm of four known tomato DNA dilutions. The number of cycles at threshold level was converted to relative quantities (RQ) with the highest expression set to one using the delta-CT formula RQ = E(minC T – sampleC T ) [51]. For maximum accuracy and reliability, RQ was divided by a normalization factor derived from the geometric mean of three reference genes, Gapdh, Actin, and DnaJ-like protein, to generate normalized relative quantities (NRQ) [51], [52]. Stability of the reference transcripts was validated using geNorm, and normalization factors were calculated in the geNorm applet [51]. Relative expression change was calculated by calibrating treated (infected) samples to the mean NRQ of at least three control replicates within each experiment. Data are presented as fold change in defense gene expression in infected tomato plants relative to mock-inoculated control plants. Each experiment was replicated at least three times.
Experiments with extracellular polysaccharide (EPS I)
EPS I was extracted from R. solanacearum strain UW551 and extensively purified using a modification of a described protocol [9]. Bacterial cells were scraped from the surface of CPG agar medium, resuspended in water to an O.D600 of 1.0 and centrifuged for 10 min twice at 8000 rpm. The cell-free supernatant was lyophilized and redissolved in 10 ml distilled water. EPS I was precipitated overnight at −20°C using 4 vol acetone and 20 mM NaCl and redissolved in DNaseI buffer (50 mM Tris, 1 mM MgCl2). DNaseI (Roche Diagnostics, Indianapolis, IN) was added to a final concentration of 0.1 g/ml and the solution was incubated at 37°C for 1 h and extracted once with phenol, followed by successive extractions with chloroform until no interphase was visible. The aqueous layer was dialyzed extensively against distilled water. Pure EPS was recovered from the dialysate by overnight precipitation with 3 vol ethanol at −20°C. The purified EPS was air-dried, dissolved in 250 µl distilled water and frozen in aliquots until use. EPS was quantified by the Elson-Morgan assay for hexosamine sugars using N-acetylgalactosamine as the standard [53], [54]. Protein content was estimated using the Pierce BCA assay kit (ThermoScientific, Rockford, IL) with BSA as the standard, and nucleic acid content was estimated micro-spectrophotometrically. Tomato gene expression in response to purified EPS was determined 24 h after injection of 20 µg EPS directly into the vascular system through the cut petiole. Control plants were injected with sterile water. Each EPS experiment contained 10 plants per treatment and experiments were repeated four times.
ROS detection in tomato stem tissue
ROS accumulation in stems of susceptible cv. Bonny Best and horizontally resistant line H7996 was monitored 48 h to 72 h after infection of plants with R. solanacearum UW551 or UW551ΔepsB via cut petiole as described above. Water inoculated plants served as controls. Plants were cut horizontally through the mid-stem and left at room temperature to release the first wave of wounding-related oxidative burst. After 15 min, a fresh cross section (1 mm thick, 5 mm diameter) was removed from the stem and incubated in the dark with 5 µl of a 50 µM dihydrorhodamine 123 solution (DHR123, AnaSpec Inc., Fremont, CA) for 30 min to allow ROS in the tissue to oxidize non-fluorescent DHR 123 to the fluorescent rhodamine 123. Fluorescence was observed with a Leica MZ FLIII fluorescence stereomicroscope using 480/40 nm (excitation) and a 510 nm barrier filter. Another stem section was used to quantify R. solancearum populations as described above to ensure comparison of fluorescence between samples containing similar bacterial populations. Each experiment contained 6 to 8 plants per treatment, and the experiment was repeated three times.
Reactive oxygen species (ROS) detection in tomato leaves
ROS accumulation in host tissue was detected by endogenous peroxidase-dependent in situ histochemical staining with 3,3′-diaminobenzidine (DAB, Sigma, USA) using a slightly modified protocol [55]. Oxidation of DAB by ROS creates a visible brown precipitate in the host tissue. Eight tomato leaves from 28-day old tomato plants of BW-susceptible cv. Bonny Best and horizontally resistant line Hawaii7996 were infused with 1×109 CFU/ml R. solanacearum cells or water as control. Three leaves were cut at the petiole 48 h after inoculation and immediately immersed in DAB solution (1 mg/ml in water, pH 3.8). Leaves were incubated for 18 h in the dark at room temperature, and then bleached in boiling 96% ethanol for 10 min, cleared and stored in 70% ethanol until imaging. The fourth leaf was used to quantify R. solancearum populations in the leaf. Three disks (5 mm diameter) per leaf were pooled, ground in sterile water, serially diluted and plated on CPG solid medium in triplicate. Colonies were counted after 48 h incubation at 28°C. The experiment was repeated five times with comparable results.
Data analysis
The log2 of NRQs of each plant defense gene tested was used to analyze differences in gene expression caused by infection with R. solanacearum compared to untreated water controls. The log2 of fold change was used to compare gene expression elicited by strains GMI1000 and UW551, as well as expression elicited by strains UW551 and UW551ΔepsB [56]. Data were analyzed by ANOVA using the GLM procedure. Specific comparisons of least-square means were evaluated for significance using Turkey's HSD adjusted P-values (Table S2). Gene expression levels elicited by purified EPS I were compared using a 2-tailed t-test. Repeated measures ANOVA using the PROC mixed method was used to compare disease progress curves of diverse strains in resistant and susceptible tomato plants. These analyses were conducted in SAS version 9.1 (SAS Institute, Cary, NC). A P-value of <0.05 was considered statistically significant.
Supporting Information
Virulence of wild-type Ralstonia solanacearum strain UW551 and EPS-deficient mutant UW551Δ epsB on resistant and susceptible tomato plants. 21-day-old susceptible (cv. Bonny Best) and horizontally resistant (H7996) tomato plants were inoculated A: by pouring bacteria onto the soil to a final concentration of about 1×108 CFU/g soil or B: with 2000 cells via the cut petiole of the first true leaf followed by incubation in a 28°C growth chamber. Plants were rated daily over 14 days on a disease index scale from 0 to 4 where 0 indicated healthy and 4 indicated 100% wilted. Each point represents the mean disease index for three independent experiments each with 16 plants per treatment.
(TIF)
Primers used in the real-time qRT-PCR analysis of defense-related tomato genes.
(DOC)
ANOVA results for gene expression elicited by R. solanacearum strain GMI1000 or UW551 in BW-susceptible tomato cultivar Bonny Best and horizontally resistant line.
(DOC)
Acknowledgments
We gratefully acknowledge statistical advice from Ting-Li Lin (University of Wisconsin Statistical Consulting Service). We thank Timothy Denny and Mark Schell (University of Georgia) for providing the ΔepsB construct and their insightful comments.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by a Deutsche Forschungsgemeinschaft (DFG) Postdoctoral Fellowship to AM and by United States Department of Agriculture (USDA) Cooperative State Research, Education, and Extension Service Plant Biosecurity project 2006-04560, the USDA-Agricultural Research Service Floral and Nursery Crops Research Initiative and by the University of Wisconsin College of Agricultural and Life Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jones J, Dangl J. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Zipfel C. Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol. 2009;12:414–420. doi: 10.1016/j.pbi.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Strange RN, Scott PR. Plant disease: a threat to global food security. Annu Rev Phytopathol. 2005;43:83–116. doi: 10.1146/annurev.phyto.43.113004.133839. [DOI] [PubMed] [Google Scholar]
- 4.Denny T. Plant pathogenic Ralstonia species. In: Gnanamanickam SS, editor. Plant-Associated Bacteria. New York: Springer Press; 2006. [Google Scholar]
- 5.Hanson PM, Wang J, Licardo O, Haudin, Mah S, et al. Variable reaction of tomato lines to bacterial wilt evaluated at several locations in southeast Asia. Hort Science. 1996;31:143–146. [Google Scholar]
- 6.Thoquet P, Olivier J, Sperisen C, Rogowsky P, Laterrot H, et al. Quantitative trait loci determining resistance to bacterial wilt in tomato cultivar Hawaii7996. Mol Plant-Microbe Interact. 1996;9:826–836. [Google Scholar]
- 7.Wang JF, Olivier J, Thoquet P, Mangin B, Sauviac L, et al. Resistance of tomato line Hawaii7996 to Ralstonia solanacearum Pss4 in Taiwan is controlled mainly by a major strain specific locus. Mol Plant-Microbe Interact. 2000;13:6–13. doi: 10.1094/MPMI.2000.13.1.6. [DOI] [PubMed] [Google Scholar]
- 8.Denny TP, Baek S. Genetic evidence that extracellular polysaccharide is a virulence factor of Pseudomonas solanacearum. Mol Plant-Microbe Interact. 1991;4:198–206. [Google Scholar]
- 9.McGarvey JA, Denny TP, Schell MA. Spatial-temporal and quantitative analysis of growth and EPS I production by Ralstonia solanacearum in resistant and susceptible tomato cultivars. Phytopathology. 1999;89:1233–1239. doi: 10.1094/PHYTO.1999.89.12.1233. [DOI] [PubMed] [Google Scholar]
- 10.Orgambide G, Montrozier H, Servin P, Roussel J, Trigalet-Demery D, et al. High heterogeneity of the exopolysaccharides of Pseuodomonas solanacearum strain GMI1000 and the complete structure of major polysaccharide. J Biol Chem. 1991;266:8312–8321. [PubMed] [Google Scholar]
- 11.Alvarez AM. Integrated approaches for detection of plant pathogenic bacteria and diagnosis of bacterial diseases. Annu Rev Phytopathol. 2004;42:339–366. doi: 10.1146/annurev.phyto.42.040803.140329. [DOI] [PubMed] [Google Scholar]
- 12.Fegan M, Prior P. How complex is the “Ralstonia solanacearum species complex”? In: Allen C, Prior P, Hayward AC, editors. Bacterial Wilt Disease and the Ralstonia solanacearum species complex. St. Paul, MN: APS Press; 2005. pp. 449–461. [Google Scholar]
- 13.Schell MA. Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory network. Annu Rev Phytopathol. 2000;38:263–292. doi: 10.1146/annurev.phyto.38.1.263. [DOI] [PubMed] [Google Scholar]
- 14.Genin S, Boucher C. Ralstonia solanacearum: Secrets of a major pathogen unveiled by analysis of its genome. Mol Plant Pathol. 2002;3:111–118. doi: 10.1046/j.1364-3703.2002.00102.x. [DOI] [PubMed] [Google Scholar]
- 15.Denny TP. Involvement of bacterial polysaccharides in plant pathogenesis. Annu Rev Phytopathol. 1995;33:173–197. doi: 10.1146/annurev.py.33.090195.001133. [DOI] [PubMed] [Google Scholar]
- 16.Saile E, McGarvey JA, Schell MA, Denny TP. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology. 1997;87:1264–1271. doi: 10.1094/PHYTO.1997.87.12.1264. [DOI] [PubMed] [Google Scholar]
- 17.Swanson J, Yao J, Tans-Kersten J, Allen C. Behavior of Ralstonia solanacearum Race 3 biovar 2 during latent and active infection of geranium. Phytopathology. 2005;95:136–146. doi: 10.1094/PHYTO-95-0136. [DOI] [PubMed] [Google Scholar]
- 18.Penacortes H, J JF, Willmitzer L. Signals involved in wound-induced proteinase-inhibitor-II gene expression in tomato and potato plants. Proc Natl Acad Sci USA. 1995;92:4106–4113. doi: 10.1073/pnas.92.10.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, Roger T, Bender CL, Schaller A, He SY, et al. Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 2003;36:485–499. doi: 10.1046/j.1365-313x.2003.01895.x. [DOI] [PubMed] [Google Scholar]
- 20.Block A, Schmelz E, O'Donnell PJ, Jones JB, Klee HJ. Systemic acquired tolerance to virulent bacterial pathogens in tomato. Plant Physiol. 2005;138:1481–1490. doi: 10.1104/pp.105.059246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smart CD, Myers KL, Restrepo S, Martin GB, Fry WE. Partial resistance of tomato to Phytophthora infestans is not dependent upon ethylene, jasmonic acid, or salicylic acid signaling pathways. Mol Plant-Microbe Interact. 2003;16:141–148. doi: 10.1094/MPMI.2003.16.2.141. [DOI] [PubMed] [Google Scholar]
- 22.Tornero P, Gadea J, Conejero V, Vera P. Two PR-1 genes from tomato are differentially regulated and reveal a novel mode of expression for a pathogenesis-related gene during the hypersensitive response and development. Mol Plant-Microbe Interact. 1997;10:624–634. doi: 10.1094/MPMI.1997.10.5.624. [DOI] [PubMed] [Google Scholar]
- 23.Danhash N, Wagemakers CA, Kan JAv, deWit PG. Characterization of four chitinase cDNAs obtained from Cladosporium fulvum-infected tomato. Plant Mol Biol. 1993;22:1017–1029. doi: 10.1007/BF00028974. [DOI] [PubMed] [Google Scholar]
- 24.Agrios G. St. Paul: American Phytopathological Society Press; 2005. Plant Pathology. [Google Scholar]
- 25.Chen YY, Lin YM, Chao TC, Wang J-F, Liu AC, et al. Virus-induced gene silencing reveals the involvement of ethylene-, salicylic acid- and mitogen-activated protein kinase-related defense pathways in the resistance of tomato to bacterial wilt. Physiol Plantarum. 2009;136:324–335. doi: 10.1111/j.1399-3054.2009.01226.x. [DOI] [PubMed] [Google Scholar]
- 26.Ho F-I, Chen Y, Lin Y, Cheng C, Wang J-F. A tobacco rattle virus-induced gene silencing system for a soil-borne vascular pathogen Ralstonia solanacearum. Botanical Studies. 2009;50:413–424. [Google Scholar]
- 27.Pfund C, Tans-Kersten J, Dunning FM, Alonso JM, Ecker JR, et al. Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol Plant-Microbe Interact. 2004;17:696–706. doi: 10.1094/MPMI.2004.17.6.696. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y, Chou I, Wang J, Ho F, Chu Y, et al. Transposon mutagenesis reveals differential pathogenesis of Ralstonia solanacearumon tomato and Arabidopsis. Mol Plant-Microbe Interac. 2008;21:1261–1270. doi: 10.1094/MPMI-21-9-1261. [DOI] [PubMed] [Google Scholar]
- 29.Hirsch J, Deslandes L, Feng DX, Balague C, Marco Y. Delayed symptom development in ein2-1, an Arabidopsis ethylene-insensitive mutant, in response to bacterial wilt caused by Ralstonia solanacearum. Phytopathology. 2002;92:1142–1148. doi: 10.1094/PHYTO.2002.92.10.1142. [DOI] [PubMed] [Google Scholar]
- 30.Deslandes L, Pileur F, Liaubet L, Boucher C, Arlat M, et al. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci USA. 2002;99:2404–2409. doi: 10.1073/pnas.032485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernandez-Blanco C, Feng DX, Hu J, Sanchez-Vallet A, Deslandes L, et al. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Cell. 2007;19:890–903. doi: 10.1105/tpc.106.048058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katagiri F. A global view of defense gene expression regulation – a highly interconnected signaling network. Curr Opin Plant Biol. 2004;7:506–511. doi: 10.1016/j.pbi.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 33.van Loon LCV, Rep M, Pieterse CMJ. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- 34.Gabriel DW, Allen C, Schell M, Denny TP, Greenberg JT, et al. Identification of open reading frames unique to a select agent: Ralstonia solanacearum Race 3 Biovar 2. Mol Plant-Microbe Interact. 2006;19:69–79. doi: 10.1094/MPMI-19-0069. [DOI] [PubMed] [Google Scholar]
- 35.D'Haeze W, Holsters M. Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol. 2004;12:555–561. doi: 10.1016/j.tim.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 36.Aslam SN, Newman M, Erbs G, Morrissey KL, Chinchilla D, et al. Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr Biol. 2008;18:1078–1083. doi: 10.1016/j.cub.2008.06.061. [DOI] [PubMed] [Google Scholar]
- 37.Romanenko AS, Lomovatskaya LA, Shafikova TN, Borovskii GB, Krivolapova NV. Potato cell plasma membrane receptors to ring rot pathogen extracellular polysaccharides. J Phytopathol. 2003;151:1–6. [Google Scholar]
- 38.De Pinto MC, Lavermicocca P, Evidente A, Corsaro MM, Lazzaroni S, et al. Expopolysaccharides produced by plant pathogenic bacteria affect ascorbate metabolism in Nicotiana tabacum. Plant Cell Physiol. 2003;44:803–810. doi: 10.1093/pcp/pcg105. [DOI] [PubMed] [Google Scholar]
- 39.Romeiro RS, Kimura O. Induced resistance in pepper leaves infiltrated with purified bacterial elicitors from Xanthomonas campestris pv. vesicatoria. J Phytopathol. 1997;145:495–498. [Google Scholar]
- 40.Makino S, Ikegami S, Kano H, Sashihara T, Sugano H, et al. Immunomodulatory effects of polysaccharides produced by Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1. J Dairy Sci. 2006;89:2873–2881. doi: 10.3168/jds.S0022-0302(06)72560-7. [DOI] [PubMed] [Google Scholar]
- 41.Mazmanian S, Kasper D. The love-hate relationship between bacterial polysaccharides and the host immune system. Nature Reviews Immunology. 2006;6:849–858. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- 42.Sun W, Dunning FM, Pfund C, Weingarten R, Bent AF. Within-species flagellin polymorphism in Xanthomonas campestris pv. campestris and its impact on elicitation of Arabidopsis FLS2-dependent defenses. Plant Cell. 2006;18:764–779. doi: 10.1105/tpc.105.037648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimault V, Prior P. Bacterial wilt resistance in tomato associated with tolerance of vascular tissue to Pseudomonas solanacearum. Plant Pathology. 1993;42:589–594. [Google Scholar]
- 44.Wang JF, Hanson P, Barnes JA. Worldwide evaluation of an international set of resistance sources to bacterial wilt in tomato. In: Prior P, Allen C, Elphinstone J, editors. Bacterial Wilt: Molecular and Ecological Aspects. Berlin: Springer Press; 1998. pp. 269–275. [Google Scholar]
- 45.Boucher C, Barberis P, Trigalet A, Demery D. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J Gen Microbiol. 1985;131:2449–2457. [Google Scholar]
- 46.Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature. 2002;415:497–502. doi: 10.1038/415497a. [DOI] [PubMed] [Google Scholar]
- 47.Milling A, Meng F, Denny TP, Allen C. Interactions with hosts at cool temperatures, not cold tolerance, explain the unique epidemiology of Ralstonia solanacearum Race 3 biovar 2. Phytopathology. 2009;99:1127–1134. doi: 10.1094/PHYTO-99-10-1127. [DOI] [PubMed] [Google Scholar]
- 48.Hendrick CA, Sequeira L. Lipopolysaccharide-defective mutants of the wilt pathogen Pseudomonas solanacearum. Appl Environ Microbiol. 1984;48:94–101. doi: 10.1128/aem.48.1.94-101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalogeraki VS, Winans SC. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene. 1997;188:69–75. doi: 10.1016/s0378-1119(96)00778-0. [DOI] [PubMed] [Google Scholar]
- 50.Tans-Kersten J, Guan Y, Allen C. Ralstonia solanacearum pectin methylesterase is required for growth on methylated pectin but not for bacterial wilt virulence. Appl Environ Microbiol. 1998;64:4918–4923. doi: 10.1128/aem.64.12.4918-4923.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034.0031–0034.0011. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hellemans J, Mortier G, Paepe Ad, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levvy GA, McAllan G. The N-acetylation and estimation of hexosamines. Biochem J. 1959;73:127–132. doi: 10.1042/bj0730127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elson LA, Morgan WTJ. A colorimetric method for the determination of glucosamine and chondrosamine. Biochem J. 1933;26:1824–1829. doi: 10.1042/bj0271824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giovanini MP, Puthoff DP, Nemacheck JA, Mlttapalli O, Saltzmann KD, et al. Gene-for-gene defense of wheat against the Hessian Fly lacks a classical oxidative burst. Mol Plant-Microbe Interact. 2006;19:1023–1033. doi: 10.1094/MPMI-19-1023. [DOI] [PubMed] [Google Scholar]
- 56.Rieu I, Powers SJ. Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell. 2009;21:1031–1033. doi: 10.1105/tpc.109.066001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Virulence of wild-type Ralstonia solanacearum strain UW551 and EPS-deficient mutant UW551Δ epsB on resistant and susceptible tomato plants. 21-day-old susceptible (cv. Bonny Best) and horizontally resistant (H7996) tomato plants were inoculated A: by pouring bacteria onto the soil to a final concentration of about 1×108 CFU/g soil or B: with 2000 cells via the cut petiole of the first true leaf followed by incubation in a 28°C growth chamber. Plants were rated daily over 14 days on a disease index scale from 0 to 4 where 0 indicated healthy and 4 indicated 100% wilted. Each point represents the mean disease index for three independent experiments each with 16 plants per treatment.
(TIF)
Primers used in the real-time qRT-PCR analysis of defense-related tomato genes.
(DOC)
ANOVA results for gene expression elicited by R. solanacearum strain GMI1000 or UW551 in BW-susceptible tomato cultivar Bonny Best and horizontally resistant line.
(DOC)