Abstract
Trinucleotide expansions cause disease by both protein- and RNA-mediated mechanisms. Unexpectedly, we discovered that CAG expansion constructs express homopolymeric polyglutamine, polyalanine, and polyserine proteins in the absence of an ATG start codon. This repeat-associated non-ATG translation (RAN translation) occurs across long, hairpin-forming repeats in transfected cells or when expansion constructs are integrated into the genome in lentiviral-transduced cells and brains. Additionally, we show that RAN translation across human spinocerebellar ataxia type 8 (SCA8) and myotonic dystrophy type 1 (DM1) CAG expansion transcripts results in the accumulation of SCA8 polyalanine and DM1 polyglutamine expansion proteins in previously established SCA8 and DM1 mouse models and human tissue. These results have implications for understanding fundamental mechanisms of gene expression. Moreover, these toxic, unexpected, homopolymeric proteins now should be considered in pathogenic models of microsatellite disorders.
Keywords: bidirectional expression, neurodegeneration, translation initiation
Translation of mRNA into protein is an exquisitely regulated, almost error-free process. Well-established rules of translational initiation have been used as a cornerstone in biology to understand gene expression and to predict the consequences of disease-causing mutations (1). For microsatellite expansion disorders, mutations within or outside ATG-initiated ORFs are thought to cause disease either by protein gain-of-function, protein loss-of-function, or RNA gain-of-function mechanisms (2, 3).
Microsatellite expansion mutations that express polyglutamine (polyGln) expansion proteins include Huntington disease (Huntingtin, HD), spinal bulbar muscular atrophy, and spinocerebellar ataxia types 1, 2, 3, 6, 7, and 17. Since the discovery of these CAG·CTG expansion mutations, efforts to understand disease mechanisms have focused on elucidating the molecular effects of the polyGln proteins expressed from these loci. Although these polyGln expansion proteins bear no similarity to each other apart from the polyGln tract, a hallmark of these diseases is protein accumulation and aggregation in nuclear or cytoplasmic inclusions. Surprisingly, although the polyGln expansion proteins are widely expressed in the CNS and other tissues, only restricted populations of neurons are vulnerable in each disease (3).
Myotonic dystrophy type 1 (DM1) and type 2 (DM2) are the best-characterized examples of RNA-mediated expansion disorders (2). The mutation causing DM1 is a CTG-repeat expansion located in the 3′ UTR of the dystrophia myotonica-protein kinase (DMPK) gene. Although DM1 can be clinically more severe than DM2, the discovery of the DM2 mutation and several mouse models provide strong support that many features of these diseases result from RNA gain-of-function effects in which the dysregulation of RNA-binding proteins is mediated by the expression of CUG and CCUG transcripts (4). Additionally, RNA gain-of-function effects have been reported for CGG and CAG expansion RNAs (5, 6).
Both RNA and protein mechanisms appear to operate in spinocerebellar ataxia type 8 (SCA8) because the CTG·CAG expansion mutation is expressed in both the CUG (ataxin 8 opposite strand, AXN8OS) and CAG (ataxin 8, ATXN8) directions. ATXN8 expansion transcripts express polyGln protein from an ATG-initiated ORF, and both polyGln protein (7) and AXN8OS CUGEXP transcripts (8) accumulate in affected cells.
Results
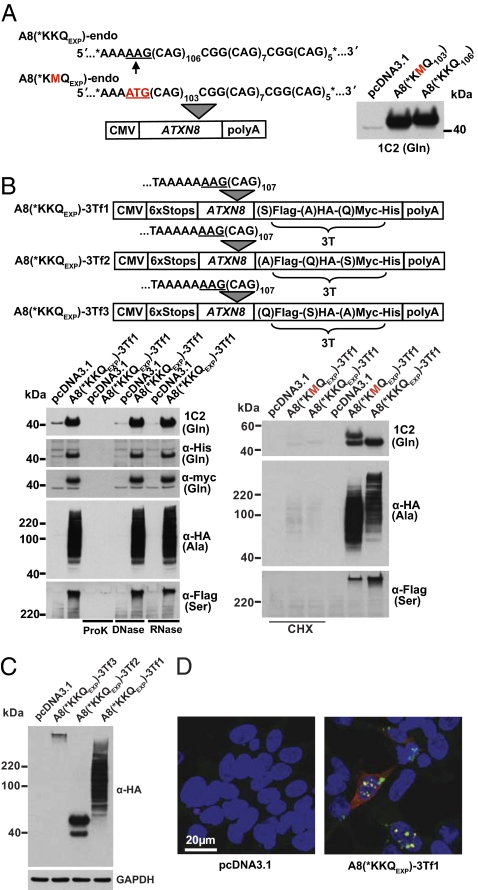
To understand the role of the ATXN8 polyGln protein in SCA8, we mutated the only ATG initiation codon located 5′of the CAG expansion on an ATXN8 (A8) minigene. Unexpectedly, we found this mutation did not prevent expression of the polyGln expansion protein in transfected cells (Fig. 1A). Sequence analysis showed that neither full-length nor spliced transcripts, which are expressed at approximately equal ratios from the (−)ATG minigene, are predicted to contain an AUG initiation codon (Fig. S1A). To test if non-ATG translation also could occur in other frames, a triply-tagged A8 minigene, A8(*KKQexp)-3Tf1, was generated by inserting a 6X STOP codon cassette (two stops in each frame) upstream of the CAGexp and three different C-terminal tags to monitor protein expression in all frames [i.e., CAG glutamine (Gln); AGC serine (Ser); and GCA alanine (Ala)] (Fig. 1B). Surprisingly, although transcripts generated from this tagged construct were confirmed to lack initiator AUG codons (Fig. S1B) by RT-PCR, tagged polyGln, polyAla, and polySer proteins were expressed (Fig. 1B) in transfected cells.
Fig. 1.
Non-ATG translation in HEK293T cells. (A) Immunoblot (Right) from A8-transfected cells (Left) with or without the ATG start codon. (B) (Upper) Modified A8 constructs with the 6X STOP cassette and 3′ tags in each frame [A8(*KKQEXP)-3Tf1]. (Lower) Immunoblots of lysates with or without proteinase K, DNase I, RNase I (Left) and of cells treated with cycloheximide (CHX) (Right). An ATG start codon in the polyGln frame variably results in the generation of a second, higher molecular weight band, and this sequence change also can affect polyAla migration and relative levels of polySer. (C) Immunoblots of A8(*KKQEXP)-3Tf1, A8(*KKQEXP)-3Tf2, and A8(*KKQEXP)-3Tf3 lysates show that polyAla, polyGln, and polySer are expressed at relatively high, intermediate, and low levels, respectively. The additional lower molecular weight polyGln band expressed from A8(*KKQEXP)-3Tf2 is caused by sequence heterogeneity after repeat contraction in Escherichia coli. (D) IF staining of polyGln (α-His/cy3), polyAla (α-HA/cy5), and polySer (α-FLAG/FITC).
The polyGln expansion proteins migrated at one or more discrete molecular weights, polyAla as a high molecular weight smear with a faint laddering pattern seen on light exposures, and polySer at the top of polyacrylamide gels in SDS (Fig. 1B) or 8 M urea (Fig. S2A). As expected, these proteins were degraded by proteinase K, were not affected by RNase I or DNase I, and were not detected with addition of cycloheximide (Fig. 1B). The relative levels of polyGln expressed with and without an ATG codon are similar (Fig 1 A and B). To compare the relative levels of the polyGln, polyAla, and polySer, each was tagged with the same HA epitope. The protein blot shown in Fig. 1C indicates that polyAla is expressed at the highest levels, followed by polyGln and then polySer. Immunofluorescence (IF) shows these proteins can be expressed simultaneously in a single cell and that relative levels in individual cells can vary dramatically (Fig. 1D and Fig. S2B). Consistent with previous reports, the polyGln protein is localized primarily within the nuclear aggregates (3), the polyAla protein is primarily diffuse when in the cytoplasm and aggregated when nuclear (9, 10), and the polySer protein forms both nuclear and cytoplasmic aggregates (9). Additionally, an ATG start codon in the polyGln frame variably resulted in an additional higher molecular weight band, suggesting that translational initiation occurs at the ATG and one or more additional sites in some sequence contexts [compare polyGln for A8(*KMQEXP)-endo and A8(*KMQEXP)-3Tf1 (Fig. 1 A and B)].
To determine if this repeat-associated non-ATG (RAN) translation is affected by sequence context, we modified the A8(*KKQEXP)-3Tf1 construct by removing 90 bp of the ATXN8 sequence so that the 6X STOP cassette was almost adjacent to the CAGEXP and by adding a seventh STOP immediately upstream of the polyGln, polyAla, or polySer frames (Fig. S3). These constructs also expressed polyGln and polyAla but only low levels of polySer, with the exception that a TAG stop immediately preceding the glutamine frame prevented translation of polyGln but not of polyAla or polySer.
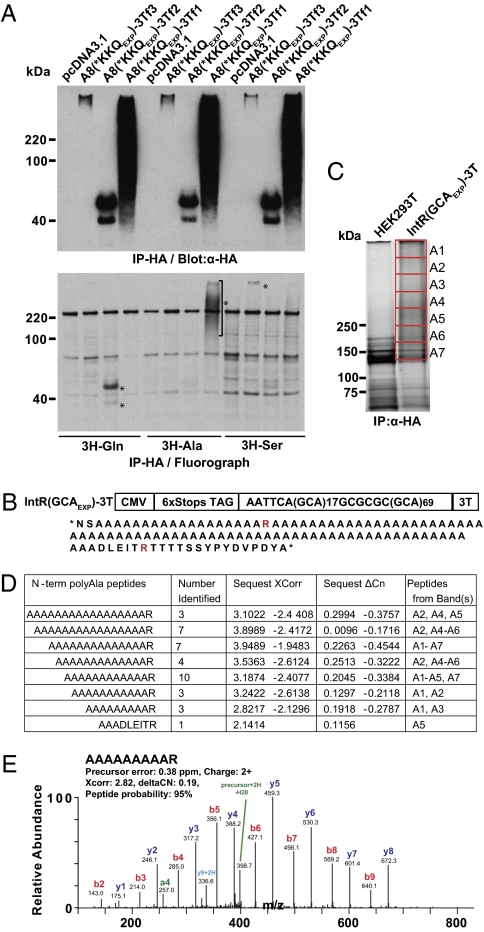
Because these results were completely unexpected, we used several approaches to establish the identity of these homopolymeric proteins. First, each protein was detected with one or more C-terminal epitope tags (myc, His, and HA for Gln; HA for Ala; and HA and Flag for Ser), and the polyGln protein was detected with a monoclonal antibody (1C2) specific to polyGln expansion tracts (Fig. 1) (11). Second, [3H]-Gln, [3H]-Ala, and [3H]-Ser were preferentially incorporated into proteins immunoprecipitated with tags in the polyGln, polyAla, and polySer frames, respectively (Fig. 2A). HEK293T cells transfected with triple-tagged constructs containing the HA-tag in the Ala [A8(*KKQEXP)-3Tf1], Gln [A8(*KKQEXP)-3Tf2], or Ser [A8(*KKQEXP)-3Tf3] frames were grown with [3H]-Gln, [3H]-Ala, or [3H]-Ser amino acids, respectively. Immunoprecipitations were performed using α-HA antibody, separated by PAGE on duplicate gels and detected by either immunoblot or fluorography. Fig. 2A Upper shows that all three proteins in each set were pulled down by immunoprecipitation, and the corresponding fluorograph (Fig. 2A Lower) shows that [3H]-Gln was preferentially incorporated into the ∼40-kDa proteins with the HA tag in the polyGln frame. Similarly, [3H]-Ala, and [3H]-Ser were preferentially incorporated into proteins immunoprecipitated with tags in the polyAla and polySer reading frames, respectively.
Fig. 2.
Protein labeling and MS. (A) Protein blot (Upper) and fluorograph (Lower) of [3H]-Gln, [3H]-Ala, or [3H]-Ser after immunoprecipitation with α-HA of cells transfected with A8(*KKQEXP)-3Tf1, A8(*KKQEXP)-3Tf2, A8(*KKQEXP)-3Tf3, or empty vector. (B) (Upper) Diagram of IntR(GCAEXP)-3T construct with CGCGCG interruption. (Lower) Predicted sequence of polyAla protein with arginine interruption. (C) Preparative deep-purple gel showing slices digested with trypsin for MS. (D) N-terminal polyAla peptides identified by MS with varying numbers of alanine. (E) Representative spectrum of N-terminal peptide AAAAAAAAAR with matched b-ions (red) and y-ions (blue).
Third, we used MS to confirm that RAN translation results in the expression of a polyAla expansion protein. PolyAla was selected for MS because (i) polyAla-specific antibodies are not available, and (ii) the putative polyAla protein is expressed at sufficiently high levels required for MS in transfected cells. An arginine residue was introduced into the recombinant protein so that trypsin digestion of the N terminus would generate smaller peptide fragments of suitable size for MS (Fig. 2B). IntR(GCAEXP)-3T lysates were separated by PAGE, and MS was performed on proteins isolated from gel slices A1–A7 (Fig. 2C). Associated mass spectra were submitted for database searching against a human protein database and a list of all possible polyAla proteins in which translation could begin before or within the repeat and for which initiation would allow the possible inclusion of an N-terminal methionine residue. MS/MS identified a series of N-terminal peptides with varying numbers of alanines [(A)9–17R and AAADLEITR] (Fig. 2 D and E). No peptides containing N-terminal methionine were detected, suggesting that translation initiation in cells occurs without incorporating an N-terminal methionine or that it is removed rapidly by methionine aminopeptidase or endopeptidase activity (12). Additionally, the predicted C-terminal digestion fragment (TTTTSSYPYDVPDYA) was identified (Fig. S4). In summary, these results demonstrate that RAN translation results in polyAla expression in transfected cells and that these proteins run as a broad smear on SDS-PAGE.
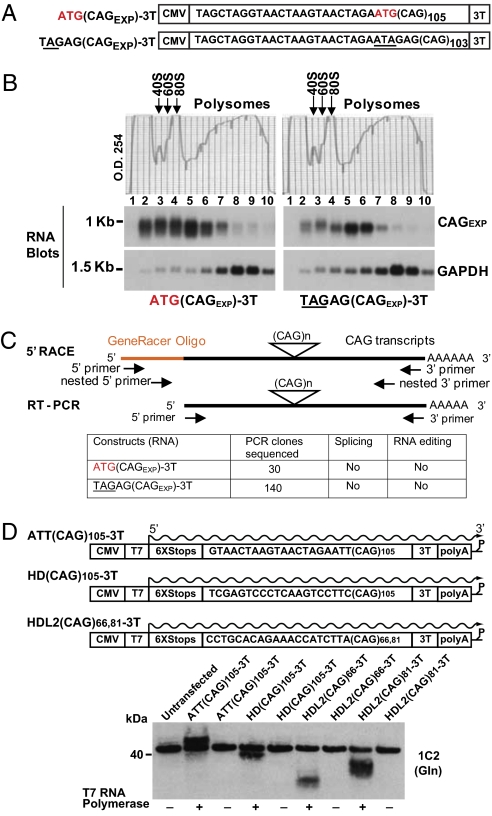
To test if transcripts undergoing RAN translation are modified or edited, resulting in the introduction of AUG initiation codons, we isolated and characterized mRNA from actively translating polyribosomes isolated from cells transfected with (CAGEXP)-3T constructs with or without an ATG initiation codon (Fig. 3A). Northern analysis showed that transcripts expressed from both the (+)ATG and (−)ATG constructs migrated at the predicted size (∼700 nt) and cosedimented with light polyribosomal fractions (Fig. 3B). Further characterization of ribosome-bound CAGEXP transcripts from fraction six by 5′ RACE and RT-PCR show that (i) transcription initiated within a few bp of the predicted transcription start site and (ii) the sequence predicted by the DNA was found in the corresponding transcripts and that no upstream AUG initiation codons were introduced by RNA splicing or editing among 140 independently isolated clones (Fig. 3C). To rule out independently the possibility that +ATG transcripts might be generated from these plasmids by a cryptic promoter or alternative splicing, RNA transfections were performed. We transcribed RNA in vitro from the ATT(CAGEXP) construct and two additional linearized non-ATG constructs and transfected the RNA into cells. Consistent with previous results, RAN translation of polyGln protein also occurred with cell-free–generated RNA (Fig. 3D).
Fig. 3.
Polysome-associated transcripts and RNA transfections. (A) (CAGEXP)-3T constructs with or without an ATG initiation codon. (B) Polyribosome profiles from transfected cells. (Upper) OD 254. (Lower) RNA blots with relative levels of CAG transcripts. (C) (Upper) 5′ RACE and RT-PCR strategy used to characterize (+)AUG and (−)AUG transcripts from polysome fraction #6. (Lower) Table summarizing sequencing results from cloned RT-PCR products. (D) In vitro transcription and RNA transfection into HEK293T cells. (Upper) Constructs used to produce capped, polyadenylated mRNAs that extend from the T7 promoter to the PvuII site (P). (Lower) Immunoblots following RNA transfections.
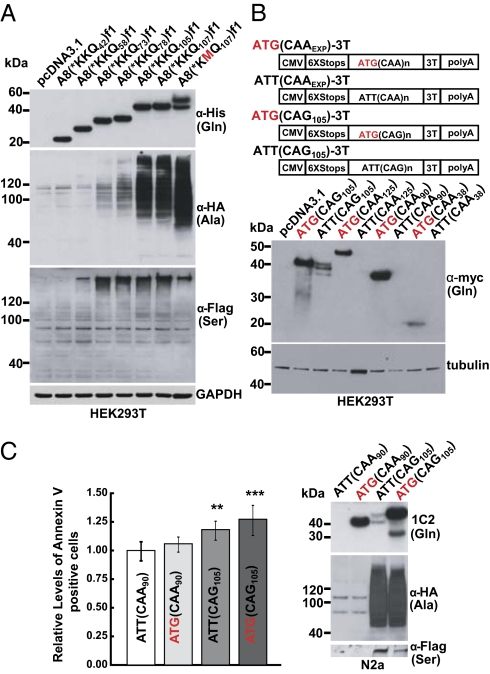
To test if RAN translation depends on repeat length, constructs containing 15–107 CAGs were examined. Constructs with 42–107 CAGs express polyGln by RAN translation, but constructs with 15 or 20 CAGs did not (Fig. 4A and Fig. S5 A and B). PolyAla was robustly expressed with 105 and 107 CAGs, moderately with 73 and 78 CAGs, and not with 42 and 58 CAGs. PolySer was detected with 58–107 repeats but not with 42 repeats (Fig. 4A). Thus, RAN translation is length dependent, and longer repeat tracts are associated with the simultaneous expression of multiple proteins.
Fig. 4.
Repeat length, sequence motif, and toxicity. (A) Immunoblots of lysates after transfection with A8(*KKQEXP)-3Tf1 or A8(*KMQEXP)-3Tf1 with varying repeat lengths. (B) (Upper) Triple-tagged constructs containing a CAA or CAG repeat tract with or without ATG. (Lower) Corresponding immunoblot. (C) (Left) Relative levels of annexin-V–positive N2a cells transfected with [ATT(CAG105)-3T, ATG(CAG105)-3T] compared with polyGln only [ATG(CAA90)-3T] and negative [ATT(CAA90)-3T] controls. (Right) Corresponding immunoblots. **P < 0.01; ***P < 0.001].
Because both repeat length and secondary hairpin structures are associated with CAG and several other disease-causing microsatellite expansions, we compared RAN translation of polyGln expansion proteins expressed from constructs containing hairpin-forming CAG and non–hairpin-forming CAA repeats. Cells transfected with CAG expansion constructs with or without ATG start codons expressed polyGln (Fig. 4B). In contrast, polyGln was expressed only from the CAA expansion constructs with an ATG start codon, suggesting that hairpin formation is required for RAN translation. All constructs were confirmed to express transcripts by RT-PCR (Fig. S5C). Because CUGEXP transcripts also form hairpin structures, we tested CTGEXP constructs and show that RAN translation also occurs in all three frames (polyleucine, polyAla, and polycystine) (Fig. S5D).
Next, we addressed if RAN-translation products trigger apoptosis and therefore could be implicated in disease pathogenesis. Murine neuroblastoma (N2a) cells transfected with ATT(CAG105)-3T and ATG(CAG105)-3T, which express polyGln, polyAla, and polySer, showed significant increases in annexin-V staining (13), compared with control cells (Fig. 4C and Fig. S5E). These results indicate that the products of RAN translation can cause apoptosis.
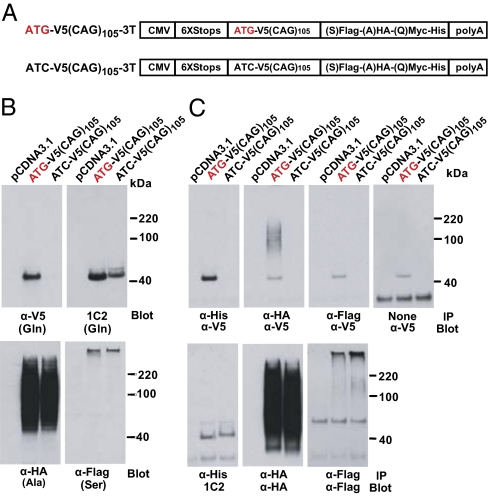
Because most disease-causing CAG·CTG expansions are found in larger polyGln ORFs, we tested if RAN translation in the polyAla and polySer frames would still occur in the presence of an ATG-initiated polyGln ORF (Fig. 5 A and B). PolyAla and polySer proteins were expressed irrespective of the polyGln ORF. The absence of the V5-tag on the polyGln from the (−)ATG V5 construct demonstrates RAN translation of polyGln initiated downstream of the V5 tag and close to, or within, the repeat. Immunoprecipitation using antibodies to 3′epitopes followed by immunoblot with α-V5 showed only a small fraction of polyAla protein has undergone frame shifting from the ATG-initiated V5-polyGln frame (Fig. 5C). Although previously frame shifting has been suggested to result in hybrid polyGln-polyAla and polyGln-polySer proteins in spinocerebellar ataxia type 3 (SCA3) and Huntington disease (14, 15), our data demonstrate that frame shifting is rare, and an out-of-frame ATG initiation codon is not required for polyAla or polySer expression.
The potential for RAN translation in other disease-relevant sequence contexts was investigated using constructs with 20 bp of 5′ flanking sequence upstream of the CAG repeat from the HD, Huntingtin-like 2 (HDL2), SCA3, or DM1 loci (Fig. S6A). These constructs showed robust polyGln and polyAla and variable polySer expression with the highest non-ATG polySer translation for A8(*KKQEXP) and HDL2 (Fig. S6B). RT-PCR confirmed that each construct expressed unspliced transcripts with ATG-initiated ORFs only in the glutamine and serine frames for A8(*KMQEXP) and DM1 constructs, respectively.
Fig. 5.
RAN translation in the presence of an ATG-initiated ORF. (A) Constructs with 5′ V5 tag in glutamine frame and 3′ tags. (B) Corresponding protein blots. (C) Protein blots after immunoprecipitation with antibodies to 3′ epitope tags in polyGln (α-His), polyAla (α-HA), and poly-Ser (α-Flag) frames probed for the 5′ epitope tag with α-V5 (Upper) or 1C2, α-HA, α-Flag (Lower).
To understand better the conditions required for expression of these homopolymeric expansion proteins, we performed lentiviral transductions of HEK293T cells and mouse brain. Similar to the transfections described above, RAN translation of polyGln and polyAla also occurs in lentiviral-transduced cells and intact mouse brain (Fig. S6 C–E). Taken together, these data demonstrate that RAN translation can occur when the transgene is integrated into the genome and that CAG expansions located in a variety of sequence contexts and under a variety of conditions can express homopolymeric expansion proteins in the absence of an ATG-start codon.
Next, rabbit reticulocyte lysates (RRLs) were used to test if non-ATG translation also occurs in a cell-free system. In contrast to cells, RAN translation in RRLs was limited. Only HDL2 produced polyGln without an ATG, none of the constructs generated detectable polyAla, and the highest levels of non-ATG–initiated polySer were from the HD and SCA3 constructs (Fig. S7A). Moreover, RAN translation in RRLs, but not in cells, is substantially affected by mutating previously reported alternative initiation codons (ATT and ATC) (16, 17) (Fig. S7 B–D), indicating that sequence requirements for RAN translation in RRLs are less permissive than in cells. Next, we used constructs that undergo non-ATG translation in RRLs to test if N-terminal methionine incorporation still occurs in the absence of an AUG initiation codon. We showed that polyGln expressed from constructs lacking initiator and internal methionine codons incorporated S35-methionine (Fig. S7E). Additionally, in vitro translation using S35-labelled Met-tRNAiMet indicates that in RRLs non-ATG translation is initiated with a tRNAiMet (Fig. S7F). The incorporation of an N-terminal methionine in RRLs in vitro is not surprising, because previously documented alternative initiation codons (ATT, ATC) are used. However, RAN translation in cells may use a different initiation mechanism, because we were not able to detect N-terminal methionine for any of the polyAla proteins using MS.
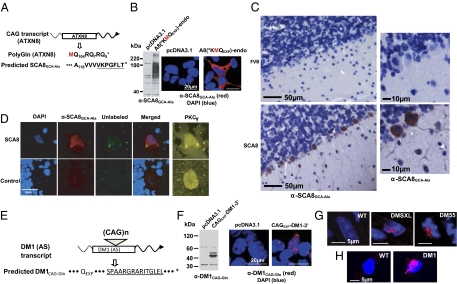
To determine if novel homopolymeric proteins are expressed in vivo, we developed peptide antibodies to putative RAN-translated SCA8-polyAla (SCA8GCA-Ala) and DM1 polyGln (DM1CAG-Gln) proteins (Fig. 6 A and E). The specificity of both the α-SCA8GCA-Ala and α-DM1CAG-Gln antibodies was demonstrated by Western blot and IF detection in cells expressing recombinant SCA8GCA-Ala and DM1CAG-Gln proteins but not in control cells (Fig. 6 B and F). Consistent with the possible role for RAN-translated proteins in SCA8, we detected α-SCA8GCA-Ala immunostaining in Purkinje cell soma and dendrites throughout the cerebellum in an established SCA8 mouse model (7) (Fig. 6C). Similarly, α-SCA8GCA-Ala staining was found reproducibly in the remaining cerebellar Purkinje cells of postmortem samples from two patients with SCA8 (e.g., Fig 6D). For myotonic dystrophy, IF staining of DM1 mice (18) which express CUGEXP (Fig. S8A) and CAGEXP transcripts (Fig. S8 B and C) show rare (2.32 ± 2.04%) but reproducible α-DM1CAG-Gln nuclear aggregates in cardiac myocytes (Fig. 6G and Fig. S9 A and B) and more frequent α-DM1CAG-Gln staining in leukocytes (10.68 ± 3.66%) (Fig. S9C). The DM1 polyGln aggregates colocalize with caspase-8 (Fig. S9D), which is an early indicator of polyGln-induced apoptosis (19). Immunohistochemical staining of paraffin-embedded tissue with the polyGln-specific 1C2 antibody confirms staining in leukocytes in cardiac tissue from mice containing a CTG expansion of 55 repeats but not in control mice with 20 CTG repeats (Fig. S9E). In samples from patients with myotonic dystrophy, α-DM1CAG-Gln inclusions were found at low frequency in myoblasts (50–70 CTG·CAG repeats) (Fig. 6H) and skeletal muscle and at higher frequency in blood (Fig. S10).
Fig. 6.
In vivo evidence for RAN-translated SCA8-polyAla and DM1-polyGln. (A) ATXN8 ATG-polyGln ORF and putative non-ATG SCA8GCA-Ala protein. Underlined peptides used for antibody generation. (B) Antibody validation: α-SCA8GCA-Ala detection of recombinant protein in A8(*KMQEXP)-endo transfected cells by protein blot (Left) and IF (Right). (C) Immunohistochemical staining of SCA8 and control mouse cerebellum (FVB) using α-SCA8GCA-Ala. (D) In SCA8 human samples, α-SCA8GCA-Ala antibody shows consistent and specific staining (red-cy3) of surviving human SCA8 cells but not control Purkinje cells. Colabeling with α-PKCγ antibody (cy5, yellow) independently stains Purkinje cells and confirms their presence in both samples. (E) (Upper) DM1 CAG antisense transcript. (Lower) Predicted non-ATG–initiated polyGln protein. Underlined peptide used for antibody generation. (F) Antibody validation: α-DM1CAG-Gln detects recombinant protein with endogenous DM1 polyGln C-terminus (CAGEXP-DM1-3’) by protein blot (Left) and IF (Right). (G) IF using α-DM1CAG-Gln (cy3, red) detects DM1CAG-Gln protein in cardiomyocytes of DM1 mice with 55 (DM55), and >1,000 (DMSXL) CTGs but not in cardiomyocytes of control (WT) mice. (H) Staining with α-DM1CAG-Gln (cy3, red) in DM1 but not in control (WT) myoblasts.
Discussion
Our understanding of the molecular basis of disease has been built on studying the expected effects of mutations on the functions of their corresponding genes. For microsatellite expansion disorders, cell culture and animal models have been developed to test specific hypotheses based on the prediction that CAGEXP mutations located in polyGln ORFs express protein only in the polyGln frame and that expansions located in noncoding regions do not encode proteins (2, 3). We demonstrate that these canonical rules of translation do not apply for CAG·CTG expansions and that in the absence of an ATG codon expanded CAG and CTG trinucleotide repeats often express homopolymeric expansion proteins in all three frames. RAN translation occurs in transfected and transduced HEK293T and N2a cells. In contrast, non-ATG translation is less frequent in RRLs, suggesting that RRLs may not recapitulate what is happening inside cells. The production of polyGln protein after RNA transfections in cells indicates that cellular factors and not promoter issues affect RAN translation.
While initiation at specific alternative codons has been reported previously (16, 20), our findings show translational initiation at CAG·CTG expansion sites in cells is highly permissive. Data showing that hairpin-forming CAG and CUG repeats undergo RAN translation are consistent with previous reports that hairpin structures affect translational initiation (20). Hairpin structures are thought to allow translational initiation at suboptimal sites by delaying the 40S ribosomal subunit long enough to allow efficient interaction between the Met-tRNAi anticodon and the AUG or non-AUG start site (21). Additionally, hairpins can recruit initiation factors and ribosomal subunits to internal ribosome entry sites (IRESs) (1). Some IRESs, such as the one in the cricket paralysis virus, can facilitate translation initiation without eIFs or tRNAiMet (1). These IRES hairpins function as a tRNAiMet to initiate translation at non-AUG codons including GCA (22). This function could explain why translation in the polyAla (GCA) frame appears to initiate at multiple sites within the repeat. IRES hairpins form and are stabilized with the help of IRES translation-associated factors (ITAFs) (1), and at least one ITAF (CUGBP1) is known to bind to CAG and CUG repeats (23). The apparent requirement for hairpins, the initiation from non-AUG codons, and the association of repeat transcripts with a known ITAF suggests that RAN translation may involve an IRES-like mechanism. Differences in repeat length required for the accumulation of polyGln, polyAla, and polySer proteins may reflect differences in protein stability and/or repeat length required for efficient initiation in each frame.
The discovery of RAN translation raises the possibility that polyAla and polySer proteins contribute to the pathogenesis of some of CAG polyGln diseases and that homopolymeric proteins contribute to diseases previously thought to involve primarily RNA gain of function (e.g., DM1). In SCA8, specific staining for the SCA8GCA-Ala expansion protein is found in cerebellar Purkinje cells. In DM1, staining for the DM1CAG-Gln expansion protein is found in heart, skeletal muscle, and myoblasts. Further investigation will be required to determine which microsatellite expansions undergo RAN translation and which RAN-translated proteins contribute to disease. Our results indicate polyAla and polySer proteins are more likely to be expressed from CAG·CTG expansions exceeding 70 repeats, suggesting the possibility that RAN-translated proteins may contribute to the anticipation, the earlier onset, and increased disease severity associated with longer repeat lengths.
Recently, much of the genome (24) and a growing number of expansion mutations (25) have been shown to be transcribed bidirectionally. Given that CAGEXP and CUGEXP transcripts can express proteins without an ATG, and that both expansion transcripts are reported to cause RNA gain-of-function effects (6, 25), the molecular pathology of microsatellite disorders may be far more complex than currently appreciated. Additionally, these results raise the possibility that other repetitive sequences in the genome also undergo RAN translation and contribute to proteome diversity.
Materials and Methods
Details of cloning, custom antibodies, and molecular techniques are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank patient families for participating; G. Nelsestuen, L. Anderson, L. A. Higgens, C. Pearson, H. Orr, and N. Sonenberg for helpful discussions; J. Day (University of Minnesota, Minneapolis; AR057220) for providing tissue (P30 AR057220); J. Henriksen and the University of Minnesota BioNet Core for providing tissue (P30 CA77598 and P50 CA101955) and digital imaging; the National Institute of Neurological Disorders and Stroke for providing tissue (P01NS058901 and R01NS040389); and the Muscular Dystrophy Association for funding.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013343108/-/DCSupplemental.
References
- 1.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 3.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 4.Liquori CL, et al. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 5.Jin P, et al. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moseley ML, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 8.Daughters RS, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oma Y, Kino Y, Sasagawa N, Ishiura S. Intracellular localization of homopolymeric amino acid-containing proteins expressed in mammalian cells. J Biol Chem. 2004;279:21217–21222. doi: 10.1074/jbc.M309887200. [DOI] [PubMed] [Google Scholar]
- 10.Shoubridge C, Cloosterman D, Parkinson-Lawerence E, Brooks D, Gécz J. Molecular pathology of expanded polyalanine tract mutations in the Aristaless-related homeobox gene. Genomics. 2007;90:59–71. doi: 10.1016/j.ygeno.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Trottier Y, et al. Cellular localization of the Huntington's disease protein and discrimination of the normal and mutated form. Nat Genet. 1995;10:104–110. doi: 10.1038/ng0595-104. [DOI] [PubMed] [Google Scholar]
- 12.Mogk A, Schmidt R, Bukau B. The N-end rule pathway for regulated proteolysis: Prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17:165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- 14.Toulouse A, et al. Ribosomal frameshifting on MJD-1 transcripts with long CAG tracts. Hum Mol Genet. 2005;14:2649–2660. doi: 10.1093/hmg/ddi299. [DOI] [PubMed] [Google Scholar]
- 15.Davies JE, Rubinsztein DC. Polyalanine and polyserine frameshift products in Huntington's disease. J Med Genet. 2006;43:893–896. doi: 10.1136/jmg.2006.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peabody DS. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- 17.Touriol C, et al. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol Cell. 2003;95:169–178. doi: 10.1016/s0248-4900(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 18.Gomes-Pereira M, et al. CTG trinucleotide repeat “big jumps”: Large expansions, small mice. PLoS Genet. 2007;3:e52. doi: 10.1371/journal.pgen.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.U M, et al. Extended polyglutamine selectively interacts with caspase-8 and -10 in nuclear aggregates. Cell Death Differ. 2001;8:377–386. doi: 10.1038/sj.cdd.4400819. [DOI] [PubMed] [Google Scholar]
- 20.Kozak M. Context effects and inefficient initiation at non-AUG codons in eukaryotic cell-free translation systems. Mol Cell Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kochetov AV, et al. AUG_hairpin: Prediction of a downstream secondary structure influencing the recognition of a translation start site. BMC Bioinformatics. 2007;8:318. doi: 10.1186/1471-2105-8-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Domier LL, McCoppin NK, D'Arcy CJ. Sequence requirements for translation initiation of Rhopalosiphum padi virus ORF2. Virology. 2000;268:264–271. doi: 10.1006/viro.2000.0189. [DOI] [PubMed] [Google Scholar]
- 23.Fox JT, Stover PJ. Mechanism of the internal ribosome entry site-mediated translation of serine hydroxymethyltransferase 1. J Biol Chem. 2009;284:31085–31096. doi: 10.1074/jbc.M109.035576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katayama S, et al. RIKEN Genome Exploration Research Group;; Genome Science Group (Genome Network Project Core Group);; FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 25.Batra R, Charizanis K, Swanson MS. Partners in crime: Bidirectional transcription in unstable microsatellite disease. Hum Mol Genet. 2010;19(R1):R77–R82. doi: 10.1093/hmg/ddq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.