Abstract
Background
The Antarctic krill Euphausia superba is a keystone species in the Antarctic food chain. Not only is it a significant grazer of phytoplankton, but it is also a major food item for charismatic megafauna such as whales and seals and an important Southern Ocean fisheries crop. Ecological data suggest that this species is being affected by climate change and this will have considerable consequences for the balance of the Southern Ocean ecosystem. Hence, understanding how this organism functions is a priority area and will provide fundamental data for life history studies, energy budget calculations and food web models.
Methodology/Principal Findings
The assembly of the 454 transcriptome of E. superba resulted in 22,177 contigs with an average size of 492bp (ranging between 137 and 8515bp). In depth analysis of the data revealed an extensive catalogue of the cellular chaperone systems and the major antioxidant proteins. Full length sequences were characterised for the chaperones HSP70, HSP90 and the super-oxide dismutase antioxidants, with the discovery of potentially novel duplications of these genes. The sequence data contained 41,470 microsatellites and 17,776 Single Nucleotide Polymorphisms (SNPs/INDELS), providing a resource for population and also gene function studies.
Conclusions
This paper details the first 454 generated data for a pelagic Antarctic species or any pelagic crustacean globally. The classical “stress proteins”, such as HSP70, HSP90, ferritin and GST were all highly expressed. These genes were shown to be over expressed in the transcriptomes of Antarctic notothenioid fish and hypothesized as adaptations to living in the cold, with the associated problems of decreased protein folding efficiency and increased vulnerability to damage by reactive oxygen species. Hence, these data will provide a major resource for future physiological work on krill, but in particular a suite of “stress” genes for studies understanding marine ectotherms' capacities to cope with environmental change.
Introduction
The Southern Ocean is an important breeding and/or foraging location for a wide range of charismatic megafauna such as whales, seals, penguins and other sea birds including albatrosses. Whilst these species fascinate the public, they represent the apex of a complex food chain, the keystone species of which is the Antarctic krill Euphausia superba [1]. This shrimp-like crustacean is not only a major prey item for these animals, but is also a significant consumer, grazing on the phytoplankton bloom in the austral summer and on algae under the sea ice in winter. In a more global context, it has been suggested to be the most abundant eukaryotic species in the World's Oceans [2] existing in large schools or swarms with densities that may be between 10,000–30,000 individual animals per square metre [3], [4]. Current catches are 125,000–150, 000 tonnes per year [5], but at its peak in 1982, krill fisheries comprised 13% of the global annual catches of crustaceans [6]. Hence, E. superba with a circumpolar distribution plays not only a pivotal role in the Antarctic ecosystem, but also significantly impacts on the economy of the Southern Ocean fishing industry [1], [7].
Research on krill has a long history, reaching back to the “Discovery” expeditions of the 1920/1930s that sought to understand the drivers of fluctuating populations of baleen whales which fed on krill and which were then harvested for their oil [8]. More recently, research efforts have also focussed on stock estimates and distributions, again with regard to fisheries exploitation [7]. Whilst catches have remained relatively stable and never reached the maximum TAC (total allowable catch) defined by the regulatory body the Commission for the Conservation of Antarctic Marine Living Resources (CCAMLR), there is growing concern about decreasing krill stocks and any consequential effects on both the Southern Ocean ecosystem and the fisheries industry [9].
An historical analysis of krill numbers in the southwest Atlantic sector (which contains over half the total krill stocks of the Southern Ocean) [10], [11] indicates that there has been a significant decline in population densities since the 1970's which correlates with the extent of winter sea ice cover. The ice provides an essential habitat and food supply via ice algae for these animals over the winter period [10], [12]. The southwest Atlantic sector of the Southern Ocean includes the Antarctic Peninsula, which is experiencing some of the most rapid rises in sea water temperature on the planet [13] with a 1°C increase in the surface layers of the Bellingshausen Sea in 50 years [14], [15] and reduced winter sea ice duration [16]. This continued and rapid temperature rise has profound implications, not only for krill stocks, but also for broader food web interactions in the Southern Ocean [10], [17]–[22] and for the fishing industry [7], [9], [23].
There is a considerable body of scientific knowledge about the general biology and ecology of krill, in particular growth, biochemical composition and reproduction. One of the main drivers for understanding krill biology has been the fisheries' requirements for stock forecasting and conservation measures, but this is now joined by concerns over climate change effects and the requirement to take a more holistic over-view to understand food web structures [7], [24], [25]. However, there is very little knowledge of processes at the tissue, cellular or molecular level. As a consequence, and given the pivotal role of this key species in the Southern Ocean ecosystem, detailed physiological and molecular studies are critical if we are to understand this species and its responses to a changing environment. Such studies would provide fundamental data for a deeper understanding of life histories, as well as important parameters for energy budget calculations and models [26].
Working with wild free living krill is beyond the capability of current technologies. Therefore cellular, tissue and molecular data can only be derived from either individuals recently sampled from the sea, with the concommitant stresses involved or from laboratory manipulations of aquarium held animal stocks. Hence a fundamental issue for the future progression of these studies will be the linking of laboratory developed assays with wild caught sampling [27], [28]. The physiology of E. superba has been studied in a number of laboratory based experiments for periods of up to 5 months using non-specialised facilities [25], [26], [29]. Some of these studies have highlighted the diversity of problems encountered when working on a pelagic species, such as krill adapted to living in swarms with social aggregations [29], [30]. These studies have emphasized the necessity of understanding stress responses in this species as an a priori to improve experimental protocol and also to provide a baseline laboratory physiological metric for such comparative field sampling [26], [30] as such molecular analyses can provide a more detailed description of animal condition over observational physiology approaches [31]. To date there are only 3095 ESTs from E. superba in the public databases, but the classical “stress genes” are largely absent from these gene sets [32], [33].
Here we describe the transcriptome of E. superba, the first 454 generated data for a pelagic Antarctic species or any pelagic crustacean globally. Analysis here focuses on chaperone genes, in particular the typical “stress” genes of the Heat Shock Protein (HSP) family. This data will provide a major resource for future physiological work on krill, but in particular a suite of “stress” genes for studies understanding marine ectotherms capacities to cope with environmental change, such as future elevated sea water temperatures, ocean acidification or water freshening etc.
Results and Discussion
The non-normalised krill libraries were subjected to a full 454 run that yielded 943,817 reads. After cleaning the data and removing small reads, 699,248 reads containing 205,888,141 bases with an average read size of 293 bp were entered into Newbler [34] for assembly. These assembled into the 22,177 contigs (261,280 reads) which were used for the further analysis. Because the aim of this project was to conduct a preliminary characterisation of the krill transcriptome, with, as described below, an emphasis on chaperones and stress-related genes for future analyses there was a requirement for longer sequences of good quality which would enable us to distinguish between gene family members. Hence the descriptive analysis presented here utilised only the contigs produced by the assembly. Whilst the singletons potentially contain useful lowly expressed sequences, they also contain a substantial proportion of artefacts derived from cDNA synthesis, sequencing and contamination [35]. However these will be retained within our databases and utilised when more targeted pyrosquencing experiments (using both 454 and short reads) will generate an enlarged transcriptome. The contigs ranged in size from 137 bp to 8515 bp, with an average size of 491.9 bp. 48 contigs were greater than 3kb and 94 contigs comprised more than 300 reads, with the largest contig of 8515 bp comprising the most reads with 740 sequences. Self BLAST of this dataset produced only 625 matches with a value of e−100, indicating a low level (<3%) of redundancy in the assembly of the reads. The contigs contained 41,470 repeat sequence motifs in coding and/or UTR regions (microsatellites), of which 339 comprised over 7 exact repeat units (Table S1). There were 27,776 SNPs/INDELS present in 3,980 contigs designated as high confidence by the Newbler program (Table S2), although a further circa 25,000 SNPs were identified at lower confidence level as defined by Newbler [34] Hence these provide significant resources for researchers in allied fields and will fuel investigations into, for example, population structure and gene flow dynamics in this crucial pelagic species.
BLAST sequencing similarity searching of the GenBank non-redundant database produced matches against 5563 of the contigs using a <1e−10 cut off value (representing 25% of the total number of contigs). Although this seems rather low, the number of matches is higher when compared to those of other non-model marine invertebrates which have recently been subjected to 454 transcriptome analyses (c.f. 12% in the blue mussel Mytilus galloprovincialis, 17% in the Antarctic bivalve Laternula elliptica and 23.9% in larvae of the coral Acropora millipora [35]–[37]. This is due to the phylogenetic position of E. superba within the Arthropoda, a phylum which contains a number of species that have been subjected to whole genome sequencing programmes. The prime example of these is the fruit fly Drosophila melanogaster and the insect vectors of human disease such as Anopheles gambiae (www.vectorbase.org). Of these, the Drosophila genes, in particular, are well annotated (www.flybase.org). However, E. superba is a marine invertebrate and more closely related (within the sub-phylum Eucrustacea, super-order Eucarida) are a number of commercially important seafood species, such as the Pacific white shrimp Litopenaeus vannamei, the black tiger shrimp Penaeus monodon and the fleshy prawn Fenneropenaeus chinensis. These have been subjected to several medium-scale EST projects in the past and comprise the majority of the 33,813 nucleotide entries and 400,793 ESTs for the Eucarida in the public database (www.ncbi.nlm.nih.gov) (as of 15/05/10). BLAST sequence similarity searches against these more morphologically and physiologically similar species produces a greater number of matches (35–43%) (Table 1) and some of these, particularly the immune-related genes, are well annotated and characterised [c.f. 38]–[42]. To date E.superba is poorly represented in the database with only 3095 ESTs [32]–[33], of which over 80% were represented in the transcriptome presented here (Table 1). The E. superba mitochondrial genome has been almost completely sequenced on two occasions with the aim of producing markers for exploitation and management of krill fisheries resources [43], [44]. Whilst a previous 454 run on a non-model species, Mytilus galloprovincialis produced enough mitochondrial reads to enable alignment of this new data against the relevant mitochondrial sequence [36], this was not possible on the krill data, as only 8 mitochondrial genes (cytochrome c oxidase, subunits 1, 2 and 3, ATP synthase subunit 6 and NADH dehydrogenase subunits 1,2,4 and 5) were present in our dataset, which may be due to the fact that non-normalised libraries were produced from a single source of krill material. Sequencing of the transcriptomes of krill subjected to different treatments will almost certainly enhance the identification of mitochondrial genes.
Table 1. Representation of Eucarida species in the public databases (at 15/07/10).
| Species | No of ESTs in database | BLASTn matches | tBLASTx matches | ||||
| krill 454 | ESTs | % match | krill 454 | ESTs | % match | ||
| Euphausia superba | 3095 | 2177 | 2567 | 83 | 2560 | 2547 | 82 |
| Fenneropenaeus chinensis | 10511 | 556 | 1706 | 16 | 1759 | 3717 | 35 |
| Penaeus monodon | 35690 | 968 | 10464 | 29 | 3141 | 15400 | 43 |
| Litopenaeus vannamei | 162755 | 1412 | 24482 | 15 | 4893 | 63145 | 39 |
Number of species-specific ESTS and the number of krill ESTs that match a corresponding number of Eucarida ESTs.
To obtain an overall view of the krill transcriptome and also as part of the contig verification process, all contigs comprising over 450 reads were analysed in-depth. These most commonly expressed sequences with an associated database match represented a varied mix of functional groups (Table 2). The largest contig, with a length of 8515bp also contained the highest number of reads (740). BLAST sequence similarity searching of this contig produced a match with an E-value of 0.0 against the shrimp, Litopenaeus vannemei β-1,3-glucan binding protein (βGBP-HDL) [39] (Table 2). The full length of this 1454 amino acid protein was represented in the krill contig. Not surprisingly, given the E-value, there was strong conservation at the amino acid level with 41.2% identity and 59.4% similarity between the two putative proteins. There was also conservation of the precursor N-terminal domain and processing sites for the mature peptide. However, out of the 4 O-glycosylation and 5 N-glycosylation sites found to be preserved between the two Penaeid shrimp and crayfish βGBP-HDL proteins, only 2 and 1 respectively were present in the krill sequence, indicating potential differences in post translational modification [39]. Functional characterisation of the shrimp protein revealed a dual function: binding of β-glucans trigger the activation of this pattern-recognition protein and the prophenol-oxidase system, which is a central component of the shrimp innate immunity defence system, but βGBP-HDL is also involved in lipid transport [45], [46]. Whilst the definition as to which of these functions predominates is not possible within the confines of this descriptive study, the second most common sequence was fatty acid synthase, a key enzyme in the production of fatty acids. The Antarctic environment is subject to extreme seasonality with huge phytoplankton blooms during the austral summer [47]–[49] and strong seasonal physiology is known for many Antarctic species [50], [51]. The krill samples used for this transcriptome were caught during the summer and hence during the period when the animals were taking advantage of the ready food supply to fuel the seasonal increase in metabolism, growth and reproduction [52], [53]. This latter process is also supported by the presence of vitellogenin, a major reproductive protein.
Table 2. Most commonly expressed sequences with associated BLAST matches.
| Contig ID | Length (bp) | No of reads | Description | Species | Common name | E-value |
| 00790 | 8515 | 740 | β-1,3-glucan binding protein | Litopenaeus vannemei | Pacific white shrimp | 0.0 |
| 00604 | 7491 | 689 | Fatty acid synthase | Aedes aegypti | Yellow fever mosquito | 0.0 |
| 00865 | 5235 | 643 | Myosin heavy chain | Aedes aegypti | Yellow fever mosquito | 6.4 e-253 |
| 01872 | 2994 | 617 | Ribonucleoside diphosphate reductase | Aedes aegypti | Yellow Fever mosquito | 0.0 |
| 00704 | 462 | 596 | Ribosomal protein L10 | Callinectes sapidus | Blue crab | 1.0 e-70 |
| 00239 | 279 | 576 | Myosin light chain | Litopenaeus vannemei | Pacific white shrimp | 3.0 e-16 |
| 00111 | 2061 | 568 | ATP synthase | Bombyx mori | Silk worm | 0.0 |
| 02321 | 2341 | 534 | Eukaryotic translation initiation factor 5A | Penaeus monodon | Black tiger prawn | 9.0 e-63 |
| 00248 | 1639 | 525 | NADH dehydrogenase sub-unit 4 | Euphausia superba | Antarctic krill | 0.0 |
| 00826 | 1750 | 502 | Calreticulin | Fenneropenaeus chinensis | Fleshy prawn | 1.5 e-152 |
| 00709 | 2128 | 499 | Elongation factor 1γ | Artemia salina | Brine shrimp | 1.4 e-152 |
| 01202 | 4840 | 493 | Vigilin | Culex quinquefasciatus | mosquito | 3.8 e-257 |
| 00995 | 505 | 492 | 60s ribosomal protein L5 | Ixodes scapularis | Blacklegged tick | 1.0 e-45 |
| 20909 | 849 | 490 | Dehydrogenase, glyceraldehyde dephosphate | Pleocyemata sp. | Lobster | 1.5 e-122 |
| 19460 | 1984 | 484 | Chaperonin | Nasonia vitripennis | Jewel wasp | 0.0 |
| 00477 | 800 | 483 | Vitellogenin | Pandalopsis japonica | Shrimp species | 4.9 e-17 |
| 00056 | 867 | 483 | Ferritin peptide | Fenneropenaeus chinensis | Fleshy prawn | 4.7 e-71 |
| 00581 | 860 | 479 | ATP dependant RNA helicase | Caligus rogercresseyi | Sea louse | 3.1 e-181 |
| 21501 | 2124 | 474 | Cyclin A | Penaeus monodon | Black tiger prawn | 4.7 e-155 |
| 18212 | 417 | 464 | Ribosomal protein L1 | Lonomia obliqua | Caterpillar | 1.4 e-40 |
| 19379 | 1095 | 458 | Death-associated protein-like | Penaeus monodon | Giant tiger prawn | 3.2 e-16 |
| 00914 | 1918 | 456 | Protein disuphide isomerase | Scylla paramamosain | Green mud crab | 1.4 e-162 |
| 01103 | 1039 | 455 | Acidic ribosomal protein P0 | Spodoptera frugiperda | Fall armyworm | 4.0 e-115 |
The majority of the remainder of the putative genes were clearly related to this seasonal physiology and an active metabolism with presence of transcripts involved in DNA replication and RNA metabolism (ribonucleoside diphosphate reductase and vigilin respectively), protein synthesis (elongation factors and ribosomal genes), energy production (dehydrogenase, glyceraldehyde dephosphatase), cell division (cyclin A) and growth (myosin heavy and light chains). The relatively high levels of vigilin may potentially be linked to those of vitellogenin, as although it is suggested to play a general role in RNA metabolism [54], it has been specifically shown to stabilise vitellogenin transcripts in some species [55]. The seemingly odd presence of an apoptosis-related gene (contig19379) is almost certainly linked to the processes involved in ecdysis and moulting. Again, this is a process with increased activity in the austral summer, being linked to nutritional status and environmental conditions [56]. Overall, this initial survey of the pyrosequencing data substantiates a previous molecular study, which indicated that krill sampled in the summer were far more active than those sampled in winter [33].
However, of most interest to our research, was the presence of genes involved in chaperone functions and the stress response in the most commonly expressed sequences (calreticulin, chaperonin, protein disulphide isomerase and ferritin) (Table 2). Also, whilst not in this listing, the heat shock proteins HSP90 and HSP70 were also highly represented in the transcriptome with 417 and 307 reads respectively. To futher understand these systems and provide a baseline for future studies on the environmental stress response of E. superba, a number of putative genes were examined in more detail. These sequences were identified from the BLAST sequence similarity results as being involved in either chaperone or antioxidant functions. Where possible, the more commonly known names of the chaperone genes have also been linked to the new nomenclature as designated by the HUGO Gene Nomenclature committee and NCBI [57].
The cellular chaperone systems
The cellular chaperones can be divided into two main categories: the cytosolic and the endoplasmic reticulum (ER) systems. Many of these proteins are expressed ubiquitously in the normal cell state to aid in the folding of native polypeptides and their translocation to different cellular compartments [58], [59]. However, during the stress response, they may be up-regulated to further assist mis-folded proteins to attain or regain their native states and also target degraded proteins and regulate their removal from the cell, thus preventing the formation of cytotoxic aggregates [60]–[62]. Of these, the best known are the cytosolic genes, as these include the HSPs, specifically HSP70 which has long been associated with the cellular stress response (CSR) [58].
Chaperones do not act in isolation, efficient protein folding in the cell is often achieved by the cellular equivalent of a production line, with upstream chaperones capturing and transferring elongating, nascent chains to more specialised systems. In the cytosol, there are two main pathways involving either HSP70 or prefoldin [61]. The HSP70 route is non-specific, whilst prefoldin seems to specifically act in conjunction with the chaperonin that contains T-complex polypeptide 1 (CCT) proteins. In the ER, the route is, again, either via an HSP70 gene family member: BiP (binding protein) or calreticulin/calnexin. Numerous components of all of these chaperone pathways are present in the E. superba dataset. These will be described below, of which, given our research interests, the HSPs will be characterised in most detail.
The HSP cytoplasmic chaperones
HSP70s
The heat shock proteins are a large family of proteins which are named according to their molecular weight in kiloDaltons. The most studied of these are the 70kD heat shock proteins (HSP70s). There are two main forms of these 70kD proteins, the heat shock cognate (HSC70) which is expressed constitutively and an inducible form (HSP70) which is normally expressed in response to external stimuli [58]. Whilst HSP70 is the “classical” stress gene, the expression levels and regulation of these gene family members in Antarctic species is of particular interest for two reasons: protein folding is more difficult at low temperatures and in the limited examples studied to date, Antarctic species show enhanced expression of these genes [63]–[66] with one adaptation to the cold being the permanent expression of the inducible form in the fish [63], [64], [66] and also the lack of the classical heat shock response (HSR) [65]–[69].
In the krill assembly, 16 contigs, comprising a total of 1,026 reads (coverage ranging from 2–323 reads per contig) produced the best sequence matches to HSP70 using BLAST searching tools. Ten sequences showed the most sequence similarity to the classic inducible form of HSP70. The alignment of the 9 non-redundant sequences (Figure S1) showed that only contig 02253 was complete. The nucleotide sequence of this contig putatively codes for a protein of 668 amino acids. The percentage identity of the deduced amino acid sequence of contig 02253 compared with orthologues from other crustaceans is very high, between 78–84%, confirming the remarkable conservation of this family. As could be expected, this percentage is even higher when you only consider comparisons within the super order Eucarida (>81%) or outside the class Malacostraca (<79%). More detailed analysis of this full length protein sequence reveals the presence of three signature motifs of the HSP70 family: IDLGTTYSCV (amino acids 9–18), IFDLGGGTFDVSIL (amino acids 196–209), IVLVGGSTRIPKIQKL (amino acids 334–349). Interestingly, at the C-terminus there are four repetitions of a tetrapeptide sequence (GGMP) (Figure S1). Certain of these repetitive peptides have antigenic properties [70] and are also implicated in the association between HSP70 and HSP90 in a multichaperone complex. The ATP/GTP active site is also identifiable via the motif: AEAYLGAT (amino acids 133–138) and a potential bi-partite nuclear targeting sequence KRKHKKDPADNKR at amino acids 252–264. The terminal sequence motif of the HSP70 family identifies their cellular location, with the EEDV sequence motif at the end of contig 02253 attesting to its cytoplasmic localisation [71], whereas HDEL and PEAEYEEAKK characterise members localised to the endoplasmic reticulum and mitochondria respectively.
With regard to the other contigs in the alignment, 19890 and 19889 could potentially be the product of a duplication of HSP70 as the amino acid translations are very similar to that of 02253. The differences between these 3 sequences are possibly explained by the multiple origin of the animals used and therefore constitute allelic variants. A fourth sequence, contig 06573, was relatively short (423bp) and closely resembles the previously described contigs. However, it may represent another form due to the signature situated at amino acid 189 (STGQ) (Figure S1). Whilst this sequence is at the end of the read and therefore the quality is less assured, these supplemental amino acids are also present in the HSPs of other crustaceans. For example, there is a short succession of amino acids intercalated at a similar position (amino acid 191 in the alignment in Figure S2) in the two HSP70 isoforms of the hydrothermal vent shrimp, Rimicaris exoculata, as well as in the crab, Dromia personata and the oysters Ostrea edulis and Crassostrea gigas (Figure S2). Whether contig 06573 represents a duplication of the HSP70 gene in E. superba is difficult to exactly determine without the full length sequence and extensive PCR amplification between sequence fragments, which is planned in a future study. However, there is evidence of a duplication of HSP70 genes in Antarctic species; in the molluscs Laternula elliptica, Nacella concinna, but also the crustacean Paraceradocus gibber [65], [69], [72] and other crustacean species known by their capacity to respond to thermal shock namely, Rimicaris exoculata [73], Palaemonetes varians [74] and Macrobrachium rosenbergii [75] amongst others.
Of the other contigs represented in the HSP70 alignment (Figure S1), contig 20245 (and 20197, which is not shown in the alignment as it is an exact duplicate sequence of 20245) is clearly different to those described above, although this is not possible in comparisons with 06573 as there is no overlapping sequence. On BLAST sequence similarity searching these two sequences represent the most likely candidates for the constitutive form of HSC70 (HSPA8), but at 531bp and 163bp respectively accurate designation was difficult.
The overall conclusions from examining the contigs listed above, permits the observation that E. superba possesses numerous forms of HSP70. The designation of specific isoforms coding for inducible or constitutive expression patterns is a delicate issue, based on sequence alone. Only by following the expression of these variant molecules under different regimes and environmental stresses will enable the true definition of their role.
HSP90
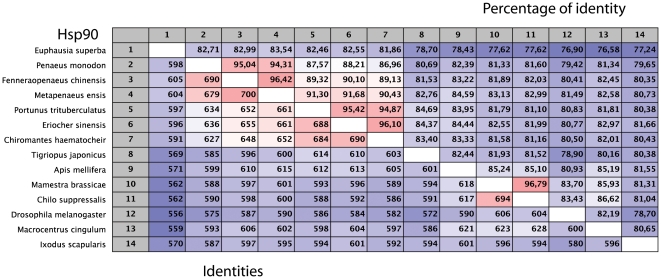
Six contigs showed high sequence similarity to HSP90. The full length sequence, with the exception of 4 amino acids from the N-terminal region was constructed from 4 overlapping contigs 00022, 00026, 02405 and 02406 (Figure S3). This sequence of 2151 nucleotides coded for a protein of 717 amino acids containing the characteristic signatures for the HSP90 protein: NKEIFLRELISNSSDALDKIR (amino acids: 28–48), LGTIAKSGT (amino acids: 95–103), IGQFGVGFYSAYLVAD (amino acids: 119–134), IKLYVRRVFI (amino acids: 346–355) and GVVDSEDLPLNISRE (amino acids: 372–386) [76]. The presence of the pentapeptide MEEVD at the extreme C-terminal indicated that this E. superba EusHSP90-1 is cytosolic and is also implicated in the binding of known co-chaperone molecules.
Two other contigs showed high sequence similarity with HSP90 (10288 and 00025), but there were sufficient amino acid differences present (Figure S3) to suggest that a second HSP90 gene exists in E. superba (designated EusHSP90-2). Two cytoplasmic HSP90s exist in human (HSP90 α and β) and have been reported in other vertebrates. In the invertebrates, only one protein form has been characterised, although in Mytilus galloprovincialis it may be the product of either one or two genes [77] and only one form has been cloned in the crustaceans Peneaus monodon and Metapenaeus ensis [78], [79]. Recently, two different HSP90s have been identified in the crab Portunus trituberculatus [80]. Analysis of the two ptHSP90s showed that both were most similar to the vertebrate HSP90α and are the result of a duplication of the orthologous vertebrate HSP90α gene [80]. Comparisons were made between these two crab sequences and the E. superba sequences across a common region of 128 amino acids. EusHSP90-1 shows most similarity to the HSP90-1 of Portunus (86.7% amino acid identity) (79.7% with ptHSP90-2 and 74.2% with EusHSP90-2). Likewise EusHSP90-2 is also most close in terms of identity to ptHSP90-1 (78.1%) than to the second form (76.6%) of ptHSP90. It should be noted that the percentage identity of the two forms of HSP90 in Portunus is 86%.
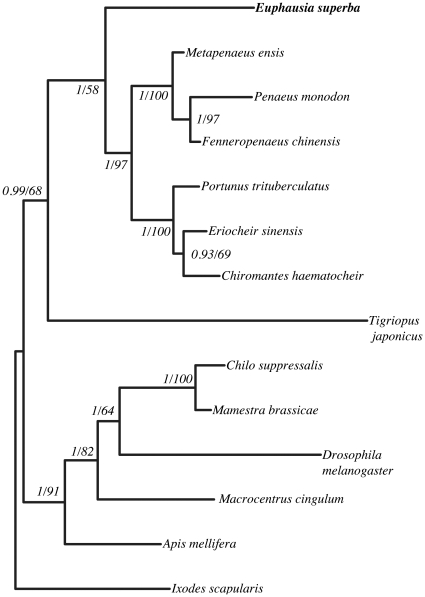
These observations lead to the hypothesis that the Crustacean HSP90-1 genes derive from a common ancestor and that the sequences described here constitute orthologues. However, the percentage identities between the different forms of HSP90 underline that both forms of the crustacean gene are closest in terms of sequence similarity with the vertebrate α form. Hence these molecules are the result of an independent duplication event and the high variability between the paralogues observed in the Euphausia indicate that this phenomenon was very ancient in this taxa. This hypothesis is in accord with the results observed in the Peneids, which at the phylogenetic level are intermediate between the Euphausia and the Brachyura and only appear to have a single HSP90 gene [78], [79]. The HSP90 sequences from a number of invertebrates belonging to the Panarthropoda were aligned (Figure S4) and show the conservation of the protein in this taxon (Figure 1). Bayesian and Maximum Likelihood analyses using the Chelicerate Ixodes scapularis as an outgroup generated congruent trees positioning krill as the sister group of the decapods (Figure 2). The resulting tree structure is coherent with regard to the positions of the different taxons [81], [82], even if certain nodes are weakly resolved, i.e. the base of the Eucrustacea (Figure 2).
Figure 1. Percentage amino acid identities between HSP90 genes from the Panarthropoda.
For accession numbers see Figure 2.
Figure 2. Phylogeny based on Bayesian analysis and maximum likehood of the amino acid data set of HSP90 sequences from the pancrustaceans (14 taxa, 689 characters).
Ixodes scapularis was assigned as outgroup. Trees obtained by Bayesian analysis and by maximum likelihood analysis of the amino acid dataset were fully congruent. Numbers at nodes are posterior probabilities and bootstrap values (based on 100 replicates) respectively obtained from the analysis of the amino acid dataset. Sequence accession numbers are: Ixodes scapularis: XM_002414763; Apis mellifera: FJ713701; Macrocentrus cingulum: EU570066; Drosophila melanogaster: NM_079175; Mamestra brassicae: AB251894; Chilo suppressalis: AB206477; Tigriopus japonicus: EU831278; Chiromantes haematocheir: AY528900.1; Eriocheir sinensis: EU809924; Portunus trituberculatus: FJ392027; Fenneropenaeus chinensis: EF032650; Penaeus monodon: FJ855436; Metapenaeus ensis: EF470246; Euphausia superba: contig00022. Only the ptHSP90-1 and EusHSP90-1 are used in this analysis.
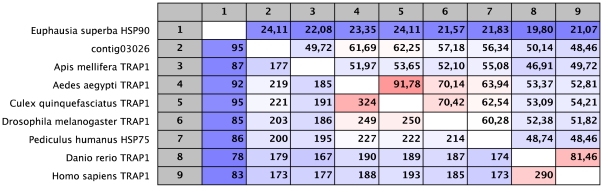
The last sequence with high sequence similarity to HSP90 (contig 03026) putatively codes for the terminal 354 amino acids. However, this shows considerable differences with EusHSP90-1 (24% identity over the region concerned) and does not contain the cytosolic signature EEVD at the extreme C-terminal. Further analysis of the BLAST results indicated that this sequence is actually more similar to TRAP1 (Tumour Necrosis Factor (TNF) Receptor-Associated Protein 1) or HSP75, even though some of the database matches have been designated as HSP90 (Aedes aegypti and Culex quiquifasciatus). This example demonstrates some of the confusion over the naming of these genes, as in Homo sapiens, where all of these variant names have now been potentially designated as HSPC5 [57]. The alignment and percentage identity calculation between the putative E. superba sequences and TRAP1/HSP75 genes from different invertebrates and vertebrates showed significant conservation of this molecule in different taxa (Figure 3 and Figure S5). This homologue of the HSP90s is localised in the mitochondria [83], [84]. Although the precise function of these molecules is yet to be determined, they are implicated in cellular stress resistance pathways and therefore are involved in the general functioning of the cell cycle, cellular differentiation and apoptosis. It is evident that the mitochondria play an essential role in cellular homeostasis under stressful conditions as well as the control of intracellular Ca2+, energy generation and apoptosis (see also section on other identified chaperones including mitochondrial genes). Despite their sequence similarity to the HSP90 genes, the TRAP1 family exhibit significant functional differences [83]. They are particularly studied for their role in cancer defence and represent targets of interest in anti-cancer therapies [85].
Figure 3. Percentage amino acid identities of TRAP1 sequences.
These are from a number of invertebrates and vertebrates with the EusHSP90-1 and contig03026. Accession numbers:. Aedes aegypti TRAP1: AAD29307; Culex quinquefasciatus TRAP1: XM_001861228; Drosophila melanogaster TRAP1: AAD29307; Pediculus humanus HSP75: XP_002425720; Danio rerio TRAP1: AAI4468; Apis mellifera TRAP1: XP_623366; Homo sapiens TRAP1: Q12931.
The prefoldin cytoplasmic pathway
The other major cytosolic chaperone system involves prefoldin. This chaperone is the first stage in a pathway that largely acts on correct folding and polymerization of the cytoskeletal proteins, actin and tubulin [86]. It is a multimeric protein comprised of 6 subunits; 2 are α-like (designated 3 and 5) and the remainder are β-like (assigned numbers 1, 2, 4 and 6). This protein processes nascent polypeptides and then passes them onto CCT for further processing. This second chaperone is more complex comprising 16 subunits in 2 stacked rings, each with 8 polypeptide subunits: alpha, beta, gamma, delta, epsilon, eta, theta and zeta. All subunits for both complexes were present in our data set with the exception of CCT theta (Table 3).
Table 3. Components of the prefoldin cytosolic chaperone system leading to tubulin polymerisation.
| Putative gene designation | Contig ID | Length (bp) | No of reads | Description |
| Prefoldin | 02592 | 544 | 24 | sub-unit 1: Caligus clemensi (sea louse) 2.4e−23 |
| 14357 | 325 | 4 | sub-unit 2: C1BJW5: Osmerus mordax (rainbow smelt) 7.6e−23 | |
| 14161 | 407 | 7 | sub-unit 3: Q5RCG9: Pongo abelii (Sumatran orangutan) 7.2e−31 | |
| 12250 | 446 | 15 | sub-unit 4: Oncoryhnchus mykiss (rainbow trout) 4.3e−11 | |
| 06004 | 1045 | 62 | sub-unit 5: Culex quinquifasciatus (mosquito) 7.7e−37 | |
| 01685 | 499 | 39 | sub-unit 6: Ixodes scapularis (blacklegged tick) 1.6e−20 | |
| Chaperonin that contains T-complex polypeptide 1 (CCT) | 19460 | 1984 | 484 | CCT1:α subunit: C3Y2C5: Branchiostoma floridae (Florida lancelet) 4.5e−210 |
| 00135 | 1864 | 268 | CCT2: β subunit: B5X2M8: Salmo salar (Atlantic salmon) 1.7e−187 | |
| 02814 | 1917 | 247 | CCT3: γ subunit: B0WSS5: Culex quinquifasciatus (mosquito) 3.6e−199 | |
| 00117 | 1994 | 388 | CCT4: δ subunit: C3XS21: Branchiostoma floridae (Florida lancelet) 1.1 e−192 | |
| 21580 | 626 | 278 | CCT5: ε subunit: Q68FQ0: Rattus norvegicus(rat) 3.3e-79 mid portion of gene | |
| 00119 | 202 | 149 | CCT5: ε subunit: C0HBB4: Salmo salar (salmon) 4.2e−17 3′ end of gene | |
| 00120 | 204 | 51 | CCT5: ε subunit: Identical clone to contig 00120 | |
| 04098 | 1804 | 349 | CCT6: η subunit: Q95V46: Artemia sanfranciscana (brine shrimp) 3.5e−212 | |
| CCT7: θ subunit: not present | ||||
| 00149 | 1785 | 376 | CCT8: ζ subunit: B0W8W8: Culex quinquifasciatus (mosquito) 4.0e-202 | |
| Tubulin-specific co-factors | 03429 | 1127 | 19 | tbca: B0W943: Culex quinquifasciatus (mosquito) 7.4e−14 |
| 02234 | 2032 | 62 | tbcb: Q5E951: Bos taurus (bovine) 5.8e-61 | |
| tbcc: not present | ||||
| 05722 | 958 | 8 | tbcd: Q5ZI87: Gallus gallus (chicken) 7.0e−53 | |
| 02424 | 1104 | 14 | tbcd: Q8BYA0: Mus musculus (mouse) 5.0e−40 | |
| 07816 | 635 | 4 | tbce: Q28EJ7: Xenopus tropicalis (pipid frog) 4.6e−21 | |
CCT action is not exclusively restricted to actin and tubulin, as it has also been shown to bind approximately 15% of newly synthesized proteins sized between 30–60kDa and so may play a more general role in the cytoplasm [87]. However, as part of its primary role, CCT may pass on the cytoskeletal folding intermediates for further processing and in the case of tubulin, this can lead to the production of microtubules. This is achieved via tubulin cofactors (designated A–E), all of which are also present in our dataset with the exception of subunit c; subunit d is present in the form of 2 non-overlapping clones, hence the dual entry (Table 3). The tubulin co-factors act on α and β-tubulin folding intermediates to generate polymerizable heterodimers for microtubule growth and centrosome generation, but also confer a certain level of stability within these structures and hence directly influence cell division processes as well as providing a chaperone function [88].
Analysis shows that all major elements of the two cytosolic chaperone systems were present in the E. superba data set. Contigs 18509 and 13343 showed strong sequence similarity to members of the HSP70 family and are almost certainly molecular chaperones. However a closer analysis and alignment with database entries for HSP70 isoforms 4 and to members of the HSP105/110 (Figure S6), shows that with these partial sequences, it is impossible to precisely identify which group (HSPA14, HSPH1, HSPH2 or HSPH3) these fragments actually belong to. Similarly, contig 06563 showed a match with high probability to HSP70 protein 14 (HSPA14), but with only 34% identity to the mouse protein, it is either not highly conserved between species or is a different, potentially novel family member. Only by comprehensive analysis of all HSP70 family members will the exact designation of vertebrate orthologues be possible in non-model species, a non-trivial task.
Similarly, HSPH1 (contig 18509) is another member of the HSP70 family and whilst it has been shown to prevent the aggregation of proteins under severe stress, it also interacts with other HSPs, inhibiting HSPA8/HSC70 ATPase and chaperone activity. In addition, a number of other cytosolic chaperones and co-chaperones were present, some of which appear to have mainly chaperone functions (e.g. HSPa4l (contig09322)), whilst others interact with, and control the action of other chaperones. Of particular note were contigs with high sequence similarity to the HSP70 co-chaperones DnaJ, chaperonin 10 (contig 01709) and HSPBP1 (contig06138). This latter protein interacts with HSPA1A, an HSP70 family member and has been shown to inhibit the action of HSPA1A and therefore impact on degradation of target proteins. The DnaJ proteins are an extensive gene family which directly stimulate the ATPase activity of the HSP70 chaperones, but also determine the activity of HSP70s by stabilising their interactions with substrate proteins [89]. All members of this gene family contain a J domain, but they have been divided into 3 main groups depending on what other domains are present in the protein. In humans, so far there have been 4 type A, 14 type B and 22 type C DnaJ proteins described [57]. In the E. superba dataset 18 contigs potentially comprising at least 13 different family members over all three groups were present (Table 4), indicating a similar complement of these genes in krill compared to vertebrates. Of the full data set, only 2 small heat shock proteins were identified in contigs 05090 and 05374. These are difficult to define as they are not very conserved between species. Finally, three non-overlapping contigs were identified with high sequence similarity to Hop. This is the gene name for the HSC70/HSP90 organising protein and mediates the association between these two chaperones. However, as stated previously, the cytosolic chaperone system with the prevalence of HSP70 gene family members is not the only cellular chaperone system or pathway.
Table 4. Putatively identified HSP40 genes in E. superba.
| Contig ID | Length (bp) | No of reads | Putative assignment | Closest match |
| 04109* | 1336 | 29 | Dnaja1 | Q0P4H4: Xenopus tropicalis (toad) 2.2e−117 |
| 02240 | 1358 | 30 | Dnaja3 | Q16R72: Aedes aegypti (mosquito) 6.9 e−138 |
| 14139 | 589 | 5 | Dnaja3 | D2A3L8: Tribolium castaneum (red flour beetle) 2.0e−155 |
| 09733 | 412 | 5 | Dnajb1 | Q28F52: Xenopus tropicalis (toad) 1.5e−9 |
| 02096 | 1169 | 23 | Dnajb11 | B0WE51: Culex quinquefasciatus (mosquito) 1.6e−115 |
| 09747 | 1095 | 23 | Dnajc2 | Q6NWJ4: Danio rerio (zebrafish) 3.6e−63 |
| 01898 | 1254 | 28 | Dnajc3 | B7P6D4: Ixodes scapularis (tick) 4.1e−87 |
| 08765 | 603 | 5 | Dnajc7 | Q48AJ9: Tetraodon nigroviridis (pufferfish) 1.4e−61 |
| 13604 | 454 | 8 | Dnajc7 | B6VFL2: Bombina orientalis (toad) 4.5e−28 |
| 11977 | 1179 | 11 | Dnajc9 | Q5EAY1: Xenopus laevis (toad) 5.7e−45 |
| 05667 | 1086 | 11 | Dnajc11 | B7PLX9: Ixodes scapularis (tick) 9.4e−71 |
| 06410 | 441 | 3 | Dnajc13 | D3ZNI6: Rattus novegicus (rat) 4.5e−17 |
| 08550 | 1726 | 11 | Dnajc16 | B7QBO3: Ixodes scapularis (tick) 1.5e−36 |
| 16253 | 542 | 5 | Dnajc19 | A8Q605: Brugia malayi (nematode) 1.7e−13 |
| 00224 | 632 | 6 | Dnajc21 | O62360: Caenorhabditis elegans (nematode) 3.1e−28 |
| Fragments less than 400bp | 16579 (266, 5); 14282 (310, 2); 10787 (399, 4); | |||
Only contigs with reads above 400bp in length are annotated. * denotes full length sequences. Small fragment data includes size in bp and number of reads (in brackets).
The endoplasmic reticulum chaperone systems
The ER has its own protein quality control mechanisms and acts, in particular, on secretory proteins. Newly synthesized secreted and membrane-bound proteins are folded and assembled in the ER and failure to achieve correct conformation results in in situ tagging, retention and degradation, all still within the ER. This comprehensive quality checking is achieved via two main pathways involving either the HSP70 BiP complex or the calnexin/calreticulin system [90].
The ER BiP pathway
This system recognises the presence of unfolded regions on proteins containing hydrophobic residues and plays an important role in the folding and post-translational modification of non-glycosylated proteins. BiP is an HSP70 family member, often referred to as GRP78 (HSPA5). Studies involving immunoglobulin binding assays have shown that this protein is part of a multiprotein complex which also includes GRP94 (HSPC4), CaBP1, protein disulphide isomerase (PDI), an HSP40 co-chaperone, cyclophillin B, Erp72, GRP170, UDP-glucosyltransferase and SDF2-L1 [90]. Of the genes in this listing, only BiP and GRP74 were specifically identified in the E. superba dataset (Table 5). GRP94 is one of most abundant ER proteins and on sequence similarity is thought to be a homologue of HSP90. An additional number of contigs showed high sequence similarity to members of the PDI family (Table 5), none of which are implicated in the BiP complex described above, but have been shown to localise to the ER and have chaperone activity. They also contain variable numbers of thioredoxin domains, which contribute to the disulphide isomerase activity, involved in the disulphide exchange reactions in post translational modification [91].
Table 5. Components of the ER chaperone system.
| Gene | Contig ID | Length (bp) | No of reads | Description |
| BiP (GRP78) | 01477 | 2341 | 259 | B0W934: Culex quinquifasciatus (mosquito) 2.7e−264 |
| GRP74 | 01266 | 1940 | 41 | A5LGG7: Crassostrea gigas (oyster) 5.3e−197 |
| Calreticulin | 00826 | 1750 | 502 | B8K275: Fenneropenaeus chinensis (Fleshy prawn) 1.5e−152 |
| Calnexin | 08163 | 1065 | 16 | A5LGG8: Crassostrea gigas (oyster) 3.4e−59 |
| PDI | 00914 | 1918 | 456 | Erp60: B4QC45: Drosophila simulans (fly) 1.1e-144. Contains 2 thioredoxin domains. |
| PDI | 05765 | 1293 | 53 | TXNDC5: Thioredoxin domain containing protein 5: Q8NBS9: Homo sapiens(human) 1.9e−87. Contains 3 thioredoxin domains. |
| PDI | 01765 | 2182 | 124 | PDIA6: Q15084: Homo sapiens (human) 2.7e−120. Contains 2 thioredoxin domains. |
| PDI | Fragments less than 400bp: 13243 (352,5); 05245 (344,2); 11325 (330, 4); 13589 (280, 3). | |||
Small fragment data includes size in bp and number of reads (in brackets).
The ER Calnexin/Calreticulin pathway
This second pathway affects the folding and post-translational modification of virtually all glycosylated, secreted or integral membrane proteins that pass through the ER. Although calnexin and calreticulin are some of the most abundant chaperones in the ER, they are either not detected, or present in trace amounts in the BiP complex described above. Evidence suggests that these two networks are spatially separated and that as secreted proteins mature they are transported inside the ER from one network (BiP) onto calnexin/calreticulin [90]. These two genes are putatively represented by a full length contig for calreticulin (00826) and a partial 3′ clone (08163) for calnexin. Whilst these two genes share a high degree of amino acid similarity and functionality (Ca2+ binding, lectin-like activity and recognition of mis-folded proteins), they have distinct cellular localisations which affect their mode and sphere of action. Calnexin is an integral membrane protein, whilst calreticulin is a luminal protein and thus more mobile. In addition to their role as chaperones, they also play a significant role in ER Ca2+ binding and storage. Hence they are also involved in Ca2+ homeostasis and ER-dependent Ca2+ signalling [92] and therefore are potential candidates for understanding responses to stress both in terms of protein turnover, but also cellular signalling.
Other identified chaperones including mitochondrial genes
In addition to the various cytosolic chaperones identified, a number of other contigs showed high sequence similarity to more specialised protein folding systems. For example, the MESD chaperone (contig 01700) specifically assists in the folding of β-propeller/EGF molecules within low density lipoprotein receptors (LDLRs) and the nuclear chaperone Asf1 (contig 05542) facilitates histone deposition and histone exchange and removal during nucleosome assembly and disassembly. In contrast, contig13343, in spite of having an N-terminal ATPase domain, has many amino acid differences with the classical HSP70s and to date, most research has indicated a primary role in the mammalian immune response.
Similar to the ER, the mitochondria also has its own chaperone system. Two co-chaperones were putatively identified: HscB (contig 12123) involved in iron-suphur cluster assembly and contig 16412 which showed high sequence similarity to GrpE, the main HSP70 co-chaperone in the mitochondria and equivalent to the cytosolic DnaJ [93]. As regards the main HSP70 family member in this organelle, this is GRP75 (HSPA9) (contig 10241). This protein is not induced by heat, but by other forms of stress e.g. glucose deprivation and oxidative injury. Indeed over expression of GRP75 in cell lines has indicated a primary role in inhibition of ROS accumulation under stressful conditions [94].
This latter example exemplifies the multifunctional role that chaperone proteins play in the cell. Whilst they are documented as being one of the major groups of proteins involved in protein folding and the cellular stress response [95], our understanding of their role is becoming increasingly complex. Not only do they assist in the de novo folding of nascent proteins (estimated to be in excess of 20% of all cellular proteins), a requirement that increases under stressful conditions, but also in interactions with signal transduction proteins controlling cell homeostasis, proliferation, differentiation and cell death [96]. Hence they are proposed as important multifunctional hubs in cellular networks [97]. As such, even subtle environmental perturbations are proposed as being able to profoundly affect not only chaperone production, but also the functions of the whole cell network [97]. Whilst all major components of the various chaperone systems were identified in this study, despite extensive searches, transcription factors controlling the expression of many of these were not present in the dataset. Sequences with strong sequence similarity to either the Heat Shock Factor (HSF) or the Hypoxia Inducible Factor (HIF) transcription factors were not identified. The reason for this may simply be timing, in that this data represents a single time point and these transcripts had decayed by the time the RNAs were extracted.
Clearly, with regard to the stress response, some of the important interactions to review will be those with the major antioxidant proteins involved in combating the production of ROS, which directly impact on protein functioning.
Antioxidants
The examination of these enzymes, in terms of their structure and function is closely allied to our interest in heat shock proteins and the cellular stress response and may be especially important for Antarctic species that have evolved at constant low temperatures for millions of years [72], [98]. Sequence similarity searches were carried out for several of the best characterised members of this functional group with good sequence coverage in our data set. In this respect, catalase, a major antioxidant enzyme previously described in the clam [99] is only represented by a single contig (08109) (match: 6.5e−22) matching the very 3′ end of the gene (from amino acids 425 onwards). This small amount of predicted amino acid sequence does not really allow for effective annotation or comparisons with other species.
Ferritin
In contrast, the full length sequence of a putative ferritin gene is present in contig 00056, which is also one of the more highly expressed sequences in the data set (Table 2). This heteromeric protein comprises 2 subunits: a heavy (H) and a light (L) chain. The heavy chain is ubiquitous and contains the catalytic ferroxidase centre which is responsible for the oxidation of iron and allows iron uptake and release. The light chain is catalytically inactive [100]. The sequence described here is the heavy chain variant, containing the feroxidase motif. In line with previous findings with other crustacean ferritins, the E. superba sequence shares more amino acid sequence identity with the human (53.6%) rather than the Drosophila heavy chain (35.9% identity at the amino acid level) substantiating the theory of differential evolution of the insect ferritins [101]. This protein is well described as being expressed in response to oxidative stress and hypoxia, but interestingly, it has also be shown to be elevated in response to freezing in the marine snail (Littorina littorea) [102]. This may explain the relatively high expression levels in this Antarctic crustacean, although it has also been shown to play an immune role in marine invertebrates [38], [103].
Glutathione S-transferase (GSTs)
The numbers of these genes present in each species varies widely from 16 in human, through 28 in Anopheles gambiae, 37 in Drosophila to 48 in Arabidopsis [104]–[106]. GSTs have been classified into families, the largest of which is the cytosolic family comprising alpha, mu, omega, pi, sigma, theta and zeta. Other family members include a mitochondrial form (designated kappa), membrane bound microsomal forms and some with restricted distributions such as phi, tau and lambda in plants and epsilon and delta in insects.
In the E. superba dataset 20 contigs were identified with high sequence similarity to GSTs. Of these, 3 were most likely to be the related prostaglandin D synthases (00275, 02350 and 02637), with contig 09884 most closely matching the mitochondrial kappa form. The remaining were assigned as cytosolic forms and further designated into putative gene family members, based on sequence similarity searches (Table 6). Of the 10 contigs which contained either full length or almost full-length sequences, it was possible to differentiate 8 genes putatively belonging to the omega, sigma, delta, theta and mu families. Of the latter, 4 contigs showed significant matches against the mu family, but three of these (08776, 10471 and 03844) are potentially allelic variants, sharing approximately 85% identity and 93% amino acid similarity. This is in contrast to contig 06543, which also showed sequence similarity to the mu family, but shared only 64.7% identity to the other putative family members and therefore was designated as a separate gene. Previous examination of the E. superba ferritin gene, showed this to be evolutionarily more similar to the vertebrate, than the insect lineage. The reverse is true of the GSTs, with the identification of a putative delta GST. Recently a delta GST has been described in the Chinese mitten crab [107] and the data presented here substantiate these findings that the delta GSTs are also present in the Crustacea and not restricted to the more traditional insect taxonomic groupings.
Table 6. Putatively identified GST genes in E. superba.
| Contig ID | Length (bp) | No of reads | Putative assignment | Closest match |
| 02590* | 1069 | 45 | delta | A9QUN5: Blattella germanica (German cockroach) 7e−38 |
| 03298 | 1065 | 16 | omega | B0VZ90: Culex quinquifasciatis (Southern house mosquito) 2.6e-47. 5′ end missing |
| 07558* | 976 | 14 | omega | D1MAK0: Anopheles cracens (mosquito) 2.4e−44 |
| 08955* | 950 | 22 | sigma | O18598: Blattella germanica (German cockroach) 3.1e−17 |
| 06543* | 848 | 88 | mu | C3KHT9: Anoplopoma fimbria (sable fish) 9.2e−65 |
| 10471* | 820 | 19 | mu | B8JIS5: Danio rerio (zebrafish) 4.7e-66 |
| 04853 | 627 | 19 | sigma | C3V9U8: Chironomus tetans (midge) 3.1e−14. Mid portion of gene. |
| 03844 | 603 | 93 | mu | Q0GZP5: Cyprinus carpio (common carp) 2.5e−56. Partial 5′ end. |
| 08403 | 531 | 4 | theta | Q5PY76: Aedes aegypti (yellow fever mosquito) 7.0e−33. Partial 5′ end. |
| 08776 | 417 | 7 | mu | B8J188: Danio rerio (zebrafish) 2.5e-35. Partial 5′ end. |
| Fragments less than 400bp | 20423 (400, 6); 09683 (268, 4); 12330 (263, 2); 03758 (243, 8); 10951 (243, 13); 20077 (153, 9) | |||
Only contigs with reads above 400bp in length are annotated. * denotes full length sequences.
Surveying the potential functionality of these proteins in E. superba, there are potential clues from the Insecta. The sigma class in mammals function as prostaglandin synthases, but in Drosophila, these have been shown to be primarily active in the detoxification of oxidative damage [108]. Also the Drosophila delta GSTs are involved in combating oxidative stress and metabolism of endogenously formed lipid peroxidation products [109]. It will be interesting to further characterise the function of these sequences in E. superba in the light of lipids as a major energy store for Antarctic marine species and the enhanced levels of dissolved oxygen and the potential for increased ROS damage in Southern Ocean species.
Superoxide dismutase: a novel duplication event
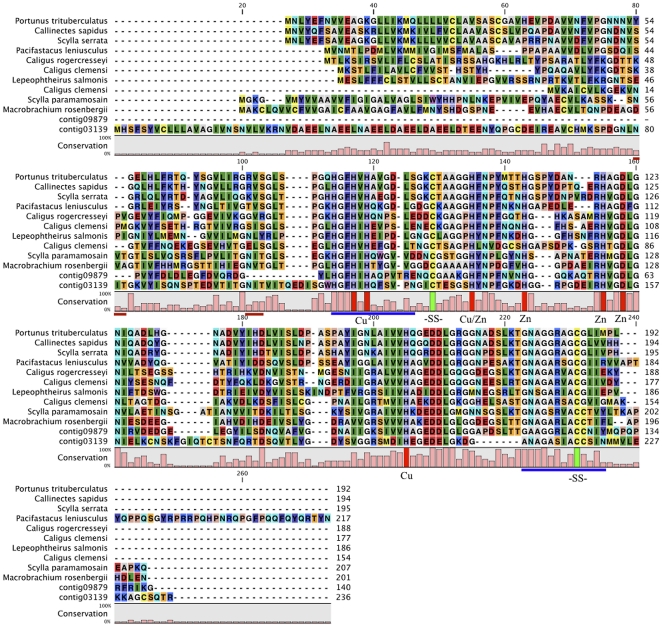
Superoxide dismutases are important antioxidant enzymes. Three distinct types have been identified in eukaryotes depending on the metal ions found at the active site: iron SOD (Fe-SOD) (found only in prokaryotes and plants to date), manganese SOD (Mn-SOD) and copper/zinc SOD (Cu,Zn SOD). Surprisingly searches of the E. superba dataset identified 4 putative SOD genes (Table 7). Two putative Mn-SOD transcripts were clearly differentiated via the manganese superoxide dismutase signature (DVWHHAYY) and then further defined using BLAST sequence similarity searching into the mitochondrial and cytosolic forms (Table 7). Analysis of the Cu,Zn SODs was more complex. An alignment of the two putative Cu,Zn SOD protein sequences from E. superba with those of other Eucrustacea identified the conserved amino acid residues for binding Cu and Zn and the signature motifs of Cu,Zn SODs in both genes (Figure 4). There are two forms of this gene documented in a variety of species [110]: an intracellular and an extracellular form [111]. However, the two E. superba sequences did not contain either the consensus header or consensus tail sequences (KAVCVL and GVIGT respectively) associated with the intracellular form [110]. Sequence analysis showed that the two E. superba transcripts shared only 32.9% identity and hence were not the result of alternative splicing, as has been found in Caenorhabditis elegans [112]. Duplicated Cu,Zn SOD variants have been identified in Xenopus laevis [113], but the genome of this species is known to be tetraploid and hence this situation may not be unexpected. There is very little data on the genome evolution of the Eucrustacea, but, to date, there is no evidence of extra genome duplications in this taxa and therefore the two transcripts in E. superba potentially represent a novel species-specific gene duplication.
Table 7. Identification of SOD genes via sequence similarity searches.
| Contig | Gene | BLAST sequence similarity match | |||||
| Accession number | Organism | Common name | P | Expect | % id | ||
| 00295 | cytMn-SOD | C3W7U6 | Cancer pagurus | Rock crab | 1189 | 2.3e-119 | 75 |
| 06804 | mtMn-SOD | Q3Y596 | Macrobrachium rosenbergii | Giant fresh water prawn | 814 | 1.2e-79 | 70 |
| 03139 | Cu,Zn-SOD | Q09JE3 | Argas monolakensis | Mono lake bird tick | 291 | 3.3e-24 | 41 |
| 09879 | Cu,Zn-SOD | Q45Q33 | Macrobrachium rosenbergii | Giant fresh water prawn | 328 | 3.9e-28 | 56 |
Figure 4. Multiple sequence alignment of the two putative E. Superba Cu-Zn superoxide dismutases with those of other Eucrustacea.
Signature motifs for Cu-Zn superoxide dismutases are indicated by the blue lines under the consensus. The four amino acid residues required for binding copper and four amino acids required for binding zinc are indicated by arrows, as well as two cysteines residues (SS) for the disulphide bridge [110]. Putative N-glycosylation sites for contig03139 only are indicated by the red lines under the consensus and the predicted signal peptide (again for contig03139 only) is marked in italics and underlined. Accession numbers: Portunus trituberculatus: C8XTB0; Callinectes sapidus: Q9NB64; Scylla serrata: A5Y446; Pacifastacus leniusculus: Q9XYS0; Caligus rogercresseyi: C1BQG7; Caligus clemensi: 1: C1C108, 2: C1C2E3; Lepeophteirus salmonis: C1BSR9; Scylla paramamosain: D2DSH2; Macrobrachium rosenbergii: Q45Q33.
It is not possible, given the data presented here to determine why this gene duplication event, and a requirement for additional antioxidant enzymes, has occurred in E. superba. It is suggested that animals living in constant cold temperatures may be more vulnerable to damage by reactive oxygen species, due to slow cell and protein turnover rates and the consequent accumulation of oxidised proteins [99], [114], however examination of the databases shows a complex pattern of Cu,Zn duplication in the Arthropoda. Bombyx mori, the silk worm, has both intracellular and extracellular Cu,Zn genes. In contrast Culex quinquefasciatus, the Southern house mosquito, has 4 different putative genes for Cu,Zn SOD (B0WC98, B0WNS9, B0X9L3, B0VZ56), all of which are different with the latter two containing the intracellular motifs. The two extracellular variants, only show 17.2% identity and 26.9% similarity at the amino acid level, but do contain the required motifs for Cu,Zn SOD, i.e. duplication of both the intra- and extracellular forms. A more complicated story arises with 5 putative genes for SOD in Drosophila sechellia (P61854, B4HNH5, B4HMD6, B4ICF4 and B4HT88). The last accession number describes a Mn-SOD, P61854 is the intracellular form of Cu,Zn SOD whilst the other 3 genes (181, 264 and 270 amino acids respectively) all appear to code for different variants of extracellular Cu,Zn SOD. In the Eucrustacea, to date, only one form of Cu,Zn SOD has been identified with the exception of the sea louse, Caligus clemensi, which has two, but these can be clearly differentiated into an intracellular and extracellular form. In Antarctic marine species, gene duplication events have fuelled adaptations to the cold, particularly in the case of antifreeze proteins [115] and this extra SOD may be another such example. As more non-model species are sequenced the complexity of gene duplication events will become more apparent and fuel the requirement to understand such events in the light of adaptation to different habitats and life histories.
Other antioxidant genes
Additional to the more detailed analyses above, further contigs were identified with high sequence similarity to genes with antioxidant functions. Contig15252 was shown to be most similar to a copper transport protein in the Southern house mosquito (Culex quinquifasciatus) (accession number: B0X8C6). Also two contigs (02344 and 02345) comprised non-overlapping clones with sequence similarity to the ER localised hypoxia up-regulated protein (Hyou1). Whilst this latter protein is a member of the HSP70 family and therefore may play function as a molecular chaperone and participate in protein folding, its critical role is thought to be cryoprotection triggered by oxygen deprivation.
Conclusions
This paper describes, for the first time, a transcriptome of the Antarctic krill Euphausia superba using 454 pyrosequencing technologies. The most highly expressed genes are concerned with lipid and seasonal metabolism. However, the classical “stress proteins”, such as HSP70, HSP90, ferritin and GST are also highly expressed, which lead us to carry out an extensive characterisation of both the cellular chaperone system and the major antioxidant proteins (Figure 5). Knowledge of these gene networks is particularly pertinent in the study of Antarctic marine species in terms of both their adaptations to living in an extreme environment, but also to their responses to climate change. In a recent study on the transcriptomes of Antarctic notothenioid fish 177 protein families were shown to be over-expressed compared with temperate relatives, many of these were genes involved in protein biosynthesis, protein folding, lipid metabolism and antioxidants [116], similar to those described in this paper. These were hypothesized as required adaptations to living in the cold and validate some of the proposed costs of living in such an environment, such as decreased protein folding efficiency [117] and increased vulnerability to damage by reactive oxygen species [99].
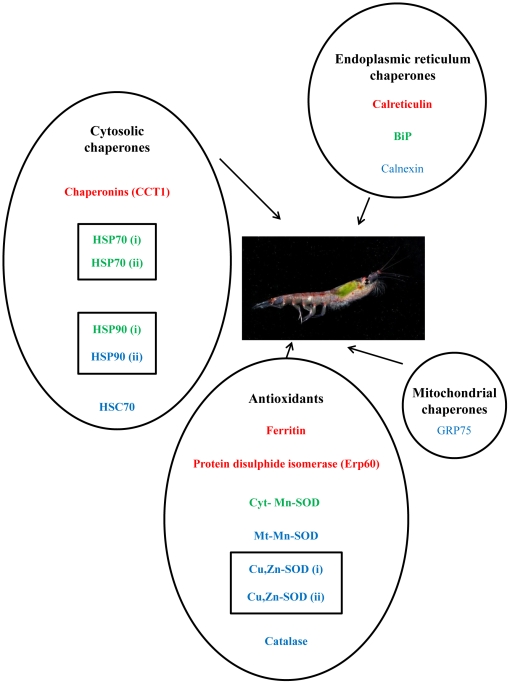
Figure 5. Summary of most highly expressed chaperone and “stress” transcripts in E. superba.
Picture of E. superba from the Bellingshausen Sea continental shelf, taken onboard RRS James Clark Ross cruise JR230 (benthic pelagic coupling cruise) by Pete Bucktrout (British Antarctic Survey). Gene names in red = 400+ copies in the transcriptome data, Green = 200–400 gene copies and blue = less than 200 gene copies. Genes labelled (i) and (ii) plus boxed in the diagram are duplicated genes and their analysis fully described in the main text.
As more Antarctic transcriptomes become available it will be possible to determine how wide-spread altered expression patterns, gene duplication events and even altered cellular networks are in Southern Ocean or indeed, polar animals. E. superba is a pelagic species and experimental work is always limited by availability and ship time. The mere act of recovering the animals from the sea via trawls is stressful and it will always be difficult to partition the stress effects of the sampling regime from the experimental changes to the transcriptome. As such, there is a need to identify not only the stress response, but also the trade-offs associated with enhanced “stress gene” production. It is already known that some Antarctic species permanently express the inducible form of HSP70 as well as other HSP70 family members [63]–[69]. In more experimentally amenable species it has been shown that there is a demonstrable energetic cost associated with the production of heat shock proteins [118] and as yet we have no knowledge of the effect of this in terms of altered transcription patterns in Antarctic species or how it will affect their response to climate change effects. Is the stenothermal nature of Antarctic marine species solely a result of adaptation to living in a cold constant environment for so long, or is it a trade-off effect of survival? Recent large-scale EST and 454 studies [37], [116] and the data described here, are starting to provide the building blocks towards more detailed studies across a range of Antarctic species with the ultimate aim of providing ecosystem level answers to the costs of life in the cold.
Materials and Methods
Specimen collection, RNA isolation and cDNA production
The animals were collected on cruise JR177. Catches were made in the vicinity of the South Orkney Islands (60.44°S to 60.53°S and 47.97°W to 48.13°W). All of the krill were taken by target fishing, mainly using a pelagic net (RMT8), based on observations of krill swarms using an EK60 echo sounder at 38 and 120KHz. The nets were towed for only a short time and, after hauling, the krill were transferred as quickly as possible to the cold room at 1–3°C for sorting. Catches were made at different times of the day (00:50, 06:00, 07:00, 14:00 and 20:30) and samples flash frozen before being stored at −80°C. RNA was isolated from whole animals using the RNeasy system with on-column DNase treatment (Qiagen) according to manufacturers instructions. RNA was initially qualified using a standard spectrophotometer and a pool of 6 animals prepared for library production. 1.5µg of pooled sample of krill total RNA was used for the library production, with the RNA purity additionally checked on an Agilent Bioanalyzer. The cDNA library was constructed using the Creator™ SMART™ kit (Clontech Laboratories Inc., Mountain View, CA, USA). In brief, total RNA was reverse transcribed to first stand cDNA coupled with (dC) tailing and CDS III primers (both containing SfiI digestion site sequences) following a step of template switching and extension by reverse transcription. The second strand cDNAs were then amplified using long-distance PCR with Advantage 2 PCR enzyme system (Clontech Laboratories Inc.). The quality of cDNAs was then visually checked by loading the cDNAs on a 1.1% agarose gel. The cDNAs were then purified using QIAquick PCR Purification Kit (Qiagen; Carlsbad, CA, USA) according to the manufacturer's instructions. The second strand cDNAs (5 µg) were then subjected to sequencing library construction and pyrosequencing using GS FLX Titanium system (Roche applied Science, Indianapolis, IN, USA) at McGill University and Genome Quebec Innovation centre (Montreal, QC, Canada) using the standard GS FLX protocol.
454 Assembly and Analysis
The initial assembly comprised 943,817 reads. Crossmatch (P. Green, unpublished) was then applied to screen for adaptor sequences and other artifacts of the pyrosequencing procedure and also vector sequences using the UniVec database (www.ncbi.nlm.nih.gov/VecScreen/UniVec.html). Stripping the masked sequence from the ends and removing reads with masked sequence in the middle as well as eliminating resulting reads below 50bp, resulted in 699,248 sequences that were entered into the Newbler program [34] for assembly. This resulted in 22,177 contigs. None of the singletons were used for further analysis. Files containing the reads have been submitted to the National Center for Biotechnology Information Short Read Archive (accession number: SRA023520). The mapping facility of Newbler was applied to the assembly to determine the number of SNPs, and Phobos [119] was used for microsatellite discovery. The contigs were then searched for sequence similarity using BLAST [120] against the genbank non-redundant database [121]. Sequence manipulation was carried out using either the EMBOSS suite of programmes [122] with clustering using ClustalW [123] or CLC Mainworkbench [124]. Alignments were produced and displayed in CLC Mainworkbench [124]. Putative N-glycosylation sites in the Cu,ZN SODs were predicted using the NetGlyc 1.0 Server [125] and signal peptide using the SignalP 3.0 server [126] using both neural networks and hidden Markov models [127] and accessed via the CBS server [128].
Supporting Information
Putative translations and amino acid alignment of the 9 E. superba contigs with sequence similarity to HSP70.
(PDF)
Alignment of the deduced amino acid sequences of the E. superba contigs 02253 (full length) and 06573 (partial) with those of other Eucrustacea. Sequence accession numbers are: Balanus amphitrite: Q86MC3; Artemia sanfranciscana: Q95V47; Moina macropoda: ACB11341; Daphnia magna: ACB11340; Tigriopus japonicus: B8PT12; Pachygrapsus marmoratus: ABA02164; Homarus americanus: ABA02165; Penaeus monodon: AAQ05768; Metapenaeus ensis: Q1HGN3; Litopenaeus vannamei: AAT46566; Rimicaris exoculata-3: D2SPE2; Rimicaris exoculata-2: ACL52279; Rimicaris exoculata-1: ABF85673; Palaemonetes varians: ACR77532; Mirocaris fortunata: A1XQQ5; Macrobrachium rosenbergii: Q6S4R6; Portunus trituberculatus: D2DWR3; Portunus sanguinolentus: A8KCI1; Eriocher sinensis: B5AMI7; Calinectes sapidus: Q194W6; Scylla paramamosain: B3VKG9.
(PDF)
Putative translations and amino acid alignment of the 6 E. superba contigs with sequence similarity to HSP90.
(PDF)
Amino acid alignment of HSP90 genes from the Panarthropoda. Sequence accession numbers are: Ixodes scapularis: XM_002414763; Apis mellifera: FJ713701; Macrocentrus cingulum: EU570066; Drosophila melanogaster: NM_079175; Mamestra brassicae: AB251894; Chilo suppressalis: AB206477; Tigriopus japonicus: EU831278; Chiromantes haematocheir: AY528900.1; Eriocheir sinensis: EU809924; Portunus trituberculatus: FJ392027; Fenneropenaeus chinensis: EF032650; Penaeus monodon: FJ855436; Metapenaeus ensis: EF470246; Euphausia superba: contig00022. Only the ptHSP90-1 and EusHSP90-1 are used in this alignment.
(PDF)
Amino acid alignment of TRAP1 sequences. These are from a number of invertebrates and vertebrates with the EusHSP90-1 and contig03026. Sequence accession numbers are: Aedes aegypti TRAP1: AAD29307; Culex quinquefasciatus TRAP1: XM_001861228; Drosophila melanogaster TRAP1: AAD29307; Pediculus humanus HSP75: XP_002425720; Danio rerio TRAP1: AAI4468; Apis mellifera TRAP1: XP_623366; Homo sapiens TRAP1: Q12931.
(PDF)
Amino acid alignment of E. superba additional putative HSP70 family members with HSP105 and APG HSP70 family members. Accession numbers: Tigriopus japonicus HSP105: ACA03526; Apis mellifera HSP70Cb: XP_623199.1; Nasonnia vitripennis: XP_001607146; Pediculus humanus HSP105: XP_002431004; Xenopus laevis HSP105: NP_001086692; Callithrix jacchus HSP105-2: XP_002748999; Callithrix jacchus HSP70-4: XP_002744640; Mus musculus HSP70-4-APG2: Q61316; Mus musculus HSP70-4-APG1: P48722; Homo sapiens HSP70-4: ABM69040.
(PDF)
Microsatellite repeats found in excess of 7 copies per repeat unit in E. superba data.
(XLS)
Variant nucleotides (SNPs/INDELS) found in E.superba data.
(XLS)
Acknowledgments
We would like to thank Geraint Tarling from BAS for providing the krill samples from the James Clark Ross Cruise JR177 and also thank the crew for their support on sample collection during that cruise. Paul Seear at BAS performed the krill RNA extractions. Thanks also go to Pascal Marquis and Ken Dewar at McGill University Quebec who carried out the 454 runs. We would also like to thank Bela Tiwari of the NERC environmental Bioinformatics Centre (NEBC), CEH, Wallingford for valuable discussions on 454 software, analysis pipelines and processes and Phil Trathan (BAS) for critical reading of the introduction.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding for the 454 run provided by Livestock Gentech: an Alberta Ingenuity Centre at the University of Alberta. JYT benefits from funding provided by la Région Bretagne (France) and also l'Institut Paul-Emile Victor (IPEV). The analyses were completed by the team from the British Antarctic Survey within the Ecosystems Adaptations and Physiology Work Package. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Murphy EJ, Cavanagh RD, Johnston NM, Hofmann EE, editors. Integrating Climate and Ecosystem Dynamics (ICED): Report of the Southern Ocean Food Web Modelling Workshop, 16–18 April 2008, Virginia, USA. 2010. Available: http://www.iced.ac.uk/documents/Foodwebs Modelling Report-final.pdf. Accessed 12/12/2010.
- 2.Nicol S. Antarctic Krill - Changing Perceptions of Its Role in the Antarctic Ecosystem. 1994. pp. 144–166. In: Antarctic Science - Global Concerns. Edited by Hempel G. Berlin 33: Springer-Verlag Berlin.
- 3.Macaulay MC, English TS, Mathisen OA. Acoustic Characterization of Swarms of Antarctic Krill (Euphausia superba) from Elephant Island and Bransfield Strait. J Crustac Biol. 1984;4:16–44. [Google Scholar]
- 4.Tarling GA, Klevjer T, Fielding S, Watkins J, Atkinson A, et al. Variability and predictability of Antarctic krill swarm structure. Deep-Sea Res I. 2009;56:1994–2012. [Google Scholar]
- 5.Commission for the Conservation of Antarctic Marine Living Resources. Available: www.ccamlr.org/. Accessed 12/12/2010.
- 6.FAO. Review of the state of world marine fishery resources. 2005. FAO Fisheries Technical Paper. No. 457. Rome.
- 7.Everson I. Krill biology, ecology and fisheries. Oxford: Blackwell Science; 2000. [Google Scholar]
- 8.Siegel V. Distribution and population dynamics of Euphausia superba: summary of recent findings. Polar Biol. 2005;29:1–22. [Google Scholar]
- 9.Trathan PN, Agnew D. Climate change and the Antarctic marine ecosystem: an essay on management implications. Antarctic Sci. 2010;22:387–398. [Google Scholar]
- 10.Atkinson A, Siegel V, Pakhomov E, Rothery P. Long-term decline in krill stock and increase in salps within the Southern Ocean. Nature. 2004;432:100–103. doi: 10.1038/nature02996. [DOI] [PubMed] [Google Scholar]
- 11.Atkinson A, Siegel V, Pakhomov EA, Rothery P, Loeb V, et al. Oceanic circumpolar habitats of Antarctic krill. Mar Ecol Prog Ser. 2008;362:1–23. [Google Scholar]
- 12.Whitehouse MJ, Meredith MP, Rothery P, Atkinson A, Ward P, et al. Rapid warming of the ocean around South Georgia, Southern Ocean, during the 20th century: Forcings, characteristics and implications for lower trophic levels. Deep-Sea Res I. 2008;55:1218–1228. [Google Scholar]
- 13.IPCC: Climate change (2007) synthesis report. Contribution of work groups I, II and III to the 4th Assessment Report of the Intergovernmental Panel on Climate Change. Edited by Pachauri RK, Reisinger A. IPCC, Geneva, Switzerland: IPCC.
- 14.Gille ST. Warming of the Southern Ocean since the 1950s. Science. 2002;295:1275–1277. doi: 10.1126/science.1065863. [DOI] [PubMed] [Google Scholar]
- 15.Meredith MP, King JC. Rapid climate change in the ocean west of the Antarctic Peninsula during the second half of the 20th century. Geophys Res Lett. 2005;32(19)) [Google Scholar]
- 16.Parkinson CI. Trends in the length of the Southern Ocean sea-ice season, 1979–1999. Ann Glaciol. 2002;34:435–440. [Google Scholar]
- 17.Reid K, Croxall JP. Environmental response of upper trophic-level predators reveals a system change in an Antarctic marine ecosystem. Proc Roy Soc B. 2001;268:377–384. doi: 10.1098/rspb.2000.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forcada J, Trathan PN, Reid K, Murphy EJ. The effects of global climate variability in pup production of Antarctic fur seals. Ecol. 2005;86:2408–2417. [Google Scholar]
- 19.Forcada J, Trathan PN, Reid K, Murphy EJ, Croxall JP. Contrasting population changes in sympatric penguin species in association with climate warming. Glob Chang Biol. 2006;12:411–423. [Google Scholar]
- 20.Trathan PN, Forcada J, Murphy EJ. Environmental forcing and Southern Ocean marine predator populations: effects of climate change and variability. Phil Trans R Soc B. 2007;362:2351–2365. doi: 10.1098/rstb.2006.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trathan PN, Green C, Tanton J, Peat H, Poncet J, et al. Foraging dynamics of macaroni penguins Eudyptes chrysolophus at South Georgia during brood-guard. Mar Ecol Prog Ser. 2006;323:239–251. [Google Scholar]
- 22.Quetin LB, Ross RM. Life under Antarctic Pack Ice: A Krill Perspective. 2009. pp. 285–298. In: Smithsonian at the Poles: Contributions to International Polar Year Science - a Smithsonian Contribution to Knowledge. Edited by Krupnik I, Lang MA, Miller SE.
- 23.Kawaguchi S, Nicol S, Press AJ. Direct effects of climate change on the Antarctic krill fishery. Fisheries Manag Ecol. 2009;16:424–427. [Google Scholar]
- 24.Quetin LB, Ross RM. Behavioral and Physiological-Characteristics of the Antarctic Krill, Euphausia superba. Am Zool. 1991;31:49–63. [Google Scholar]
- 25.Teschke M, Kawaguchi S, Meyer B. Effects of simulated light regimes on maturity and body composition of Antarctic krill, Euphausia superba. Mar Biol. 2008;154:315–324. [Google Scholar]
- 26.Buchholz F. Experiments on the physiology of Southern and Northern krill, Euphausia superba and Meganyctiphanes norvegica, with emphasis on moult and growth - A review. Mar Freshw Behav Phy. 2003;36:229–247. [Google Scholar]
- 27.Clark MS, Geissler P, Waller C, Fraser KPP, Barnes DKA, et al. Low heat shock thresholds in wild Antarctic inter-tidal limpets (Nacella concinna). Cell Stress Chaperones. 2008;13:51–58. doi: 10.1007/s12192-008-0015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark MS, Peck LS. Triggers of the HSP70 stress response: environmental responses and laboratory manipulation in an Antarctic marine invertebrate (Nacella concinna). Cell Stress Chaperones. 2009;14:649–660. doi: 10.1007/s12192-009-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke A. Some observations on krill (Euphasia superba Dana) maintained alive in the laboratory. Br Antarct Bull. 1976;43:111–118. [Google Scholar]
- 30.Ritz DA, Cromer L, Swadling KM, Nicol S, Osborn J. Heart rate as a measure of stress in Antarctic krill, Euphausia superba. J Mar Biol Assoc UK. 2003;83:329–330. [Google Scholar]
- 31.Truebano M, Burns G, Thorne MAS, Hillyard, Peck LS, et al. Transcriptional response to heat stress in the Antarctic bivalve Laternula elliptica. J Exp Mar Biol Ecol. 2010;391:65–72. [Google Scholar]
- 32.De Pitta C, Bertolucci C, Mazzotta GM, Bernante F, Rizzo G, et al. Systematic sequencing of mRNA from the Antarctic krill (Euphausia superba) and first tissue specific transcriptional signature. BMC Genomics. 2008;9:45. doi: 10.1186/1471-2164-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seear P, Tarling GA, Teschke M, Meyer B, Thorne MAS, et al. Effects of simulated light regimes on gene expression in Antarctic krill (Euphausia superba Dana). J Exp Mar Biol Ecol. 2009;381:57–64. [Google Scholar]
- 34. Newbler assembly software available: www.454.com. Accessed 12/12/2010.
- 35.Meyer E, Aglyamova GV, Wang S, Buchanan-Carter J, Abrego D, et al. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics. 2009;10:219. doi: 10.1186/1471-2164-10-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Craft JA, Gilbert JA, Temperton B, Dempsey KE, Ashelford K, et al. Pyrosequencing of Mytilus galloprovincialis cDNAs: Tissue-Specific Expression Patterns. Plos One. 2009;5(1):e8875. doi: 10.1371/journal.pone.0008875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark MS, Thorne MAS, Vieira FA, Cardoso JCR, Power DM, et al. Insights into shell deposition in the Antarctic bivalve Laternula elliptica: gene discovery in the mantle transcriptome using 454 pyrosequencing. BMC Genomics. 2010;11:362. doi: 10.1186/1471-2164-11-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang JQ, Li FH, Wang ZH, Zhang XJ, Zhou Q, et al. Cloning, expression and identification of ferritin from Chinese shrimp, Fenneropenaeus chinensis. J Biotechnol. 2006;125:173–184. doi: 10.1016/j.jbiotec.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Romo-Figueroa MG, Vargas-Requena C, Sotelo-Mundoa RR, Vargas-Albores F, Higuera-Ciapara I, et al. Molecular cloning of a beta-glucan pattern-recognition lipoprotein from the white shrimp Penaeus (Litopenaeus) vannamei: correlations between the deduced amino acid sequence and the native protein structure. Dev Comp Immunol. 2004;28:713–726. doi: 10.1016/j.dci.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Cheng WT, Tung YH, Chiou TT, Chen JC. Cloning and characterisation of mitochondrial manganese superoxide dismutase (mtMnSOD) from the giant freshwater prawn Macrobrachium rosenbergii. Fish Shellfish Immunol. 2006;21:453–466. doi: 10.1016/j.fsi.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Cheng WT, Tung YH, Liu CH, Chen JC. Molecular cloning and characterisation of copper/zinc superoxide dismutase (Cu,Zn-SOD) from the giant freshwater prawn Macrobrachium rosenbergii. Fish Shellfish Immunol. 2006;21:102–112. doi: 10.1016/j.fsi.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 42.Cheng WT, Tung YH, Liu CH, Chen JC. Molecular cloning and characterisation of cytosolic manganese superoxide dismutase (cytMn-SOD) from the giant freshwater prawn Macrobrachium rosenbergii. Fish Shellfish Immunol. 2006;20:438–449. doi: 10.1016/j.fsi.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 43.Machida RJ, Miya MU, Yamauchi MM, Nishida M, Nishida S. Organization of the mitochondrial genome of Antarctic krill Euphausia superba (Crustacea : Malacostraca). Mar Biotechnol. 2004;6:238–250. doi: 10.1007/s10126-003-0016-6. [DOI] [PubMed] [Google Scholar]
- 44.Shen X, Wang HQ, Ren JF, Tian M, Wang MX. The mitochondrial genome of Euphausia superba (Prydz Bay) (Crustacea: Malacostraca: Euphausiacea) reveals a novel gene arrangement and potential molecular markers. Mol Biol Rep. 2010;37:771–784. doi: 10.1007/s11033-009-9602-7. [DOI] [PubMed] [Google Scholar]
- 45.Vargas-Albores F, Yepiz-Plascencia G. Beta glucan binding protein and its role in shrimp immune response. Aquaculture. 2000;191:13–21. [Google Scholar]
- 46.Yepiz-Plascencia G, Vargas-Albores F, Jimenez-Vega F, Ruiz-Verdugo LM, Romo-Figueroa G. Shrimp plasma HDL and beta-glucan binding protein (BGBP): comparison of biochemical characteristics. Comp Biochem Physiol B. 1998;121:309–314. doi: 10.1016/s0305-0491(98)10104-9. [DOI] [PubMed] [Google Scholar]
- 47.Clarke A. Seasonality in the Antarctic Marine-Environment. Comp Biochem Physiol B. 1988;90:461–473. [Google Scholar]
- 48.Peck LS, Convey P, Barnes DKA. Environmental constraints on life histories in Antarctic ecosystems: tempos, timings and predictability. Biol Rev. 2006;81:75–109. doi: 10.1017/S1464793105006871. [DOI] [PubMed] [Google Scholar]
- 49.Peck LS, Barnes DKA, Cook AJ, Fleming AH, Clarke A. Negative feedback in the cold: ice produces new carbon sinks in the Antarctic. Global Change Biol. 2009;16:2614–2623. [Google Scholar]
- 50.Obermuller B, Peck LS, Barnes DKA, Morley S. Seasonal physiology of Antarctic marine benthic predators and scavengers. Mar Ecol Prog Ser. 2010;415:109–126. [Google Scholar]
- 51.Hagen W. Reproductive strategies and energetic adaptations of polar zooplankton. Invertebr Reprod Dev. 1999;36:25–34. [Google Scholar]
- 52.Hagen W, Kattner G, Terbruggen A, Van Vleet ES. Lipid metabolism of the Antarctic krill Euphausia superba and its ecological implications. Mar Biol. 2001;139:95–104. [Google Scholar]
- 53.Falk-Petersen S, Hagen W, Kattner G, Clarke A, Sargent J. Lipids, trophic relationships, and biodiversity in Arctic and Antarctic krill. Can J Fish Aquatic Sci. 2000;57:178–191. [Google Scholar]
- 54.Batlle M, Marsellach F-X, Huertas D, Azorin F. Drosophila vigilin, DDP1, localises to the cytoplasm and associates to the rough endoplasmic reticulum. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbagrm.2010.10.005. doi: 10.1016/j.bbagrm.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 55.Dodson RE, Shapiro DJ. Vigilin, a ubiquitous protein with a 14 K homology domains, is the estrogen-inducible vitellogenin mRNA 3′-untranslated region-binding protein. J Biol Chem. 1997;272:12249–12252. doi: 10.1074/jbc.272.19.12249. [DOI] [PubMed] [Google Scholar]
- 56.Buchholz F. Molt cycle and growth of Antarctic Krill Euphausia superba in the laboratory. Mar Ecol Prog Ser. 1991;69:217–229. [Google Scholar]
- 57.Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, et al. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14:105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Ann Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 59.Hartl FU, Hartl-Meyer M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 60.Parsell DA, Lindquist S. The Function of Heat-Shock Proteins in Stress Tolerance - Degradation and Reactivation of Damaged Proteins. Ann Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 61.Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 62.Fink AL. Chaperone-mediated protein folding. Physiol Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- 63.Place S, Hofmann G. Constitutive expression of a stress-inducible heat shock protein gene, hsp70, in phylogenetically distant Antarctic fish. Polar Biol. 2005;28:261–267. [Google Scholar]
- 64.Place SP, Zippay ML, Hofmann GE. Constitutive roles for inducible genes: evidence for the alteration in expression of the inducible hsp70 gene in Antarctic notothenioid fishes. Am J Physiol Regul Integr Comp Physiol. 2004;287:R429–R436. doi: 10.1152/ajpregu.00223.2004. [DOI] [PubMed] [Google Scholar]
- 65.Clark MS, Fraser KPP, Peck LS. Antarctic marine molluscs do have an HSP70 heat shock response. Cell Stress Chaperones. 2008;13:39–49. doi: 10.1007/s12192-008-0014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clark MS, Fraser KPP, Burns G, Peck LS. The HSP70 heat shock response in the Antarctic fish Harpagifer antarcticus. Polar Biol. 2008;31:171–180. [Google Scholar]
- 67.Hofmann GE, Buckley BA, Airaksinen S, Keen JE, Somero GN. Heat-shock protein expression is absent in the Antarctic fish Trematomus bernacchii (family Nototheniidae). J Exp Biol. 2000;203:2331–2339. doi: 10.1242/jeb.203.15.2331. [DOI] [PubMed] [Google Scholar]
- 68.Thorne MAS, Burns G, Fraser KPP, Hillyard G, Clark MS. Transcription profiling of acute temperature stress in the Antarctic plunderfish Harpagifer antarcticus. Mar Gen. 2010;3:35–44. doi: 10.1016/j.margen.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 69.Clark MS, Fraser KPP, Peck LS. Lack of an HSP70 heat shock response in two Antarctic marine invertebrates. Polar Biol. 2008;31:1059–1065. [Google Scholar]
- 70.Engman DM, Dragon EA, Donelson JE. Human humoral immunity to HSP70 during Trypanosoma-cruzi infection. J Immunol. 1990;144:3987–3991. [PubMed] [Google Scholar]
- 71.Vayssier M, Le Guerhier F, Fabien JF, Philippe H, Vallet C, et al. Cloning and analysis of a Trinchinella britovi gene encoding a cytoplasmic heat shock protein of 72 kDa. Parasitol. 1999;119:81–93. doi: 10.1017/s0031182099004461. [DOI] [PubMed] [Google Scholar]
- 72.Clark MS, Peck LS. HSP70 heat shock proteins and environmental stress in Antarctic marine organisms: A mini-review. Mar Gen. 2009;2:11–18. doi: 10.1016/j.margen.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 73.Ravaux J, Toullec JY, Leger N, Lopez P, Gaill F, et al. First hsp70 from two hydrothermal vent shrimps, Mirocaris fortunata and Rimicaris exoculata: Characterization and sequence analysis. Gene. 2007;386:162–172. doi: 10.1016/j.gene.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 74.Cottin D, Shillito B, Chertemps T, Thatje S, Leger N, et al. Comparison of heat-shock responses between the hydrothermal vent shrimp Rimicaris exoculata and the related coastal shrimp Palaemonetes varians. J Expt Mar Biol Ecol. 2010;393:9–16. [Google Scholar]
- 75.Liu J, Yang WJ, Zhu XJ, Karouna-Renier NK, Rao RK. Molecular cloning and expression of two HSP70 genes in the prawn, Macrobrachium rosenbergii. Cell Stress Chaperones. 2004;9:313–323. doi: 10.1379/CSC-40R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gupta RS. Phylogenetic analysis of the 90 kD Heat-Shock Family of protein sequences and an examination of the relationship among Animals, Plants, and Fungi species. Mol Biol Evol. 1995;12:1063–1073. doi: 10.1093/oxfordjournals.molbev.a040281. [DOI] [PubMed] [Google Scholar]
- 77.Pantzartzi CN, Kourtidis A, Drosopoulou E, Yiangou M, Scouras ZG. Isolation and characterization of two cytoplasmic hsp90s from Mytilus galloprovincialis (Mollusca: Bivalvia) that contain a complex promoter with a p53 binding site. Gene. 2009;431:47–54. doi: 10.1016/j.gene.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 78.Wu LT, Chu KH. Characterization of heat shock protein 90 in the shrimp Metapenaeus ensis: evidence for its role in the regulation of vitellogenin synthesis. Mol Repro Dev. 2008;75:952–959. doi: 10.1002/mrd.20817. [DOI] [PubMed] [Google Scholar]
- 79.Jiang SG, Qiu LH, Zhou FL, Huang JH, Guo YH, et al. Molecular cloning and expression analysis of a heat shock protein (Hsp90) gene from black tiger shrimp (Penaeus monodon). Mol Biol Rep. 2009;36:127–134. doi: 10.1007/s11033-007-9160-9. [DOI] [PubMed] [Google Scholar]
- 80.Zhang XY, Zhang MZ, Zheng CJ, Liu J, Hu HJ. Identification of two hsp90 genes from the marine crab, Portunus trituberculatus and their specific expression profiles under different environmental conditions. Comp Biochem Physiol. 2009;150:465–473. doi: 10.1016/j.cbpc.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 81.Schram FR, Hof CHJ. Fossils and the interrelationships of major Crustacean groups. In: Edgecombe GD, editor. Arthropod Fossils and Phylogeny. New York: Columbia University Press; 1998. pp. 233–302. [Google Scholar]
- 82.Jarman SN, Elliott NG, Nicol S, McMinn A. Molecular phylogenetics of circumglobal Euphausia species (Euphausiacea : Crustacea). Can J Fish Aquatic Sci. 2000;57:51–58. [Google Scholar]
- 83.Felts SJ, Owen BAL, Nguyen P, Trepel J, Donner DB, et al. The hsp90-related protein TRAP1 is a mitochondrial protein with distinct functional properties. J Biol Chem. 2000;275:3305–3312. doi: 10.1074/jbc.275.5.3305. [DOI] [PubMed] [Google Scholar]
- 84.Yamaguchi H, Morita T, Amagai A, Maeda Y. Changes in spatial and temporal localization of Dictyostelium homologues of TRAP1 and GRP94 revealed by immunoelectron microscopy. Exp Cell Res. 2005;303:415–424. doi: 10.1016/j.yexcr.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 85.Costantino E, Maddalena F, Calise S, Piscazzi A, Tirino V, et al. TRAP1, a novel mitochondrial chaperone responsible for multi-drug resistance and protection from apoptotis in human colorectal carcinoma cells. Cancer Lett. 2009;279:39–46. doi: 10.1016/j.canlet.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 86.Martin J, Gruber M, Lupas AN. Coiled coils meet the chaperone world. Trend Biochem Sci. 2004;29:455–458. doi: 10.1016/j.tibs.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 87.Thulasiraman V, Yang CF, Frydman J. In vivo newly translated polypeptides are sequestered in a protected folding environment. Embo J. 1999;18:85–95. doi: 10.1093/emboj/18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cunningham LA, Kahn RA. Cofactor D functions as a centrosomal protein and is required for the recruitment of the gamma-tubulin ring complex at centrosomes and organization of the mitotic spindle. J Biol Chem. 2008;283:7155–7165. doi: 10.1074/jbc.M706753200. [DOI] [PubMed] [Google Scholar]
- 89.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cellular and Molecular Life Sciences. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ferrari DM, Soling HD. The protein disulphide-isomerase family: unravelling a string of folds. Biochem J. 1999;339:1–10. [PMC free article] [PubMed] [Google Scholar]
- 92.Michalak M, Parker JMR, Opas M. Ca2+ signaling and calcium binding chaperones of the endoplasmic reticulum. Cell Calcium. 2002;32:269–278. doi: 10.1016/s0143416002001884. [DOI] [PubMed] [Google Scholar]
- 93.Goswami AV, Chittoor B, D'Silva P. Understanding the Functional Interplay between Mammalian Mitochondrial Hsp70 Chaperone Machine Components. J Biol Chem. 2010;285:19472–19482. doi: 10.1074/jbc.M110.105957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu Y, Liu W, Song XD, Zuo J. Effect of GRP75/mthsp70/PBP74/mortalin overexpression on intracellular ATP level, mitochondrial membrane potential and ROS accumulation following glucose deprivation in PC12 cells. Mol Cell Biochem. 2005;268:45–51. doi: 10.1007/s11010-005-2996-1. [DOI] [PubMed] [Google Scholar]
- 95.Gross M. Emergency services: A bird's eye perspective on the many different functions of stress proteins. Curr Protein Pept Sci. 2004;5:213–223. doi: 10.2174/1389203043379684. [DOI] [PubMed] [Google Scholar]
- 96.Mayer MP, Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Korcsmáros T, Kovács IA, Szalay MS, Csermely P. Molecular chaperones: The modular evolution of cellular networks. J Biosci. 2007;32:441–446. doi: 10.1007/s12038-007-0043-y. [DOI] [PubMed] [Google Scholar]
- 98.Abele D, Puntarulo S. Formation of reactive species and induction of antioxidant defence systems in polar and temperate marine invertebrates and fish. Comp Biochem Physiol. 2004;138:405–415. doi: 10.1016/j.cbpb.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 99.Philipp E, Brey T, Portner HO, Abele D. Chronological and physiological ageing in a polar and a temperate mud clam. Mech Age Dev. 2005;126:598–609. doi: 10.1016/j.mad.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 100.Hintze KJ, Theil EC. DNA and mRNA elements with complementary responses to hemin, antioxidant inducers, and iron control ferritin-L expression. Proc Natl Acad Sci USA. 2005;102:15048–15052. doi: 10.1073/pnas.0505148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang TS, Law JH, Soderhall K. Purification and cDNA cloning of ferritin from the hepatopancreas of the freshwater crayfish Pacifastacus leniusculus. Eur J Biochem. 1996;236:450–456. doi: 10.1111/j.1432-1033.1996.00450.x. [DOI] [PubMed] [Google Scholar]
- 102.Larade K, Storey KB. Accumulation and translation of ferritin heavy chain transcripts following anoxia exposure in a marine invertebrate. J Exp Biol. 2004;207:1353–1360. doi: 10.1242/jeb.00872. [DOI] [PubMed] [Google Scholar]
- 103.Beck G, Ellis TW, Habicht GS, Schluter SF, Marchalonis JJ. Evolution of the acute phase response: iron release by echinoderm (Asterias forbesi) coelomocytes, and cloning of an echinoderm ferritin molecule. Dev Comp Immunol. 2002;26:11–26. doi: 10.1016/s0145-305x(01)00051-9. [DOI] [PubMed] [Google Scholar]
- 104.Nebert DW, Vasiliou V. Analysis of the glutathione-S-transferase (GST) family. Hum Genomics. 2004;1:460–464. doi: 10.1186/1479-7364-1-6-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ding YC, Ortelli F, Rossiter LC, Hemingway J, Ranson H. The Anopheles gambiae glutathione transferase supergene family: annotation, phylogeny and expression profiles. BMC Genomics. 2003;4:35. doi: 10.1186/1471-2164-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dixon DP, Lapthorn A, Edwards R. Plant glutathione transferases. Genome Biol. 2002;3(3):reviews3004.1-3004.10. doi: 10.1186/gb-2002-3-3-reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao D, Chen L, Qin L, Zhang H, Wu P, et al. A delta class glutathione transferase for the Chinese mitten crab Eriorcheir sinensis: cDNA cloning, characterisation and mRNA expression. Fish Shellfish Immunol. 2010;21:615–622. doi: 10.1016/j.fsi.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 108.Agianian B, Tucker PA, Schouten A, Leonard K, Bullard B, et al. Structure of a Drosophila sigma class glutathione S-transferase reveals a novel active site topography suited for lipid peroxidation products. J Mol Biol. 2003;326:151–165. doi: 10.1016/s0022-2836(02)01327-x. [DOI] [PubMed] [Google Scholar]
- 109.Sawicki R, Singh SP, Mondal AK, Benes H, Zimniak P. Cloning, expression and biochemical characterization of one Epsilon-class (GST-3) and ten Delta-class (GST-1) glutathione S-transferases from Drosophila melanogaster, and identification of additional nine members of the Epsilon class. Biochem J. 2003;370:661–669. doi: 10.1042/BJ20021287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lin YC, Vaseeharan B, Chen JC. Identification of the extracellular copper-zinc superoxide dismutase (ecCuZnSOD) gene of the mud crab Scylla serrata and its expression following beta-glucan and peptidoglycan injections. Mol Immunol. 2008;45:1346–1355. doi: 10.1016/j.molimm.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 111.Marklund SL. Human Copper-Containing Superoxide-Dismutase of High Molecular-Weight. Proc Natl Acad Sci USA. 1982;79:7634–7638. doi: 10.1073/pnas.79.24.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jensen LT, Culotta VC. Activation of CuZn superoxide dismutases from Caenorhabditis elegans does not require the copper chaperone CCS. J Biol Chem. 2005;280:41373–41379. doi: 10.1074/jbc.M509142200. [DOI] [PubMed] [Google Scholar]
- 113.Schinina ME, Barra D, Bossa F, Calabrese L, Montesano L, et al. Primary structure from amino acid and cDNA sequences of 2 Cu,Zn superoxide-dismutase variants from Xenopus laevis. Arch Biochem Biophys. 1989;272:507–515. doi: 10.1016/0003-9861(89)90246-4. [DOI] [PubMed] [Google Scholar]
- 114.Fraser KPP, Clarke A, Peck LS. Growth in the slow lane: protein metabolism in the Antarctic limpet Nacella concinna (Strebel 1908). J Exp Biol. 2007;210:2691–2699. doi: 10.1242/jeb.003715. [DOI] [PubMed] [Google Scholar]
- 115.Chen LB, DeVries AL, Cheng CHC. Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notothenioid fish. Proc Natl Acad Sci USA. 1997;94:3811–3816. doi: 10.1073/pnas.94.8.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen ZZ, Cheng CHC, Zhang JF, Cao LX, Chen L, et al. Transcriptomic and genomic evolution under constant cold in Antarctic notothenioid fish. Proc Natl Acad Sci USA. 2008;105:12944–12949. doi: 10.1073/pnas.0802432105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Privalov PL. Cold denaturation of proteins. Crit Rev Biochem Mol Biol. 1990;25:281–305. doi: 10.3109/10409239009090612. [DOI] [PubMed] [Google Scholar]
- 118.Sørensen JG, Loeschcke V. Studying stress responses in the post-genomic era: its ecological and evolutionary role. J Biosci. 2007;32:447–456. doi: 10.1007/s12038-007-0044-x. [DOI] [PubMed] [Google Scholar]
- 119.Mayer C. Phobos 3.3.10. 2006–2009, www.rub.de/spezzoo/cm/cm_phobos.htm.
- 120.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucl Acids Res. 2007;35:D21–25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rice P, Longden I, Bleasby A. EMBOSS, the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 123.Higgins JD, Gibson TJ. ClustalW – improving the sensitivity of progressive multiple sequence alignment through weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.CLC MainWorkbench software. Available: http://www.clcbio.com/. Accessed 12/12/2010.
- 125.CBS prediction server, prediction of N-linked glycosylation sites in human proteins. Available: http://www.cbs.dtu.dk/services/NetNGlyc/. Accessed 12/12/2010.
- 126.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 127.Neilsen H, Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. In: Glasgow JI, Littlejohn TG, Major F, Lathrop RH, Sankoff D, Sensen C, editors. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology (ISMB-98): June 28–July 1, 1998; Montréal, Québec, Canada. AAAI Press; 1998. pp. 122–130. [PubMed] [Google Scholar]
- 128.CBS prediction server. On-line prediction servers. Available: http://www.cbs.dtu.dk/services/. Accessed 12/12/2010.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Putative translations and amino acid alignment of the 9 E. superba contigs with sequence similarity to HSP70.
(PDF)
Alignment of the deduced amino acid sequences of the E. superba contigs 02253 (full length) and 06573 (partial) with those of other Eucrustacea. Sequence accession numbers are: Balanus amphitrite: Q86MC3; Artemia sanfranciscana: Q95V47; Moina macropoda: ACB11341; Daphnia magna: ACB11340; Tigriopus japonicus: B8PT12; Pachygrapsus marmoratus: ABA02164; Homarus americanus: ABA02165; Penaeus monodon: AAQ05768; Metapenaeus ensis: Q1HGN3; Litopenaeus vannamei: AAT46566; Rimicaris exoculata-3: D2SPE2; Rimicaris exoculata-2: ACL52279; Rimicaris exoculata-1: ABF85673; Palaemonetes varians: ACR77532; Mirocaris fortunata: A1XQQ5; Macrobrachium rosenbergii: Q6S4R6; Portunus trituberculatus: D2DWR3; Portunus sanguinolentus: A8KCI1; Eriocher sinensis: B5AMI7; Calinectes sapidus: Q194W6; Scylla paramamosain: B3VKG9.
(PDF)
Putative translations and amino acid alignment of the 6 E. superba contigs with sequence similarity to HSP90.
(PDF)
Amino acid alignment of HSP90 genes from the Panarthropoda. Sequence accession numbers are: Ixodes scapularis: XM_002414763; Apis mellifera: FJ713701; Macrocentrus cingulum: EU570066; Drosophila melanogaster: NM_079175; Mamestra brassicae: AB251894; Chilo suppressalis: AB206477; Tigriopus japonicus: EU831278; Chiromantes haematocheir: AY528900.1; Eriocheir sinensis: EU809924; Portunus trituberculatus: FJ392027; Fenneropenaeus chinensis: EF032650; Penaeus monodon: FJ855436; Metapenaeus ensis: EF470246; Euphausia superba: contig00022. Only the ptHSP90-1 and EusHSP90-1 are used in this alignment.
(PDF)
Amino acid alignment of TRAP1 sequences. These are from a number of invertebrates and vertebrates with the EusHSP90-1 and contig03026. Sequence accession numbers are: Aedes aegypti TRAP1: AAD29307; Culex quinquefasciatus TRAP1: XM_001861228; Drosophila melanogaster TRAP1: AAD29307; Pediculus humanus HSP75: XP_002425720; Danio rerio TRAP1: AAI4468; Apis mellifera TRAP1: XP_623366; Homo sapiens TRAP1: Q12931.
(PDF)
Amino acid alignment of E. superba additional putative HSP70 family members with HSP105 and APG HSP70 family members. Accession numbers: Tigriopus japonicus HSP105: ACA03526; Apis mellifera HSP70Cb: XP_623199.1; Nasonnia vitripennis: XP_001607146; Pediculus humanus HSP105: XP_002431004; Xenopus laevis HSP105: NP_001086692; Callithrix jacchus HSP105-2: XP_002748999; Callithrix jacchus HSP70-4: XP_002744640; Mus musculus HSP70-4-APG2: Q61316; Mus musculus HSP70-4-APG1: P48722; Homo sapiens HSP70-4: ABM69040.
(PDF)
Microsatellite repeats found in excess of 7 copies per repeat unit in E. superba data.
(XLS)
Variant nucleotides (SNPs/INDELS) found in E.superba data.
(XLS)