Abstract
The rat demyelination (dmy) mutation serves as a unique model system to investigate the maintenance of myelin, because it provokes severe myelin breakdown in the central nervous system (CNS) after normal postnatal completion of myelination. Here, we report the molecular characterization of this mutation and discuss the possible pathomechanisms underlying demyelination. By positional cloning, we found that a G-to-A transition, 177 bp downstream of exon 3 of the Mrs2 (MRS2 magnesium homeostasis factor (Saccharomyces cerevisiae)) gene, generated a novel splice acceptor site which resulted in functional inactivation of the mutant allele. Transgenic rescue with wild-type Mrs2-cDNA validated our findings. Mrs2 encodes an essential component of the major Mg2+ influx system in mitochondria of yeast as well as human cells. We showed that the dmy/dmy rats have major mitochondrial deficits with a markedly elevated lactic acid concentration in the cerebrospinal fluid, a 60% reduction in ATP, and increased numbers of mitochondria in the swollen cytoplasm of oligodendrocytes. MRS2-GFP recombinant BAC transgenic rats showed that MRS2 was dominantly expressed in neurons rather than oligodendrocytes and was ultrastructurally observed in the inner membrane of mitochondria. Our observations led to the conclusion that dmy/dmy rats suffer from a mitochondrial disease and that the maintenance of myelin has a different mechanism from its initial production. They also established that Mg2+ homeostasis in CNS mitochondria is essential for the maintenance of myelin.
Author Summary
The myelin sheath that surrounds the axon of a neuron acts as a biological insulator. Its major function is to increase the speed at which impulses propagate along myelinated fibers in the central nervous system, as well as the peripheral nervous system. Alterations or damage affecting this structure (demyelination) result in the disruption of signals between the brain and other parts of the body. In the rat, mutations producing demyelination have been frequently identified and characterized and have contributed to a better understanding of the genetics of myelin development, physiology, and pathology. This paper reports the molecular characterization of a recessive allele responsible for the progressive disruption of myelin that was initially observed in mutant rats, previously named demyelination (dmy). This mutation generates an additional splicing acceptor site in an intron of the mitochondrial Mg2+ transporter gene (Mrs2), resulting in the insertion of a 83-bp genomic DNA segment into the Mrs2 transcript and complete functional inactivation of the mutant allele. We firstly defined the biological function of MRS2 in mammals and demonstrated the crucial and unexpected role of MRS2 in myelin physiology. Our findings might be helpful in the development of new therapeutic strategies for demyelinating syndromes.
Introduction
Myelin is an essential component of the nervous tissue of higher vertebrates. It acts as a natural insulator of axonal segments allowing, at the same time, the maintenance of axonal integrity and the fast conduction of action potentials. It also reduces ionic currents across the axonal membrane and stabilizes the extracellular milieu within rapidly-firing axon bundles.
In the central nervous system (CNS), myelin is produced by oligodendrocytes, while in the peripheral nervous system (PNS), this function is achieved by Schwann cells. Myelination is completed within a relatively short period of time during mammalian development and requires a high rate of production and transport of different kinds of molecules, mostly proteins and lipids. In adult life, myelin is constantly remodeled and the maintenance of functional myelin sheaths requires a careful balance of de novo synthesis and turnover. It is quite clear that any event generating an imbalance in the myelination or remyelination process has the greatest chance of inducing dys- or demyelination of either the central or peripheral nervous system.
Our knowledge of the myelination process has benefited from careful observations conducted on human patients affected by one of the many defects of myelination or myelin turnover. It has also benefited from researches carried out on animal models, mostly mutant mice and rats, including those that have been induced by transgenesis or genetic engineering in ES cell lines [1], [2]. Some of these models have even allowed therapies to be developed in a preclinical setting [3]. Unfortunately, only a small number of the many genes that are directly or indirectly involved in the myelination process have been identified and only a few of these genes have been functionally annotated, for example, by the characterization of one or more mutant alleles. For this reason, any new mutation occurring spontaneously or after mutagenesis is of potential interest for unraveling the molecular mechanisms involved in myelin assembly.
In an earlier paper we reported the discovery and pathology of a rat mutation designated demyelination (symbol dmy), which is characterized by severe and progressive myelin breakdown in the CNS. We mapped the locus responsible for this myelin disorder to rat chromosome (Chr) 17, very close to the prolactin (Prl) locus, in a region homologous to human Chr 6p21.1-22.3 and mouse Chr 13 [4], [5]. Based on its pathological features, as well as its genetic localization, this demyelination syndrome appeared to be unique, with no homologue so far reported in any other mammalian species, including humans.
In this report we demonstrate that the causative gene (Mrs2) encodes a protein that is an essential component of the major electrophoretic Mg2+ influx system in mitochondria [6]. This gene has orthologues in other organisms, including lower eukaryotes and plants [7], [8]. The protein shares many of the properties of bacterial CorA and yeast Alr1 proteins but its specific involvement in the myelination process was not known or even suspected.
Results
dmy/dmy rats exhibit a phenotype with typical demyelination
The pathology of homozygous dmy/dmy rats has been reported in detail previously [4]. Mutant rats exhibit no significant differences from their control littermates until 4 weeks of age. From 5 weeks on, flaccidity of the hind limbs becomes noticeable and evolves towards complete paralysis around 7–8 weeks of age. Progressive demyelination is observed in several parts of the CNS (Figure 1), namely the corpus callosum, the capsula interna, the striatum and the cerebellar peduncle, with major effects on the ventral and lateral parts of the spinal cord. Astrogliosis, which is a major feature of myelin disorder, is observed in demyelinated areas but motor neurons remain normal and there is no sign of associated inflammation in the white matter. The dmy mutation can then be regarded as a mutation affecting the maintenance and turnover of myelin rather than its initial production: this is typical demyelination [9].
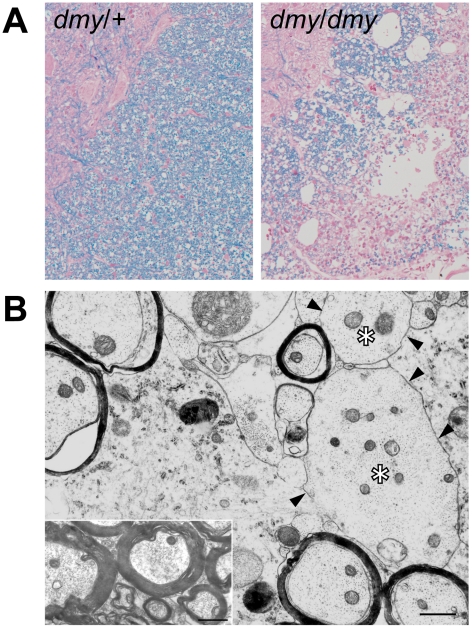
Figure 1. Demyelination in dmy/dmy rats.
A. Histopathology of the cervical part of the spinal cord of dmy/+ (left) and dmy/dmy (right) rats aged 10 weeks. Luxol fast blue-HE staining. Original magnification: ×100. B. Electron microscopy of the cervical part of spinal cord of dmy/dmy rats (10 weeks). Naked axons with demyelination (arrowheads) are indicated by asterisks. Inset: control image of the spinal cord from the age-matched wild type rat. Axons are normally myelinated. Bar = 1 µm.
The dmy syndrome is associated with a mutation in a splicing site of Mrs2, a gene encoding a mitochondrial Mg2+ channel
Out of 687 dmy/dmy mutant rats, collected from the 3,252 offspring of an intercross segregating for the dmy mutation, 23 individuals were found to carry a recombinant haplotype between the two loci that were used for the initial genetic mapping, namely; Prl (prolactin) and Hh1tts (testis-specific histone, H1t). Further investigation of these animals, using three novel informative SSLP markers, allowed us to narrow the genetic interval containing dmy down to 0.22 cM, between markers D17Kur17 and D17Got45. Within this critical section, we found no recombination between the dmy locus and either Aldh5a1 (aldhehyde dehydrogenase family 5, subfamily A1) or Mrs2 (mitochondrial 118 RNA splicing2) loci, among 687×2 = 1,374 meioses. The rat genome databases revealed that D17Kur17 and D17Got45 were at position 46.78-Mb and 47.26-Mb, respectively, on rat Chr 17, yielding a physical size of 0.48 Mb of DNA for the interval containing the dmy locus. This stretch of DNA contained 6 genes (Figure 2A).
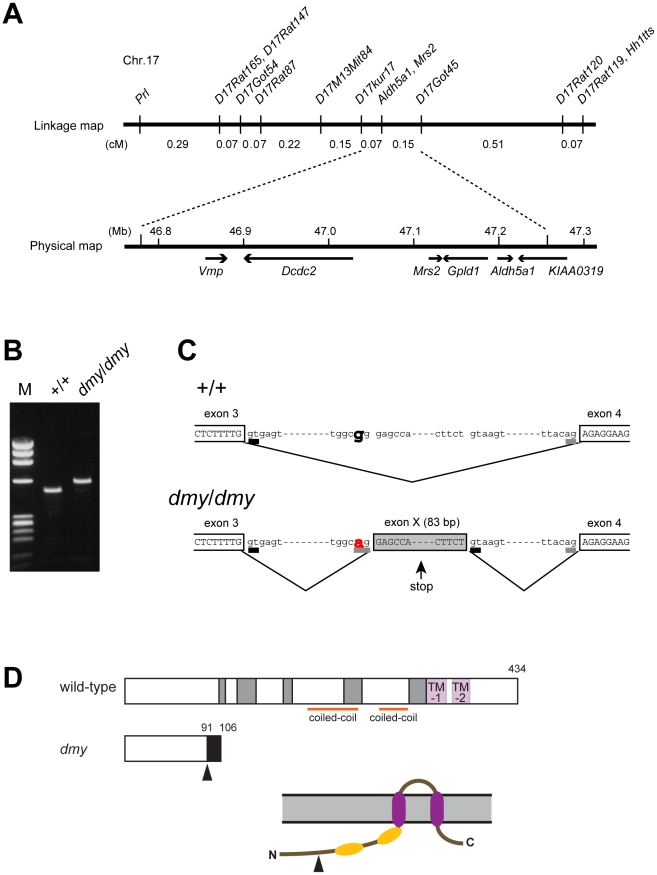
Figure 2. Positional cloning of the dmy mutation.
A. The dmy locus was localized within a 0.22-cM region of chromosome 17 between D17Kur17 and D17Got45 and no recombination was observed with SSLP markers designed from Aldh5a1 and Mrs2 genomic sequences in 1,374 informative meioses. Within the 0.48-Mb physical interval between D17Kur17 and D17Got45, harboring the dmy locus, 6 genes: Vmp (vesicular membrane protein p24), Dcdc2 (doublecortin domain containing 2), Mrs2 (MRS2 magnesium homeostasis factor (S. cerevisiae), Gpld1 (glycosylphosphatidylinositol specific phospholipase D1), Aldh5a1 (aldehyde dehydrogenase family 5, subfamily A1), and KIAA0319, were previously mapped. B. A larger RT-PCR product was obtained when amplifying the 5′ region of Mrs2 cDNAs from dmy/dmy rats with a primer set of rMrs2l-3&4 (5′-TGTACTGATCTACCCGAGTTCC-3′ and 5′-TCTGGAGTTATCACAGCCTTCA-3′). M: molecular marker, ΦX174-HaeIII digest. C. Upper: Genomic organization in the vicinity of intron 3 of the Mrs2 wild-type allele. Lower: Genomic rearrangements in the same intron 3 of the Mrs2dmy mutant allele. In the Mrs2dmy mutant allele, a novel splice acceptor site was generated as a consequence of a G-to-A transition at 177 bp downstream of the end of exon 3. An 83-bp genomic sequence (boxed in gray), downstream of the recently generated acceptor site (tggcag), is then inserted into the Mrs2 mutant transcript. This sequence contains a premature stop codon (vertical arrow), which truncates the protein almost immediately downstream of exon 3. D. Schematic representations of the wild-type and dmy MRS2 proteins. Conserved amino acid residues and transmembrane domains are indicated by grey and purple boxes, respectively. Coiled-coil regions are indicated by horizontal orange lines. The position of the dmy mutation is indicated by an arrowhead, and the additional 15 residues (GATWTPRILEECLES), indicated by a black box, are deduced to be added subsequently. Bottom: Schematic representation of the topology of MRS2. Purple: transmembrane domains, Orange: coiled-coil regions. The position of the dmy mutation is indicated by an arrowhead.
Analysis by RT-PCR of the transcription products of these 6 genes revealed that the cDNA transcribed from the Mrs2 gene was larger in dmy/dmy mutants than in the controls (Figure 2B). After sequencing, we found that the larger size of the dmy cDNA was due to the insertion of an 83 bp intronic sequence between exons 3 and 4. Comparison of the two genomic sequences revealed a G-to-A transition, 177 bp downstream of the end of exon 3 (Figure 2C, Figure S1), generating a novel splice acceptor site, which accounted for the addition of the 83bp stretch of intronic sequence to the mutant transcript. In addition, while the Mrs2 gene normally encodes a 434 amino-acid protein, the intronic insertion leads to a shorter protein (106 amino acids) due to the occurrence of a stop codon as a consequence of frame shifting within the novel pseudo-exon X. The new protein consisted of the first 91 amino acids of normal (wild-type) MRS2 protein followed by an additional 15 amino acids transcribed from the intronic stretch (Figure 2D) [10]. No nucleotide alteration was observed between normal and mutant haplotypes in the cDNA transcribed from the other 5 genes (Vmp, Dcdc2, Gpld1, Aldh5a1, and KIAA0319). These findings strongly suggested that the G-to-A mutation in intron 3 of Mrs2 in dmy/dmy rats was very likely causative of the neurological phenotype.
dmy/dmy rats exhibit morphological and biochemical features characteristic of mitochondrial deficiencies
The MRS2 protein functions as a major transporter protein (Mg2+, Ni2+ and Co2+) in yeast as well as in human cells [10], [11]. When this protein is functionally defective this leads to the “petite” phenotype in yeast and to cell death in human HEK 293 cells [11], [12]. Because mitochondrial diseases in mammals are often accompanied by elevated lactic acid, reduced ATP, increased cytochrome oxidase (COX) activity, and the morphological alteration of mitochondria [13]–[15], we measured lactic acid levels and ATP contents in the CNS and performed morphological analyses of the CNS of dmy/dmy rats.
Lactic acid concentration in the cerebrospinal fluids was significantly elevated in dmy/dmy rats when compared with normal littermates: 126±43.7 mg/dL vs 25±9.6 mg/dL (average ± SD), P<0.002 (Figure 3A). The ATP concentration was markedly reduced in dmy/dmy rats: 265±79 µM/mg vs 99±46 µM/mg (average ± SD), P<0.005 (Figure 3B). In the affected dmy/dmy rats, swollen oligodendrocytes were often observed in the white matter, showing the increased COX reaction products (Figure 3C). Ultrastructurally, their cytoplasm contained many mitochondria and Golgi apparatus-like membrane structures (Figure 3D). These findings indicated that the mitochondria of dmy/dmy rats were functionally defective.
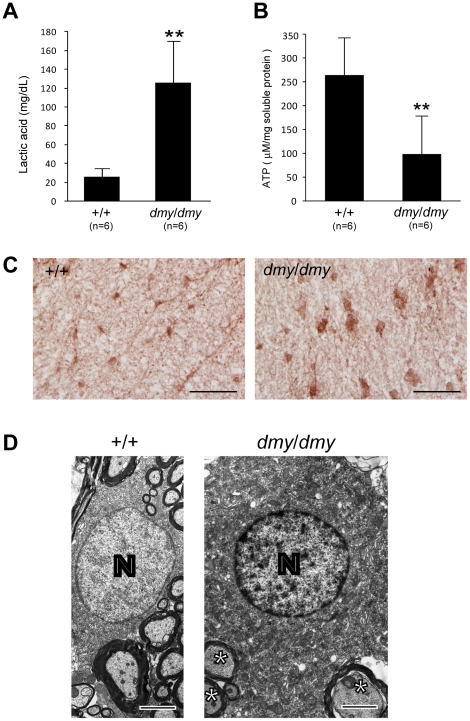
Figure 3. Biochemical and morphological abnormalities in the mitochondria of dmy/dmy mutant rats.
A. Lactic acid concentration in cerebrospinal fluid of 6–7-week-old dmy/dmy rats and age-matched wild-type rats. **, P<0.002. B. ATP levels in the brain of 6–7-week-old dmy/dmy rats and age-matched wild-type rats. **, P<0.005. C. Cytochrome oxidase staining of the spinal cords of 6–7-week-old dmy/dmy (right) and age-matched wild-type (left) rats. Swollen oligodendrocytes were often seen they showed increased COX reaction product. Bar = 50 µm. D. Electron microphotographs of a swollen oligodendrocyte in a dmy/dmy rat (right) and an oligodendrocyte in a control wild-type rat. White matter of thoracic spine at 6 weeks of age. N: Nucleus of the oligodendrocyte. Axons adjacent to the oligodendrocyte are indicated by asterisks. Bar = 2 µm.
Rescue of dmy/dmy mutant phenotypes by transgenic complementation
To ascertain that the molecular defect (i.e. G-to-A transition) observed in the dmy mutant haplotype was causative of the abnormal phenotype observed in dmy/dmy rats, we attempted to rescue the mutant phenotype by transgenic complementation. We established two independent WTC.DMY-dmy lines, expressing each Mrs2 wild-type cDNA under the control of a cytomegalovirus (CMV) promoter (Figure S2A), and found that all dmy/dmy transgenic rats exhibited a completely normal phenotype, with no paralysis of the hind limbs. Histopathological analyses demonstrated that both transgenic lines no longer exhibited any sign of demyelination of the CNS (Figure S2B). In addition, lactic acid levels of the cerebrospinal fluid of transgenic dmy/dmy rats had returned to the normal range (Figure S2C). Electron microscopic observations revealed that mitochondria of the oligodendrocytes in transgenic rats were normal in their morphology and number (). These findings confirmed that the molecular changes reported above and observed in the Mrs2 gene were indeed causative of the dmy-mutant phenotypes. For this reason we decided that the symbol of the mutant allele should, from now on, be changed to Mrs2dmy.
MRS2-GFP recombinant protein is expressed in the mitochondria
To characterize the tissues and cell types expressing MRS2 as well as the subcellular localization of this protein in the CNS, we generated a strain of rats transgenic for a recombinant MRS2-GFP BAC clone. These transgenic rats were expected to express recombinant protein under the control of the endogenous, normal Mrs2 promoter. We found that cytoplasmic dot-like MRS2-GFP signals were observed in neurons throughout the CNS. To a lesser extent, astrocytes and oligodendrocytes also exhibited occasional expression of MRS2 (Figure S3). Confocal microscopy demonstrated that MRS2 is located in the mitochondria (Figure 4A–4C). Moreover, immunoelectron microsopic examinations with anti-GFP antibody revealed that MRS2 is localized in the inner membrane of the mitochondria (Figure 4D). MRS2 expression was also observed in the myocardium, liver, testis and skeletal muscles (Figure S4).
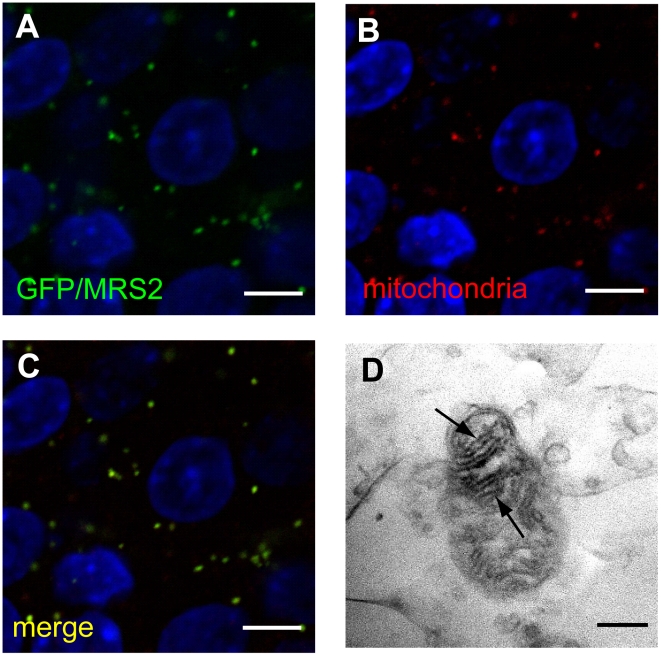
Figure 4. Expression of MRS2 protein in the mitochondria.
MRS2-GFP recombinant protein (Green) was seen in the cytoplasm of pyramidal cells (A). MRS2-GFP signals were colocalized with the mitochondria (B), as shown in the confocal image of GFP and mitochondrial immunohistochemistry (C). Nuclei were stained with DAPI (Blue). Bar: 5 µm. Immunoelectron microscopy using anti-GFP antibody revealed that MRS2-GFP signals were localized in the inner membrane of the mitochondria (arrows) (D). Bar: 200 nm.
Microglia activation and high expression of inflammatory cytokines were observed in Mrs2dmy/Mrs2dmy rats
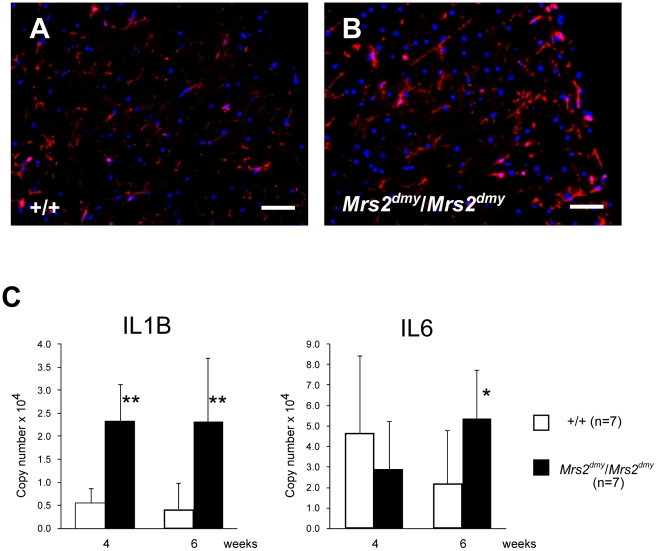
Microglial activation, characterized by cellular hypertrophy, has been reported in various dysmyelinating and demyelinating pathologies. To assess microglial activation, we performed immunohistochemistry for IBA1, a specific marker of microglia. In Mrs2dmy/Mrs2dmy rats, prolonged activation of microglia was prominently observed at 6–7 weeks of age (Figure 5A and 5B), the stage at which clinical symptoms such as flaccid paralysis were commonly observed. Expression levels of proinflammatory cytokines, such as Il1b and Il6, were also significantly higher in Mrs2dmy/Mrs2dmy rats than in wild-type littermates at 6 weeks of age (Figure 5C).
Figure 5. Activation of microglia in the central nervous system of Mrs2dmy/Mrs2dmy rats.
Immunohistochemistry for Iba1 in the lumbar part of the spinal cord of wild-type (A) and Mrs2dmy/Mrs2dmy rats (B) at 6 weeks of age. Signals of Iba1 (AlexaFluor 546 nm; red), which is upregulated during the activation of microglia, are seen in Mrs2dmy/Mrs2dmy rats much more the wild-type control. Nucleus is stained with DAPI (blue). C, Inflammatory cytokine mRNA expression in the CNS of wild-type (□) and Mrs2dmy/Mrs2dmy (▪) rats. IL1b expression was elevated in Mrs2dmy/Mrs2dmy rats at 4 and 6 weeks of age. IL6 was elevated in Mrs2dmy/Mrs2dmy rats at 6 weeks of age. * P<0.05, ** P<0.005.
Discussion
Characterization, by positional cloning, of the molecular defect responsible for the demyelinating phenotype observed in adult dmy/dmy rats led us to incriminate a mutation in the Mrs2 gene. No mutant allele before Mrs2dmy, which we report here, has ever been reported at this locus in any mammalian species.
Mrs2 encodes an inner membrane Mg2+ channel in mitochondria and belongs to a family with orthologous copies in a wide range of species [10], [12]. Mrs2 was originally identified in yeast, and orthologous copies of this gene have been identified in a variety of organisms, including bacteria (CorA), fungi (Alr1), and plants (AtMrs2). All proteins in the family have the same substrate selectivity: they transport Mg2+, Co2+ and some other divalent cations across the mitochondrial membrane. Even if these proteins exhibit relatively low sequence similarities, they all have a few important domains at the same relative position and can functionally complement each other over a wide range of phylogenetic distances [16], [17]. In mammals, the normal protein MRS2 has two universally conserved transmembrane domains (TMs) and a conserved Gly-Met-Asn (GMN) motif close to the first TM domain that forms part of the pore and is essential for Mg2+ transport [18] (Figure 2D, Figure S5). As we demonstrated, the protein is truncated in dmy/dmy mutant rats, having lost both of its essential domains and accordingly its function of an Mg2+ transmembrane transporter. In other words, Mrs2dmy is a null allele, which is totally consistent with its recessive allelic interaction.
An MRS2 is a major transport for Mg2+ uptake into mitochondria, its function would be expected to be important, if not essential, for the maintenance of respiratory complex I and accordingly for cell viability [6], [11]. This assumption was supported by the analysis of MRS2 knock-down, mediated by shRNA in a human HEK-293 cell line, which resulted in a series of physiological changes ranging from transient reduction of Mg2+ uptake to the complete loss of mitochondrial respiratory complex I, with decreased mitochondrial membrane potential and cell death, depending on the duration of knock-down treatment [11]. However, if we consider the phenotype of our mutant rat, which is apparently limited to the myelination process with a rather long lifespan, the role of MRS2 in the maintenance of cell integrity should be reconsidered.
Considering the pathological features that appear to be characteristics of the Mrs2dmy allele on the one hand, and MRS2-specific functions, as described above on the other, it is logical to consider that the demyelinating syndrome in mutant rats results from a mitochondrial disease. This assertion is supported by the observation of an elevated rate of lactic acid in the cerebrospinal fluid, reduced ATP in the brain, increased COX activity, and the morphological alteration of mitochondria, which is generally considered a major characteristic of mitochondrial diseases [13]–[15]. An increase in mitochondria is characteristic of cells with reduced respiratory capacity [19]. The association of mitochondrial dysfunction with demyelination (or leukodystrophy) has been already reported in Leigh syndrome and mitochondrial DNA depletion syndrome [20]–[23]. The tissues most frequently affected in these mitochondrial diseases are the cerebrum, peripheral nerves, and skeletal muscles, presumably because cells of these tissues require more energy than any other cells in the body. Unfortunately, the detailed pathophysiological mechanism(s) leading to demyelination in these diseases has not yet been unraveled. We consider that our mutant rat could be an interesting tool for investigating this matter.
Mitochondrial dysfunction has also been observed in multiple sclerosis (MS), one of the most common demyelination diseases, but here again many aspects of the pathophysiology require further investigation [24], [25]. This difficulty of linking gene functions with a specific syndrome is not so surprising if we consider that, according to the most recent estimates, there may be as many as 1,500 nuclear-encoded mitochondrial proteins [26] and that less than half have been identified with experimental support. Clearly, a complete protein inventory of this organelle across tissues would provide a molecular framework to relate mitochondrial biology and pathogenesis [27].
A point concerning Mrs2 gene expression in the CNS that is worth noting after our experiments and observations is that the gene in question is expressed at a higher rate in neurons than in oligodendrocytes (Figure 4, Figure S3). This was rather unexpected if we consider that oligodendrocytes are the cells actually responsible for myelination of the CNS. At this time, it remains unclear whether the demyelination in dmy/dmy rats is triggered cell-autonomously or cell-nonautonomously. Instead, it is likely that demyelination is enhanced by the surrounding cells, such as activated microglia and astroglia. At 6 weeks of age, when dmy/dmy rats began to exhibit ataxia [9], cytokine levels were elevated and microglia were activated (Figure 5), and it is considered that activated microglia cause neuronal damage through the release of potentially cytotoxic molecules, such as proinflammatory cytokines, reactive oxygen intermediates, proteinases, and complement proteins [28]. Oligodendrocytes show greater vulnerability to such molecules [29], [30]. Additionally, Kuwamura and co-workers reported prominent astrogliosis and many ED-1-positive macrophages in myelin-destroyed areas [9]. When considered together, these morphological observations led us to believe that the demyelination observed in dmy/dmy rats is probably enhanced by activated microglia and astroglia.
In summary, we identified Mrs2dmy as a loss-of-function mutation of the Mrs2 gene that normally encodes Mg2+ transporter protein of the mitochondrial inner membrane. Our observations also demonstrate that the mechanisms underlying the initial development of myelin (myelination) are different from those that are involved in its maintenance and turnover since, in Mrs2dmy/Mrs2dmy rats, myelin development is normal while its maintenance is defective. Our mutant rats also appear to be an excellent animal model, not only to evaluate the causal relationships between primary mitochondrial dysfunction and subsequent demyelination, but also for the development of therapies making use, for example, of cell transplantation.
Materials and Methods
Genetic fine mapping of dmy
Congenic strains WTC (NBRP#0020) and WTC.DMY-dmy (NBRP#0021) were both from the National BioResource Project - Rat, Kyoto University (Kyoto, Japan). (WTC.DMY-dmy × BN/SsNSlc)F1(+/dmy) rats were intercrossed to produce F2 progeny. dmy/dmy homozygotes were identified at 7–8 weeks of age, when paralysis of the hind limbs was obvious. 687 dmy/dmy rats were collected out of 3,252 F2 animals (∼21%) and used for fine mapping of the dmy locus. Simple sequence length polymorphisms (SSLPs) from the Prl (prolactin) and Hh1tts (Testis-specific histone, H1t and H4t) genes were used for genotyping as described [31]. To refine the limits of the recombinant interval between Prl and Hh1tts, two gene-specific and one anonymous SSLP markers were used: Mrs2 (5′-TCTCCCTTGCCTCTATCTCTCGTCT-3′,5′-CCTGCAGTACTGGGTAAGCCTGATG-3′), Aldh5a1 (5′-GTTAACTGCACAAGAGCAAGCCAGT-3′, 5′-GCTAATGTTAAGTCATGGGGTGAGG-3′), and D17Kur17 5′-ACCTCTTTTTGCCAGCATTG-3′, 5′-CCCTGGGATTGGTCCATA-3′).
All animal experiments were approved by the Animal Research Committee of Kyoto University and were conducted according to the Regulations on Animal Experimentation of Kyoto University.
RT-PCR and direct sequencing
Total RNA was isolated from the brain of 5-week-old animals using ISOGEN (NIPPON GENE, Tokyo, Japan). RT-PCR and direct sequencing of the PCR products were carried out as described previously [32].
Transgenic rescue and recombinant BAC transgenics
A construct containing the CMV promoter, 1.45-kb of the Mrs2 coding sequence, and SV40 polyA signal was excised from the vector (pCMV-Script; Agilent Technologies, CA, USA) and used as a transgene, which was microinjected into the pronuclei of fertilized oocytes collected from Crj:Wistar rats. Transgenic offspring founder rats were then crossed with WTC- +/dmy rats and then backcrossed again to WTC- +/dmy rats to obtain dmy/dmy homozygous and also hemizygous for the transgene (dmy/dmy, tg/-). Expression of the transgene was confirmed by RT-PCR with primers (5′-GCGAATGGAGATCCAATTTT-3′, 5′-GGGAGGTGTGGGAGGTTTT-3′) to detect SV40 polyA sequence. Brain RNA was treated with DNase I (New England BioLabs) to remove contaminating genomic DNA and then subjected to cDNA synthesis.
A rat BAC clone, CHORI-230-9K13, including the rat Mrs2 gene was modified to express MR2SL-EGFP fusion protein under the endogenous promoter by ET recombination technology [33]. Modified genomic DNA was excised from the vector and then used for in ovo transgenesis.
Quantitative PCR
Real-time PCR was performed using the Thermal Cycler Dice Real Time System (Takara Bio Inc., Otsu, Japan) with SYBR Premix Ex Taq II (Takara Bio Inc., Otsu, Japan). By monitoring amplification curves of a test sample and reference samples that contained 101–106 molecules of the gene of interest, the number of target molecules in the test sample was analyzed. The number of target molecules was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (Gapdh) as an internal control. The primers used are as follows: 5′-GCTGTGGCAGCTACCTATGTCTTG-3′ and 5′-AGGTCGTCATCATCCCACGAG-3′ for the rat Interleukin-1b (Il1b), 5′-CCACTTCACAAGTCGGAGGCTTA-3′ and 5′-GTGCATCATCGCTGTTCATACAATC-3′ for the rat interleukin-6 (Il6), 5′-GGCACAGTCAAGGCTGAGAATG-3′ and 5′-ATGGTGGTGAAGACGCCAGTA-3′ for rat Gapdh.
Electron microscopy
Perfusion fixation through the left ventricle was conducted with 4% paraformaldehyde in 0.1 M phosphate buffer (PB). Brains and spinal cords were dissected and stored in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M PB, then post-fixed with 2% osmic acid for 2 hours and embedded in epoxy resin. Ultrathin sections were double-stained with uranyl acetate and lead citrate and examined by a Hitachi H-7500 electron microscope (Hitachi, Tokyo, Japan).
Immunohistochemistry
Immunohistochemistry was performed as described previously [9]. The following primary antibodies were used: monoclonal anti-2′, 3′-cyclic nucleotide-3′-phosphodiesterase (CNPase) for oligodendrocytes (1∶1,000; Sigma, St. Louis, MO, USA), monoclonal anti-mitochondria (1∶100; Abcam, Cambridge, MA, USA), polyclonal anti-GFAP for astrocytes (1∶1,000; Dako, Carpinteria, CA, USA), polyclonal anti-Iba1 for microglia/macrophages (1∶200; Wako Pure Chemical Industries, Osaka, Japan). Cy3-conjugated anti-mouse IgG (1∶500; Jackson Laboratories) or Alexa 588-conjugated anti-rabbit IgG (1∶500; Molecular Probes) antibody was reacted. Nuclei were counterstained with DAPI (Vactor Laboratories). Signals were detected with a fluorescence microscopy (Olympus, Tokyo, Japan) or a confocal imaging system (C1Si; Nikon, Tokyo, Japan).
For immunoelectron microscopy, PFA-perfused frozen sections were incubated with rabbit antibody against fluorescent protein (1∶2,000; Molecular Probes) at 4 °C overnight. After washing in PBS, peroxidase-conjugated anti-rabbit IgG Fab fraction (Jackson Laboratories, 1∶1,000) and immunoreactions were reacted 3,3-diaminobenzidine substrate kit (Vector Laboratories), postfixed in 1% osmium tetraoxide, dehydrated in graded ethanol, and then embedded in epoxy resin. Ultrathin sections were examined by electron microscopy (Hitachi, Tokyo, Japan).
Lactic acid measurements
Cerebrospinal fluid was collected from dmy/dmy, wild-type littermates, and dmy/dmy with the normal Mrs2 transgene at 6–7 weeks of age under isoflurane anesthesia. They were then mixed with 0.8N perchloric acid to inactivate proteins. After centrifugation, lactic acid concentrations of the supernatants were measured by Determiner LA (KYOWA MEDEX Co., Ltd., Tokyo, Japan).
Cytochrome oxidase histochemistry
Frozen spinal cord sections were prepared. Then, 100 µl of freshly prepared reaction buffer [50 mM Tris/HCl (pH 7.4), 0.5 mg/ml diaminobenzidine, 20 µg/ml catalase and 0.50 mg/ml cytochrome C] was added to each section and slides were incubated for 30 min at 37°C.
ATP measurements
Rats were sacrificed by cervical dislocation and the brains were immediately excised, frozen in liquid nitrogen, and stored at −80°C until measurement. In order to release cellular ATP, frozen tissue (25 mg) was boiled for 2 min after the addition of 300 µl water containing 100 mM Tris/HCl (pH 7.75) and 4 mM EDTA. Samples were placed on ice and homogenized by sonification (micro tip, 1 s ×10 pulse). ATP concentrations were determined using the ATP bioluminescence assay kit HS II (Roche) according to the manufacturer's protocol. Data were standardized to the protein concentration which was determined by Coomassie Plus – the better Bradford assay kit (Pierce).
Statistical analysis
Statistical differences in lactic acid, ATP and mRNA expressions between wild-type and dmy/dmy rats were evaluated using the Mann-Whitney U test.
Supporting Information
Detection of the Mrs2dmy mutation. A. Chromatograms showing the Mrs2dmy G-to-A mutation. Upper: wild-type genome. Lower: Mrs2dmy/Mrs2dmy genome. The Mrs2dmy mutation disrupted AciI restriction site (GGCG) in the Mrs2dmy/Mrs2dmy genome. B. Molecular diagnosis of the Mrs2dmy mutation. In the wild type, the 349-bp PCR product amplified with primers rMrs2-31&32 (5′-AAAGTTTGACAAAGAAGGAAACG-3′ and 5′-GGGGATGGAGGGCTATGTAA-3′) is digested with AciI but not in Mrs2dmy/Mrs2dmy mutant rats. M: ΦX174-HaeIII digests.
(1.15 MB TIF)
Transgenic rescue experiment. A. Expression of the transgene in the brain of a transgenic rat. Brain cDNA from Tg-positive rats (Lanes 2 and 3) and Tg-negetive rats (Lanes 1and 4) was used as templates. Brain RNA was treated with DNaseI to remove contaminating genomic DNA. M: ΦX174 HaeIII digests. B. Histopathology of the cervical part of the spinal cord of dmy/dmy transgene-negative rats (left) and dmy/dmy transgene-positive (right) rats aged 10 weeks. Luxol fast blue-HE staining. Original magnification: ×100. C. Lactic acid concentration in cerebrospinal fluid of 6-7-week-old dmy/dmy rats and age-matched dmy/dmy Mrs2 cDNA-transgenic rats. Elevated lactic acid (126 ± 43.7 mg/dL) was reduced to normal level (22 ± 3.1 mg/dL). **, P < 0.002. D. Electron microphotograph of an oligodendrocyte in a dmy/dmy transgene-positive rat. Densely packed mitochondria (arrowheads) were found in the cytoplasm. Bar: 2μm.
(5.52 MB TIF)
MRS2 expression in the CNS of Mrs2-GFP recombinant BAC transgenic rats. MRS2 signals were mainly found in neurons (A), and occasionally in GFAP-positive astrocytes (B) and CNP-positive oligodendrocytes (C). Left: Bar: 50 μm. Center, Right: Bar: 20 μm.
(3.15 MB TIF)
MRS2 expression in Mrs2-GFP recombinant BAC transgenic rats. MRS2 signals were observed in the myocardium (A), liver (B), testis (C) and skeletal muscles (D). Bar: 50 μm.
(6.29 MB TIF)
Sequence alignment of yeast, human, and rat MRS2 proteins. Predicted transmembrane domains (TM-1, TM-2) are boxed; * indicates identical residues; : indicates conservative substitution; . indicates semiconservative substitutions. The sequence of a motif conserved in all putative magnesium transporters, G-M-N, is indicated in bold. Predicted coiled-coil regions are underlined, five regions with conserved amino acid residues (CRB-1-5; conserved residue block) are shaded grey. Arrowhead: The position of the residue affected by the dmy mutation, after which the 15 additional residues follow in mutant MRS2.
(1.38 MB TIF)
Acknowledgments
The authors are grateful to M. Yokoe for excellent technical assistance.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by grants-in-aid for Scientific Research from the Japan Society for the Promotion of Science [21300153 to TK] and a grant-in-aid for Cancer Research from the Ministry of Health, Labour, and Welfare. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Werner H, Jung M, Klugmann M, Sereda M, Griffiths IR, et al. Mouse models of myelin diseases. Brain Pathol. 1998;8:771–793. doi: 10.1111/j.1750-3639.1998.tb00200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffiths IR. Myelin mutants: model systems for the study of normal and abnormal myelination. Bioessays. 1996;18:789–797. doi: 10.1002/bies.950181005. [DOI] [PubMed] [Google Scholar]
- 3.Meyer Zu Horste G, Nave KA. Animal models of inherited neuropathies. Curr Opin Neurol. 2006;19:464–473. doi: 10.1097/01.wco.0000245369.44199.27. [DOI] [PubMed] [Google Scholar]
- 4.Kuramoto T, Sotelo C, Yokoi N, Serikawa T, Gonalons Sintes E, et al. A rat mutation producing demyelination (dmy) maps to chromosome 17. Mamm Genome. 1996;7:890–894. doi: 10.1007/s003359900263. [DOI] [PubMed] [Google Scholar]
- 5.Kitada K, Guenet JL, Serikawa T. Determination of the mouse homologous region for the rat dmy locus. J Exp Anim Sci. 2000;41:40–43. [Google Scholar]
- 6.Schindl R, Weghuber J, Romanin C, Schweyen RJ. Mrs2p forms a high conductance Mg2+ selective channel in mitochondria. Biophys J. 2007;93:3872–3883. doi: 10.1529/biophysj.107.112318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregan J, Bui DM, Pillich R, Fink M, Zsurka G, et al. The mitochondrial inner membrane protein Lpe10p, a homologue of Mrs2p, is essential for magnesium homeostasis and group II intron splicing in yeast. Mol Gen Genet. 2001;264:773–781. doi: 10.1007/s004380000366. [DOI] [PubMed] [Google Scholar]
- 8.Schock I, Gregan J, Steinhauser S, Schweyen R, Brennicke A, et al. A member of a novel Arabidopsis thaliana gene family of candidate Mg2+ ion transporters complements a yeast mitochondrial group II intron-splicing mutant. Plant J. 2000;24:489–501. doi: 10.1046/j.1365-313x.2000.00895.x. [DOI] [PubMed] [Google Scholar]
- 9.Kuwamura M, Kanehara T, Tokuda S, Kumagai D, Yamate J, et al. Immunohistochemical and morphometrical studies on myelin breakdown in the demyelination (dmy) mutant rat. Brain Res. 2004;1022:110–116. doi: 10.1016/j.brainres.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Kolisek M, Zsurka G, Samaj J, Weghuber J, Schweyen RJ, et al. Mrs2p is an essential component of the major electrophoretic Mg2+ influx system in mitochondria. Embo J. 2003;22:1235–1244. doi: 10.1093/emboj/cdg122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piskacek M, Zotova L, Zsurka G, Schweyen RJ. Conditional knockdown of hMRS2 results in loss of mitochondrial Mg2+ uptake and cell death. J Cell Mol Med. 2009;13:693–700. doi: 10.1111/j.1582-4934.2008.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiesenberger G, Waldherr M, Schweyen RJ. The nuclear gene MRS2 is essential for the excision of group II introns from yeast mitochondrial transcripts in vivo. J Biol Chem. 1992;267:6963–6969. [PubMed] [Google Scholar]
- 13.Devivo DC. The expanding clinical spectrum of mitochondrial diseases. Brain Dev. 1993;15:1–22. doi: 10.1016/0387-7604(93)90002-p. [DOI] [PubMed] [Google Scholar]
- 14.Huttemann M, Zhang Z, Mullins C, Bessert D, Lee I, et al. Different proteolipid protein mutants exhibit unique metabolic defects. ASN Neuro. 2009;1 doi: 10.1042/AN20090028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thambisetty M, Newman NJ. Diagnosis and management of MELAS. Expert Rev Mol Diagn. 2004;4:631–644. doi: 10.1586/14737159.4.5.631. [DOI] [PubMed] [Google Scholar]
- 16.Bui DM, Gregan J, Jarosch E, Ragnini A, Schweyen RJ. The bacterial magnesium transporter CorA can functionally substitute for its putative homologue Mrs2p in the yeast inner mitochondrial membrane. J Biol Chem. 1999;274:20438–20443. doi: 10.1074/jbc.274.29.20438. [DOI] [PubMed] [Google Scholar]
- 17.Zsurka G, Gregan J, Schweyen RJ. The human mitochondrial Mrs2 protein functionally substitutes for its yeast homologue, a candidate magnesium transporter. Genomics. 2001;72:158–168. doi: 10.1006/geno.2000.6407. [DOI] [PubMed] [Google Scholar]
- 18.Eshaghi S, Niegowski D, Kohl A, Martinez Molina D, Lesley SA, et al. Crystal structure of a divalent metal ion transporter CorA at 2.9 angstrom resolution. Science. 2006;313:354–357. doi: 10.1126/science.1127121. [DOI] [PubMed] [Google Scholar]
- 19.Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 20.Hung PC, Wang HS. A previously undescribed leukodystrophy in Leigh syndrome associated with T9176C mutation of the mitochondrial ATPase 6 gene. Dev Med Child Neurol. 2007;49:65–67. doi: 10.1017/s0012162207000163.x. [DOI] [PubMed] [Google Scholar]
- 22.Spinazzola A, Viscomi C, Fernandez-Vizarra E, Carrara F, D'Adamo P, et al. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat Genet. 2006;38:570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- 23.Zafeiriou DI, Koletzko B, Mueller-Felber W, Paetzke I, Kueffer G, et al. Deficiency in complex IV (cytochrome c oxidase) of the respiratory chain, presenting as a leukodystrophy in two siblings with Leigh syndrome. Brain Dev. 1995;17:117–121. doi: 10.1016/0387-7604(94)00098-i. [DOI] [PubMed] [Google Scholar]
- 24.Andrews HE, Nichols PP, Bates D, Turnbull DM. Mitochondrial dysfunction plays a key role in progressive axonal loss in Multiple Sclerosis. Med Hypotheses. 2005;64:669–677. doi: 10.1016/j.mehy.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Mahad DJ, Ziabreva I, Campbell G, Lax N, White K, et al. Mitochondrial changes within axons in multiple sclerosis. Brain. 2009;132:1161–1174. doi: 10.1093/brain/awp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez MF, Kristal BS, Chernokalskaya E, Lazarev A, Shestopalov AI, et al. High-throughput profiling of the mitochondrial proteome using affinity fractionation and automation. Electrophoresis. 2000;21:3427–3440. doi: 10.1002/1522-2683(20001001)21:16<3427::AID-ELPS3427>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 27.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dheen ST, Kaur C, Ling EA. Microglial activation and its implications in the brain diseases. Curr Med Chem. 2007;14:1189–1197. doi: 10.2174/092986707780597961. [DOI] [PubMed] [Google Scholar]
- 29.Merrill JE, Scolding NJ. Mechanisms of damage to myelin and oligodendrocytes and their relevance to disease. Neuropathol Appl Neurobiol. 1999;25:435–458. doi: 10.1046/j.1365-2990.1999.00200.x. [DOI] [PubMed] [Google Scholar]
- 30.Mitrovic B, Ignarro LJ, Montestruque S, Smoll A, Merrill JE. Nitric oxide as a potential pathological mechanism in demyelination: Its differential effects on primary glial cells in vitro. Neuroscience. 1994;61:575–585. doi: 10.1016/0306-4522(94)90435-9. [DOI] [PubMed] [Google Scholar]
- 31.Serikawa T, Kuramoto T, Hilbert P, Mori M, Yamada J, et al. Rat gene mapping using PCR-analyzed microsatellites. Genetics. 1992;131:701–721. doi: 10.1093/genetics/131.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuramoto T, Kitada K, Inui T, Sasaki Y, Ito K, et al. Attractin/mahogany/zitter plays a critical role in myelination of the central nervous system. Proc Natl Acad Sci U S A. 2001;98:559–564. doi: 10.1073/pnas.98.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detection of the Mrs2dmy mutation. A. Chromatograms showing the Mrs2dmy G-to-A mutation. Upper: wild-type genome. Lower: Mrs2dmy/Mrs2dmy genome. The Mrs2dmy mutation disrupted AciI restriction site (GGCG) in the Mrs2dmy/Mrs2dmy genome. B. Molecular diagnosis of the Mrs2dmy mutation. In the wild type, the 349-bp PCR product amplified with primers rMrs2-31&32 (5′-AAAGTTTGACAAAGAAGGAAACG-3′ and 5′-GGGGATGGAGGGCTATGTAA-3′) is digested with AciI but not in Mrs2dmy/Mrs2dmy mutant rats. M: ΦX174-HaeIII digests.
(1.15 MB TIF)
Transgenic rescue experiment. A. Expression of the transgene in the brain of a transgenic rat. Brain cDNA from Tg-positive rats (Lanes 2 and 3) and Tg-negetive rats (Lanes 1and 4) was used as templates. Brain RNA was treated with DNaseI to remove contaminating genomic DNA. M: ΦX174 HaeIII digests. B. Histopathology of the cervical part of the spinal cord of dmy/dmy transgene-negative rats (left) and dmy/dmy transgene-positive (right) rats aged 10 weeks. Luxol fast blue-HE staining. Original magnification: ×100. C. Lactic acid concentration in cerebrospinal fluid of 6-7-week-old dmy/dmy rats and age-matched dmy/dmy Mrs2 cDNA-transgenic rats. Elevated lactic acid (126 ± 43.7 mg/dL) was reduced to normal level (22 ± 3.1 mg/dL). **, P < 0.002. D. Electron microphotograph of an oligodendrocyte in a dmy/dmy transgene-positive rat. Densely packed mitochondria (arrowheads) were found in the cytoplasm. Bar: 2μm.
(5.52 MB TIF)
MRS2 expression in the CNS of Mrs2-GFP recombinant BAC transgenic rats. MRS2 signals were mainly found in neurons (A), and occasionally in GFAP-positive astrocytes (B) and CNP-positive oligodendrocytes (C). Left: Bar: 50 μm. Center, Right: Bar: 20 μm.
(3.15 MB TIF)
MRS2 expression in Mrs2-GFP recombinant BAC transgenic rats. MRS2 signals were observed in the myocardium (A), liver (B), testis (C) and skeletal muscles (D). Bar: 50 μm.
(6.29 MB TIF)
Sequence alignment of yeast, human, and rat MRS2 proteins. Predicted transmembrane domains (TM-1, TM-2) are boxed; * indicates identical residues; : indicates conservative substitution; . indicates semiconservative substitutions. The sequence of a motif conserved in all putative magnesium transporters, G-M-N, is indicated in bold. Predicted coiled-coil regions are underlined, five regions with conserved amino acid residues (CRB-1-5; conserved residue block) are shaded grey. Arrowhead: The position of the residue affected by the dmy mutation, after which the 15 additional residues follow in mutant MRS2.
(1.38 MB TIF)