Abstract
KCNQ1 encodes KCNQ1, which belongs to a family of voltage-dependent K+ ion channel proteins. KCNQ1 associates with a regulatory subunit, KCNE1, to produce the cardiac repolarizing current, IKs. Loss-of-function mutations in the human KCNQ1 gene have been linked to Jervell and Lange–Nielsen Syndrome (JLNS), a disorder characterized by profound bilateral deafness and a cardiac phenotype. To generate a mouse model for JLNS, we created a line of transgenic mice that have a targeted disruption in the Kcnq1 gene. Behavioral analysis revealed that the Kcnq1−/− mice are deaf and exhibit a shaker/waltzer phenotype. Histological analysis of the inner ear structures of Kcnq1−/− mice revealed gross morphological anomalies because of the drastic reduction in the volume of endolymph. ECGs recorded from Kcnq1−/− mice demonstrated abnormal T- and P-wave morphologies and prolongation of the QT and JT intervals when measured in vivo, but not in isolated hearts. These changes are indicative of cardiac repolarization defects that appear to be induced by extracardiac signals. Together, these data suggest that Kcnq1−/− mice are a potentially valuable animal model of JLNS.
Long QT syndrome (LQTS) is a disorder of ventricular repolarization, characterized by ECGs with prolonged QT intervals and T-wave abnormalities. LQTS can result in syncope, seizure, and sudden death as a result of torsades de pointes cardiac arrhythmias (1, 2). Two inherited forms of LQTS have been reported: Romano–Ward syndrome (RWS) (3, 4) and Jervell and Lange–Nielsen syndrome (JLNS) (5). RWS is inherited as an autosomal dominant trait of variable penetrance. JLNS is relatively rare, transmitted as a recessive trait, and is associated with deafness in addition to a long QT interval on the ECG.
At least six loci are associated with inherited LQTS, and five of the genes have been identified (6–10). Mutations in the KCNQ1 (also called KVLQT1) gene at the LQT1 locus (11p15.5) represent the most common form of LQTS (11). KCNQ1 encodes a channel-forming subunit. When expressed alone in heterologous cells, KCNQ1 (also called KvLQT1) subunits give rise to a voltage-gated potassium channel with properties unlike any endogenous current characterized to date. However, when KCNQ1 is coexpressed with KCNE1 (also called MinK or IsK), their subunits coassemble, giving rise to a conductance that recapitulates the slow component, IKs, of the delayed rectifier K+ current (12, 13). Mutations in both KCNQ1 and KCNE1 are associated with RWS and JLNS.
JLNS patients bear a profound bilateral deafness from birth that is shown to be caused by the absence of a specialized fluid, endolymph, which bathes the hair cells. The K+ concentration in endolymph is very high, and the movement of potassium into hair cells on their stimulation is essential to the transduction of sound into neural signals. Recycling of K+ from the hair cells back to the endolymph, thereby maintaining the appropriate high levels of K+ in that fluid, is crucial for normal ear function (for review, see ref. 14). In fact, the proteins of several genes associated with deafness appear to play a direct role in this recycling pathway (15–18). Ultimately, K+ are believed to be secreted into the endolymph by distinct regions of the labyrinth epithelium, namely the stria vascularis of the cochlea and the dark cells of the vestibular system. IKs is a candidate for endolymph secretion, as Kcnq1 and Kcne1 are both expressed in the marginal cells of the stria vascularis (19, 20). In addition, the vestibular dark cells have been shown to produce an IKs-like current at their apical membranes (21). A requirement for IKs is further supported by inner ear defects associated with JLNS patients (20, 22) and the phenotype of the Kcne1 knockout mice (23).
Although Kcne1−/− mice exhibit inner ear defects very similar to those seen in patients diagnosed with JLNS, the cardiac phenotype associated with loss of Kcne1 activity in mice is less clear. Drici et al. (24) demonstrated the QT interval altered when a range of heart rates were analyzed. In contrast, Kupershmidt et al. (25) found no baseline ECG abnormalities in their independently generated transgenic Kcne1 knockout mice.
In this study, we describe the production and phenotype of mice with a targeted disruption of the Kcnq1 gene. The phenotypes of the Kcnq1−/− mice resemble patients with JLNS. These mice suffer from deafness and vestibular dysfunction because of profound morphological abnormalities of the inner ear. ECG analysis revealed that Kcnq1−/− mice exhibit altered cardiac repolarization in vivo. Thus, Kcnq1 is essential for both inner ear homeostasis and normal cardiac function in mice.
Methods
Generating Kcnq1 Mutant Mice.
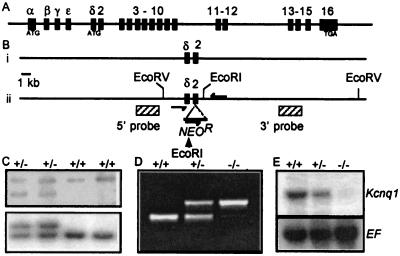
The targeting vector, pJV1, carries a 5.6-kb XbaI fragment containing 1 kb of sequences 5′ of exon 2 and 4.5 kb of sequences 3′ of the exon. Exon 2 itself is disrupted by the insertion of a PGK–Neomycin fusion. The vector also carries the diptheria toxin gene for negative selection. Forty-five micrograms of linearized vector DNA was electroporated into mouse embryonic stem cells derived from 129/Sv × 129/Sv-CP embryos (26). Geneticin-resistant colonies were analyzed by Southern blot. Cell lines correctly targeted on the 5′ end show a 12-kb EcoRI fragment in addition to the 13-kb EcoRI fragment specific to the wild-type chromosome. Clones correctly targeted on the 3′ end show an 11.5-kb EcoRV fragment in addition to the 10.2-kb band generated by the wild-type chromosome. Of 179 clones tested, 15 showed patterns indicative of the appropriate recombination events. Chimeric founders from two independent cell lines were bred to C57BL/6 females, and then heterozygous progeny from each line were interbred to generate Kcnq1+/+, Kcnq1+/− and Kcnq1−/− animals for evaluation. Genotyping of mice was done on genomic DNA by PCR using the following primers: forward primer 5′ CCAGGAGTGGGTGGTTCTAC 3′, reverse primer 5′GCCAGCACTAAAGATCTTGC 3′, Neo forward primer 5′ CGCTTCCTCGTGCTTTACG 3′. Amplification by using the forward and reverse primers gives a 240-bp product specific to the wild-type (+/+) allele. Amplification by using the Neo forward and reverse primers gives a 370-bp product specific to the null allele. Heterozygous (+/−) mice give both the 240- and 370-bp products.
RNA Analysis.
Tissues were homogenized in TRIZOL Reagent (Life Technologies, Grand Island, NY) by using a power homogenizer and RNA isolated according to Life Technologies protocols. Kcnq1 RNA was quantified by Northern blot as previously described (27).
Histological Analysis.
All animal heads were hemisectioned and fixed in 4% paraformaldehyde for 16 h. Animals between postnatal day 8 (P8) and P70 were euthanized, skinned, fixed, and then decalcified in RDO rapid decalcifier (Apex Engineering Products Corporation, Plainsfield, IL), according to the manufacturer's instructions. The tissue was embedded in paraffin, sectioned (7 μm), and counterstained with hematoxylin/eosin for histological analysis.
In Vivo ECG Recordings.
ECG measurements were obtained as previously described (28). Wild-type and Kcnq−/− mice of similar age (2–3 months) were sedated with 25 mg/kg diazepam via i.p injection and placed in a prone position in a heated and shielded box with all four extremities immersed in 3 M KCl-filled wells to reduce skin resistance. A custom-built ECG amplifier (Vibraspec, Bear Island, ME) with a high-frequency filter of 1,000 Hz and 2,000-fold gain was used to record two bipolar limb leads (leads I and II) for 5–7 min. Signals were digitized continuously at 1 kHz by using a custom-built data acquisition and analysis system (National Instruments, Austin, TX).
All ECG recordings were evaluated manually in a blind fashion. To minimize beat-to-beat variability, a signal-averaged ECG was generated from 30 sec of continuous recordings by using the R-wave peak as the fiduciary point (29). For every ECG parameter evaluated, measurements were taken from both leads, and the larger of the two values in either lead was consistently used for comparative analysis. P-wave duration was measured from the first deflection of the P-wave to the point where it rejoins the isoelectric line, whereas P-wave amplitude was measured as the difference between the maximum and minimum P-wave voltages. The area under the P-wave was measured by integrating the voltage over the duration of the P-wave. PR interval was measured from the beginning of the P-wave to the beginning of the QRS complex. QRS amplitude was measured as the difference between the maximum and minimum voltage peaks of the QRS complex, whereas QRS duration was measured from the first deflection of the Q-wave (or R-wave when the Q-wave was absent) and the end of the S-wave (defined as the point of minimum voltage in the terminal phase of the QRS complex). The QT interval was measured from the beginning of the QRS complex to the end of the T-wave, where the end of the T-wave was defined as the point where the T-wave merges with the isoelectric line. The JT interval was calculated by subtracting the QRS duration from the QT interval. The RR interval was taken as the mean RR interval over the 30-sec recording period, and the corrected QT interval (QTc) was calculated by using the following conversion formula: QTc = QT/SQRT(RR/1,000) (29). The T-wave area was measured by integrating the voltage between the end of the QRS complex (minimum QRS voltage) and the end of the T-wave. ECG parameters for one test animal were excluded from the data presented in Table 1 on the basis of Dixon's test for outliers.
Table 1.
Comparison of ECG parameters in Kcnq1+/+ and Kcnq1−/− mice
| Parameters | Kcnq1+/+ | Kcnq1−/− | P-value |
|---|---|---|---|
| RR, ms | 132 ± 18 | 128 ± 18 | 0.5 |
| Pamp, mV × 10−2 | 21 ± 5 | 29 ± 8 | <0.001 |
| P-wave duration, ms | 12 ± 1 | 13 ± 2 | 0.03 |
| P-wave area (V*ms × 10−1) | 13 ± 3 | 19 ± 6 | <0.001 |
| PR, ms | 37 ± 5 | 41 ± 4 | 0.016 |
| QRS duration, ms | 9 ± 1 | 10 ± 2 | 0.016 |
| QRS amplitude (mV × 10−2) | 148 ± 39 | 236 ± 65 | <0.001 |
| JT, ms | 60 ± 11 | 71 ± 11 | 0.006 |
| QT, ms | 69 ± 11 | 82 ± 11 | 0.003 |
| QTc, ms | 61 ± 11 | 72 ± 5 | <0.001 |
| T-wave area (V*ms × 10−1) | 71 ± 24 | 141 ± 55 | <0.001 |
All data are presented as mean ± SD. Sample sets of 16 were used for both Kcnq1−/− and Kcnq1+/+ mice. Data sets were compiled by using Student's t test.
At the termination of the ECG recordings, the mice were anesthetized with pentobarbital (50 μg/g, i.p.), weighed, and their hearts were removed via midsternal thoracotomy, rinsed with saline, and weighed. After measurement of lung and total heart weight, the atria were removed, the heart was blotted dry on filter paper (≈10 sec), and the “dry” ventricular weight was obtained.
Isolated Perfused Heart Preparation.
Mice were anesthetized with 25 mg/kg of i.p. pentobarbital. Hearts of 8 Kcnq1+/+ and 10 Kcnq1−/− mice were isolated and perfused in the Langendorff mode as described (30). Briefly, after a surgical level of anesthesia was confirmed, the heart was removed via midsternal thoracotomy. The aorta was then cannulated and the left atrium removed. Retrograde perfusion through the aorta was carried out at a constant perfusion pressure of 68 mmHg at 37°C. The flow of Thebesian veins was drained via a thin metal cannula (27 gauge) pierced through the apex of the left ventricle. To maintain a constant heart rate, the hearts were paced at a cycle length of 125 ms by using a bipolar tungsten electrode sutured onto the right atrium. The hearts were immersed in modified Tyrode's solution of 37°C and allowed to equilibrate for 15 min before ECG recordings were obtained. Krebs–Henseleit perfusion buffer containing (mM): NaCl (118), KCl (4.7), CaCl2 (2.0), MgSO4 (1.2), Na-EDTA (0.5), NaHCO3 (25), KH2PO4 (1.2), and glucose (11), was prepared at the time of the experiment and equilibrated with 95% O2 + 5% CO2 for at least 1 h before the experiment, yielding a pH of 7.4.
Volume-Conducted ECG and Monophasic Action Potential (MAP) Measurements.
The volume-conducted ECG and MAP technique was adapted from isolated rabbit heart (31). Action potential waveforms obtained with MAP electrodes are well correlated with action potential recordings obtained by the conventional microelectrode technique (32). Three Ag-AgCl electrodes were placed in a simulated Einthoven configuration at a distance of 1–2 mm from the epicardial surface. Bipolar ECG leads 1, 2, and 3 were recorded with DC-coupled differential bioamplifiers (EP Technologies, Mountain View, CA). A bipolar Ag-AgCl MAP contact electrode (tip diameter 0.25 mm) was custom built to accommodate the small mouse heart size. After stable ECG signals were obtained, the following action potential mapping protocol was carried out for each heart: first, MAP recordings were obtained from two to four epicardial sites near the base of the right and left ventricle. Great care was taken to record consistently from equivalent sites in each heart. Next, the MAP catheter was inserted into the left ventricle (LV) via the mitral valve, and MAP recordings were obtained from two to three endocardial sites of the LV septum. Finally, a small window was cut into the right ventricle (RV), and MAP recordings were obtained from two to three endocardial sites of the RV septum. MAPs were recorded with a commercially available MAP amplifier (EP Technologies). ECG and MAP signals were digitized at 2,000 samples/sec with a commercially available data acquisition system (PowerLab, A. D. Instruments, Milford, MA). ECG analysis was carried out as described for the in vivo ECG recordings. The duration of the monophasic action potential was measured at the level of 30%, 50%, 70%, and 90% repolarization [action potential duration (APD)30, APD50, APD70, APD90] and values from each site (RV epicardial, RV septum, LV epicardial, LV septum) averaged for each heart.
Results and Discussion
To inactivate Kcnq1, we generated mice with insertion of a neomycin cassette into exon 2 (Fig. 1). Exon 2 is the first exon common to all known isoforms. Heterozygous mice carrying this mutation were bred to obtain Kcnq1−/−, Kcnq1+/+, and Kcnq1+/− mice, as described in Methods. Northern analysis demonstrated that Kcnq1 mRNA was absent in the heart tissue of Kcnq1−/− mice and present at half dosage in Kcnq1+/− mice (Fig. 1E). Using the more sensitive reverse transcription–PCR analysis, we detected a Kcnq1/Neo fusion transcript. Sequence analysis of the transcript revealed that it was unlikely to make a functional protein product because of numerous stop codons contributed by Neo (data not shown). Kcnq1−/− mice were present at normal Mendelian frequency at weaning (20/77), and preliminary analyses did not indicate any increase in mortality in mice up to 4 months old. By P5, Kcnq1−/− mice had difficulty righting themselves when placed on their backs. The mice were examined at P20, and they exhibited hyperactivity characterized by rapid head bobbing and an intermittent bidirectional circling. This behavior is symptomatic of inner ear defects and is usually referred to as a shaker/waltzer phenotype (33). When dropped onto a surface, mutant mice landed on their backs, a behavior indicative of a vestibular defect (34). To ascertain auditory function of Kcnq1−/− mice, the Preyer reflex was tested on animals aged 4 weeks and older. Kcnq1−/− mice failed to demonstrate Preyer's reflex, whereas the response from Kcnq1+/− and wild-type mice was normal. Together, these results indicate that Kcnq1 expression is important for vestibular and auditory function in mice.
Figure 1.
Construction of the JV1 allele of Kcnq1, inserting the PGK–NeomycinR gene fusion into exon 2. (A) Cartoon description of the 300-kb mouse Kcnq1 gene. ATG denotes in-frame start codons for splice variant I (exon 1α) and splice variant II (exon 1δ); TGA in exon 16 represents translational stop site (43, 44). (B) Strategy for generating the JV1 allele. The wild-type (i) and mutant JV1 chromosomes (ii) are depicted. Filled rectangles indicate exons, and hatched rectangles in (ii) indicate probes used to identify mutant cell lines. (C) Identification of mutant cell lines by Southern analyses. (Upper) Cell lines correctly targeted on the 5′ end show a 12-kb EcoRI fragment (lanes 1 and 2) in addition to the 13-kb EcoRI fragment specific to the wild-type chromosome (lanes 3 and 4). (Lower) Clones correctly targeted on the 3′ end show an 11.5-kb EcoRV fragment in addition to the 10.2-kb band generated by the wild-type chromosome. (D) Detection of wild-type and mutant alleles by PCR analysis of genomic DNA of progeny from a cross of heterozygous mice. Arrows in Bii indicate relative position of primers used in this assay to generate a 370-bp product specific to the mutant allele and a 240-bp product specific to the wild-type allele. (E) RNA analysis in mutant mice. RNA was prepared from hearts of 4-week-old wild-type (+/+), heterozygous (+/−), and mutant mice (−/−) and assayed for Kcnq1 expression by Northern blot. In the blot depicted, a full-length cDNA clone of Kcnq1 was used as a probe. Identical results were obtained when blots were probed separately with cDNA clones of exons 1–10 or 11–15 (data not shown). Hybridization with a probe for mouse elongation factor (EF) confirmed equal loading of RNA in each lane.
Histological Analysis of Inner Ear Morphology.
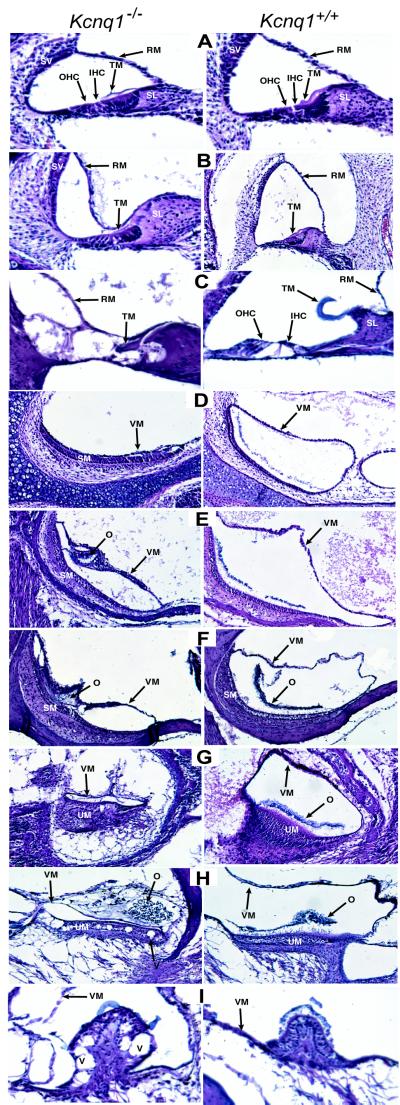
To determine the requirement of Kcnq1 gene function in endolymph production, we examined inner ear structures at P0, P3, P5, P8, P20, and P70 (Fig. 2), reasoning that an inability to maintain endolymph would be manifest by a collapse of the spaces normally containing that fluid. In the cochlea, endolymph is contained within the cochlear duct, which is bound on one side by Reissner's membrane and on the other side by the basilar membrane. The basilar membrane supports the organ of Corti, the location of the auditory hair cells. At P0 in Kcnq1−/− mice, Reissner's membrane appeared morphologically normal (Fig. 2A). However, by P3, Reissner's membrane had collapsed onto the spiral limbus and the tectorial membrane of the organ of Corti (Fig. 2B). The reduced volume of endolymph continues throughout all ages examined. By P70, Kcnq1−/− mice displayed extensive degradation of the inner and outer hair cells in all turns of the cochlea (Fig. 2C).
Figure 2.
Histological analysis of inner ear structures of Kcnq1+/+ and Kcnq1−/− mice. (A–C) Midmodular sections of cochlea at P0 (A), P3 (B), and P70 (C). Note the collapse of Reissner's membrane at P3 (B) and degeneration of inner and outer hair cells at P70 (C). (D–F) Details of the saccule at P0 (D), P8 (E), and P70
The otolith organs consist of two sac-like swellings called the saccule and utricle, which respond to linear acceleration. The most dramatic effect of the loss of Kcnq1 function was observed in the saccule, where at P0 the vestibular membrane had collapsed on the otoconia of the saccular macula, the site of the sensory hair cells (Fig. 2D). This situation is somewhat ameliorated during development, such that by P8, a small cavity is present in the saccule of Kcnq1−/− mice (Fig. 2E). Hair-cell degeneration was not observed in the saccule even at P70 (Fig. 2F). At P8, the vestibular membrane of the utricle was noticeably shrunken in Kcnq1−/− mice (Fig. 2G). The loss of endolymph was progressive, and by P70, vacuoles were observed in the sensory epithelium, suggesting hair-cell degeneration (Fig. 2H). In addition, there was a loss of connective tissue that underlies the utricle (Fig. 2 G and H).
The second set of balance organs are the fluid-filled semicircular ducts, which respond to angular acceleration. By P5, Kcnq1−/− mice showed a loss of the endolymph-filled lumen and a collapse of the vestibular membrane. The sensory hair cells of the semicircular canals are localized to the cristae ampullaris. The onset of hair-cell degeneration in the cristae was both early and severe, as indicated by the appearance of vacuoles in the sensory epithelium at P8. These were strikingly evident by P20. By P70, the cristae of Kcnq1−/− mice had undergone extensive degradation, which affected the hair cells, the transitional epithelium, and the inner core of the cristae (Fig. 2I).
The morphological phenotypes associated with Kcnq1−/− mice appear less severe than those reported for Kcne1−/− mice (23). For example, in Kcne1−/− mice, Reissner's membrane was observed lying along the lateral wall of the cochlea in a state of complete collapse, whereas in age matched Kcnq1−/− mice there was only a partial collapse. The apparent severity of the Kcne1 phenotype is particularly striking given that, whereas both Kcne1 and Kcnq1 mutant mice are expected to lack the IKs channel, only Kcnq1−/− mice will also lose the activity of KCNQ1 acting alone. The disparity between phenotypes suggests that KCNE1 function in the inner ear may not be exclusive to IKs, and KCNE1 may partner other channel subunits necessary for endolymph secretion. We also note that the severity of the Kcnq1 phenotype in the various vestibular and auditory compartments is not equivalent, indicating that the expression pattern and/or the subunit composition of endolymphatic secreting channels may be distinct in each compartment.
The deafness and collapse of the endolymphatic spaces observed in Kcnq1−/− mice is reminiscent of that reported in Nkcc1 mutant mice (18, 35, 36) and in Kcne1 knockout mice (23). Nkcc1 is expressed at high levels on the basolateral membrane of marginal cells of the stria vascularis (37) and encodes a Na-K-Cl cotransporter. This cotransporter is believed essential in the penultimate step of recycling of K+ back into the endolymph from the hair cells. Kcnq1 and Kcne1 are expressed at the apical side of the marginal cells (20). The localization of the proteins and the loss-of-function phenotypes in mice and humans thus suggests that KCNQ1 and KCNE1 play a critical role in the final step of K+ recycling and therefore in the production of the high K+ concentrations essential for inner ear function.
Analysis of Cardiac Function.
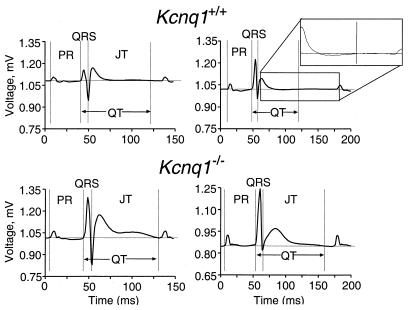
To evaluate Kcnq1 function in the murine heart, we examined ECGs recorded from Kcnq1+/+ and Kcnq1−/− mice. Examples of the ECG recordings are shown in Fig. 3. Kcnq1−/− mice displayed significant increases in most ECG parameters (Table 1), although no differences in heart rate (RR-interval) were observed. The most striking change in Kcnq1−/− mice occurred in the morphology of the T-wave. The area of the T-wave in Kcnq1−/− mice was nearly doubled relative to the controls. In almost every case during the blind trial, a mouse that displayed the extended T-wave could be correctly identified as a mutant. Interestingly, abnormal T-wave morphology is a common finding in LQT1 patients (38). Furthermore, the QT interval, QTc, and JT interval were all significantly prolonged in Kcnq1−/− mice. These data indicate that ventricular repolarization was altered in Kcnq1−/− mice. Similarly, P-wave area and duration were significantly increased in Kcnq1−/− mice, indicating that atrial repolarization was also compromised.
Figure 3.
Representative surface ECG traces (lead II) recorded in vivo from two Kcnq1+/+ mice (upper two traces) and two Kcnq1−/− mice (lower two traces). To better illustrate how the end of the T-wave was measured, the indicated portion of the ECG trace (Upper Right) was magnified as shown (Inset).
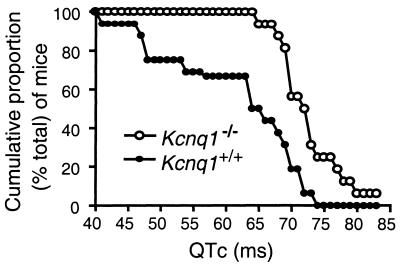
To further investigate how Kcnq1 expression influences cardiac repolarization in mice, we compared the cumulative proportions of Kcnq1+/+ and Kcnq1−/− mice for each measured QTc value (Fig. 4). Aside from the longer QTc values associated with the Kcnq1−/− genotype, this graph illustrates several salient points. First, it shows that the range of QTc values produced for the 32 mice (16 of each genotype) evaluated in this study extends from 40 to 83 ms, and that every mouse with a QTc < 65 ms was wild type (Kcnq1+/+). Second, every mouse that had a QTc > 73 ms belonged to the Kcnq1−/− genotype. Third, approximately half of the Kcnq1+/+ mice had QTc values that overlapped with those measured for the Kcnq1−/− mice. Thus, as in clinical practice, QTc values can serve as a predictor of genotype, particularly at either end of the spectrum, but the degree of overlap in the upper range of wild-type QTc values indicates that QTc alone is not sufficient to predict genotype. Hence, our data indicate that Kcnq1 expression contributes to the repolarization capabilities of the murine heart.
Figure 4.
Cumulative proportion of mice with a given QTc for Kcnq1+/+ (●) and Kcnq1−/− mice (○). (F). Note the complete collapse of the vestibular membrane at P0 (D) and partial recovery by P8 (E). (G–I) Details of the utricle at P8 (G), P70 (H), and the crista ampullaris at P70 (I). Note the collapse of the vestibular membrane at P8 (G) and vaccuoles appearing in the sensory epithelium by P70 (H, I). IHC, inner hair cells; O, otoconia; OHC, outer hair cells; RM, Reissner's membrane; SM, saccular macula; SL, spiral limbus; SV, stria vascularis; TM, tectorial membrane; UM, utricular macula; V, vaccuoles; VM, vestibular membrane.
Drici et al. (24) and Kupershmidt et al. (25) analyzed the cardiac phenotype of two independent lines of Kcne1−/− mice and reported no baseline ECG abnormalities. However, Drici et al. demonstrated an altered QT–RR relationship that translates to a longer QT at slow heart rates and a shorter QT interval at fast heart rates. In contrast, the Kcnq1−/− mice displayed significant baseline differences in most aspects of the ECG. The disparity between the two phenotypes is not surprising considering that, whereas expression of Kcnq1 remains high throughout mouse development, Kcne1 is down-regulated in adult mouse hearts (24, 25). These expression studies are thus consistent with a distinct role for Kcnq1 in adult cardiac function and with the relatively more severe phenotype associated with loss of Kcnq1 gene function compared with Kcne1 mouse mutants.
A modest slowing of conduction appeared to occur in Kcnq1−/− mice because their PR and QRS intervals were slightly longer compared with those measured from Kcnq1+/+ littermates. Interestingly, Kuperschmidt et al. (25) recently reported that when they replaced the Kcne1 coding sequence with the lacZ gene, expression of lacZ is largely restricted to parts of the conduction system in the mouse heart, thereby suggesting that IKs may play a role in the murine cardiac conduction system. However, no conduction disturbances have been reported in the two lines of Kcne1−/− mice (24).
No gross morphological defects were observed in any of the hearts, but both total heart weight (124 ± 28 and 163 ± 35 mg per Kcnq1+/+ and Kcnq1−/− hearts, respectively; P < 0.01) and dry ventricular heart weights (101 ± 19 vs. 125 ± 27 mg; P < 0.01) were increased in the Kcnq1−/− mice. No significant differences were observed in body weight (24 ± 5 and 25 ± 5 g for Kcnq1−/− and Kcnq1+/+ mice, respectively; P > 0.05). To determine whether the increase in heart weight may have contributed to the altered ECG characteristics, we checked for a correlation between heart/body weight ratio and the various ECG parameters, but none was discovered. For examples of this analysis, see supplemental data on the PNAS web site (www.pnas.org). In addition, no significant differences were observed in body weight or lung/body weight ratio (5.4 ± 1.0 mg of lung/g of body weight, n = 12, and 5.1 ± 1.0 mg/g, n = 9, for Kcnq1−/− and Kcnq1+/+ mice, respectively; P > 0.05), indicating that heart failure was also probably not responsible for the ECG changes. Possibly the mild cardiac hypertrophy of Kcnq1−/− mice is related to their hyperactivity associated with the inner ear defects or related to loss of Kcnq1 in the heart and/or other tissues where it is strongly expressed.
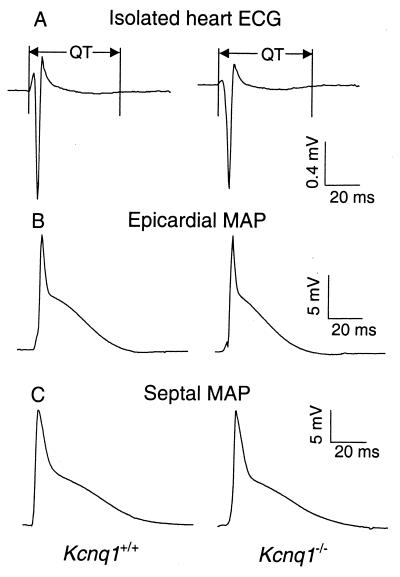
The in vivo ECG data recorded from the diazepam-sedated mice indicate there may be a higher degree of heterogeneity in cardiac repolarization in mice that lack the ability to express Kcnq1, because these mice displayed enhanced P- and T-wave areas. To evaluate whether regional differences in cardiac repolarization do exist in these mice, we recorded monophasic action potentials from four specific regions of isolated mouse hearts (see Table 2 and Fig. 5). However, no significant differences in APD30 or APD90 were observed in isolated Kcnq1−/− compared with Kcnq1+/+ mouse hearts, although the previously reported differences between murine epicardial and endocardial (septal) action potentials (39) were clearly evident (Fig. 5, compare B and C). We also measured APD50 and APD70 for each region, but these also showed no significant differences (data not shown). Consistent with the lack of APD differences in the isolated perfused heart preparation, the ECGs recorded in the same experiment also showed no differences in QRS or T-wave morphology (Fig. 5A) or QT interval (Table 2). Thus the repolarization abnormalities observed in vivo were not apparent in our in vitro preparation and analyses.
Table 2.
Comparison of ECG and action potential parameters from isolated Kcnq1+/+ and Kcnq1−/− hearts
| Parameters | Kcnq1+/+ (n = 8) | Kcnq1−/− (n = 10) |
|---|---|---|
| QT, ms | 63.6 ± 9.1 | 64.2 ± 9.6 |
| RV epicard | ||
| APD90, ms | 44.6 ± 4.5 | 43.3 ± 7.3 |
| APD30, ms | 3.3 ± 0.4 | 3.1 ± 0.7 |
| RV septum | ||
| APD90, ms | 60.0 ± 6.1 | 56.7 ± 5.2 |
| APD30, ms | 7.1 ± 1.5 | 6.7 ± 1.9 |
| LV epicard | ||
| APD90, ms | 50.3 ± 7.6 | 53.2 ± 8.3 |
| APD30, ms | 4.7 ± 0.9 | 4.1 ± 0.7 |
| LV septum | ||
| APD90, ms | 49.1 ± 9.4 | 49.3 ± 7.6 |
| APD30, ms | 7.4 ± 2.4 | 7.9 ± 1.2 |
All data are presented as mean ± SD. Data sets were compared by using Student's t test, with P > 0.05 for all APD comparisons between Kcnq1+/+ and Kcnq1−/− mouse hearts. LV, left ventricle; RV, right ventricle.
Figure 5.
Representative ECG traces (A) and MAP (B, C) traces recorded in vitro from isolated perfused hearts from Kcnq1+/+ and Kcnq1−/− mice. MAP traces were recorded from both right ventricular epicardial (B) and septal (C) surfaces.
At present, it is not immediately apparent why deletion of the Kcnq1 gene in mice (or humans) results in the observed cardiac phenotypes. The in vivo ECG results indicate repolarization deficits, but the underlying electrophysiological mechanisms responsible for these deficits remain to be elucidated. There are several possible explanations why QTc and other ECG differences were observed in vivo but not in the isolated heart preparation. First, autonomic and/or hormonal influences may have contributed to the observed phenotypes in vivo (40, 41). Another possibility is that the sedatives used (diazepam vs. pentobarbital) could have contributed to the different ECG results produced from the in vivo vs. the in vitro experiments (42). Although less likely, we cannot exclude the possibility that technical issues such as electrode placement, inherent variability in APD measurements, and perfusion conditions could also have compromised our ability to observe repolarization defects in vitro. These caveats notwithstanding, our data suggest the potentially interesting possibility that extracardiac factors are necessary to induce the cardiac phenotype seen in Kcnq1−/− mice.
The data presented here suggest that Kcnq1−/− mice represent a potentially valuable animal model for JLNS. Although JLNS and other forms of inherited LQTS represent only a minority of clinical cases, the acquired forms induced by drugs and disease are thought to affect many of the same cardiac ion channels that have been linked to congenital LQTS. Thus, the inherited form offers a model for understanding the molecular and physiological correlates of the acquired form. Future studies may seek to identify the specific patterns of Kcnq1 expression in the murine heart and to determine which current(s) rely on Kcnq1 expression in vivo.
Supplementary Material
Acknowledgments
We thank Dr. Doris Wu for her invaluable guidance in understanding inner ear defects. This work was supported in part by a PhRMA Foundation Faculty Development Grant (B.C.K.).
Abbreviations
- JLNS
Jervell and Lange–Nielsen Syndrome
- LQTS
long QT syndrome
- QTc
corrected QT interval
- MAP
monophasic action potential
- Pn
postnatal day n
- APD
action potential duration
Note Added in Proof.
Association of deafness and gastric hyperplasia with disruption of the mouse Kcnq1 gene was recently demonstrated by Lee et al. (45). The same exon was disrupted in both studies.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wang Q, Chen Q, Towbin J A. Ann Med. 1998;30:58–65. doi: 10.3109/07853899808999385. [DOI] [PubMed] [Google Scholar]
- 2.Roden D M, Spooner P M. J Cardiovasc Electrophysiol. 1999;10:1664–1683. doi: 10.1111/j.1540-8167.1999.tb00231.x. [DOI] [PubMed] [Google Scholar]
- 3.Romano C, Gemme G, Pongiglione R. Clin Pediatr. 1963;45:656–661. [PubMed] [Google Scholar]
- 4.Ward O C. J Ir Med Assoc. 1964;54:103–106. [PubMed] [Google Scholar]
- 5.Jervell A, Lange-Nielsen F. Am Heart J. 1957;54:59–78. doi: 10.1016/0002-8703(57)90079-0. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Curran M E, Splawski I, Burn T C, Millholland J M, VanRaay T J, Shen J, Timothy K W, Vincent G M, de Jager T, et al. Nat Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 7.Splawski I, Tristani-Firouzi M, Lehmann M H, Sanguinetti M C, Keating M T. Nat Genet. 1997;17:338–340. doi: 10.1038/ng1197-338. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson J L, Moss A J, Towbin J A, Keating M T. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 9.Curran M E, Splawski I, Timothy K W, Vincent G M, Green E D, Keating M T. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 10.Abbott G W, Sesti F, Splawski I, Buck M E, Lehmann M H, Timothy K W, Keating M T, Goldstein S A. Cell. 1999;97:175–187. doi: 10.1016/s0092-8674(00)80728-x. [DOI] [PubMed] [Google Scholar]
- 11.Zareba W, Moss A J, Schwartz P J, Vincent G M, Robinson J L, Priori S G, Benhorin J, Locati E H, Towbin J A, Keating M T, et al. N Engl J Med. 1998;339:960–965. doi: 10.1056/NEJM199810013391404. [DOI] [PubMed] [Google Scholar]
- 12.Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. Nature (London) 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 13.Sanguinetti M C, Curran M E, Zou A, Shen J, Spector P S, Atkinson D L, Keating M T. Nature (London) 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 14.Steel K P. Science. 1999;285:1363–1364. doi: 10.1126/science.285.5432.1363. [DOI] [PubMed] [Google Scholar]
- 15.Kubisch C, Schroeder B C, Friedrich T, Lutjohann B, El Amraoui A, Marlin S, Petit C, Jentsch T J. Cell. 1999;96:437–446. doi: 10.1016/s0092-8674(00)80556-5. [DOI] [PubMed] [Google Scholar]
- 16.Xia J H, Liu C Y, Tang B S, Pan Q, Huang L, Dai H P, Zhang B R, Xie W, Hu D X, Zheng D, et al. Nat Genet. 1998;20:370–373. doi: 10.1038/3845. [DOI] [PubMed] [Google Scholar]
- 17.Kelsell D P, Dunlop J, Stevens H P, Lench N J, Liang J N, Parry G, Mueller R F, Leigh I M. Nature (London) 1997;387:80–83. doi: 10.1038/387080a0. [DOI] [PubMed] [Google Scholar]
- 18.Dixon M J, Gazzard J, Chaudhry S S, Sampson N, Schulte B A, Steel K P. Hum Mol Genet. 1999;8:1579–1584. doi: 10.1093/hmg/8.8.1579. [DOI] [PubMed] [Google Scholar]
- 19.Sakagami M, Fukazawa K, Matsunaga T, Fujita H, Mori N, Takumi T, Ohkubo H, Nakanishi S. Hear Res. 1991;56:168–172. doi: 10.1016/0378-5955(91)90166-7. [DOI] [PubMed] [Google Scholar]
- 20.Neyroud N, Tesson F, Denjoy I, Leibovici M, Donger C, Barhanin J, Faure S, Gary F, Coumel P, Petit C, et al. Nat Genet. 1997;15:186–189. doi: 10.1038/ng0297-186. [DOI] [PubMed] [Google Scholar]
- 21.Marcus D C, Shen Z. Am J Physiol. 1994;267:C857–C864. doi: 10.1152/ajpcell.1994.267.3.C857. [DOI] [PubMed] [Google Scholar]
- 22.Schulze-Bahr E, Haverkamp W, Wedekind H, Rubie C, Hordt M, Borggrefe M, Assmann G, Breithardt G, Funke H. Hum Genet. 1997;100:573–576. doi: 10.1007/s004390050554. [DOI] [PubMed] [Google Scholar]
- 23.Vetter D E, Mann J R, Wangemann P, Liu J, McLaughlin K J, Lesage F, Marcus D C, Lazdunski M, Heinemann S F, Barhanin J. Neuron. 1996;17:1251–1264. doi: 10.1016/s0896-6273(00)80255-x. [DOI] [PubMed] [Google Scholar]
- 24.Drici M D, Arrighi I, Chouabe C, Mann J R, Lazdunski M, Romey G, Barhanin J. Circ Res. 1998;83:95–102. doi: 10.1161/01.res.83.1.95. [DOI] [PubMed] [Google Scholar]
- 25.Kupershmidt S, Yang T, Anderson M E, Wessels A, Niswender K D, Magnuson M A, Roden D M. Circ Res. 1999;84:146–152. doi: 10.1161/01.res.84.2.146. [DOI] [PubMed] [Google Scholar]
- 26.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gould T D, Pfeifer K. Hum Mol Genet. 1998;7:483–487. doi: 10.1093/hmg/7.3.483. [DOI] [PubMed] [Google Scholar]
- 28.Knollmann B C, Knollmann-Ritschel B E, Weissman N J, Jones L R, Morad M. J Physiol (London) 2000;525:483–498. doi: 10.1111/j.1469-7793.2000.t01-1-00483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell G F, Jeron A, Koren G. Am J Physiol. 1998;274:H747–H751. doi: 10.1152/ajpheart.1998.274.3.H747. [DOI] [PubMed] [Google Scholar]
- 30.Knollmann, B. C., Blatt, A. S., Horton, K., deFreitas, F., Miller, T., Bell, M., Housmans, P. R., Weissman, N. J., Morad, M. & Potter, J. D. (2001) J. Biol. Chem., in press. [DOI] [PubMed]
- 31.Behrens S, Li C, Knollmann B C, Franz M R. Pacing Clin Electrophysiol. 1998;21:100–107. doi: 10.1111/j.1540-8159.1998.tb01067.x. [DOI] [PubMed] [Google Scholar]
- 32.Franz M R, Burkhoff D, Spurgeon H, Weisfeldt M L, Lakatta E G. Eur Heart J. 1986;7:34–41. doi: 10.1093/oxfordjournals.eurheartj.a061954. [DOI] [PubMed] [Google Scholar]
- 33.Deol M S. J Med Genet. 1968;5:137–158. doi: 10.1136/jmg.5.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Llorens J, Dememes D, Sans A. Toxicol Appl Pharmacol. 1993;123:199–210. doi: 10.1006/taap.1993.1238. [DOI] [PubMed] [Google Scholar]
- 35.Deol M S. J Embryol Exp Morphol. 1963;11:493–512. [PubMed] [Google Scholar]
- 36.Flagella M, Clarke L L, Miller M L, Erway L C, Giannella R A, Andringa A, Gawenis L R, Kramer J, Duffy J J, Doetschman T, et al. J Biol Chem. 1999;274:26946–26955. doi: 10.1074/jbc.274.38.26946. [DOI] [PubMed] [Google Scholar]
- 37.Crouch J J, Sakaguchi N, Lytle C, Schulte B A. J Histochem Cytochem. 1997;45:773–778. doi: 10.1177/002215549704500601. [DOI] [PubMed] [Google Scholar]
- 38.Moss A J, Zareba W, Benhorin J, Locati E H, Hall W J, Robinson J, Schwartz P J, Towbin J A, Vincent G M, Lehmann M H, et al. Circulation. 1995;92:2929–2934. doi: 10.1161/01.cir.92.10.2929. [DOI] [PubMed] [Google Scholar]
- 39.Xu H, Guo W, Nerbonne J M. J Gen Physiol. 1999;113:661–678. doi: 10.1085/jgp.113.5.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh K B, Kass R S. Science. 1988;242:67–69. doi: 10.1126/science.2845575. [DOI] [PubMed] [Google Scholar]
- 41.Washizuka T, Horie M, Watanuki M, Sasayama S. Circ Res. 1997;81:211–218. doi: 10.1161/01.res.81.2.211. [DOI] [PubMed] [Google Scholar]
- 42.Salata J J, Jurkiewicz N K, Wang J, Evans B E, Orme H T, Sanguinetti M C. Mol Pharmacol. 1998;54:220–230. doi: 10.1124/mol.54.1.220. [DOI] [PubMed] [Google Scholar]
- 43.Paulsen M, Davies K R, Bowden L M, Villar A J, Franck O, Fuermann M, Dean W L, Moore T F, Rodrigues N, Davies K E, et al. Hum Mol Genet. 1998;7:1149–1159. doi: 10.1093/hmg/7.7.1149. [DOI] [PubMed] [Google Scholar]
- 44.Paulsen M, El Maarri O, Engemann S, Strodicke M, Franck O, Davies K, Reinhardt R, Reik W, Walter J. Hum Mol Genet. 2000;9:1829–1841. doi: 10.1093/hmg/9.12.1829. [DOI] [PubMed] [Google Scholar]
- 45.Lee M P, Ravenel J D, Hu R-J, Lustig L R, Tomaselli G, Berger R D, Brandenburg S A, Litzi T J, Bunton T E, Limb C, et al. J Clin Invest. 2000;106:1447–1455. doi: 10.1172/JCI10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.