Abstract
Mutations in LMNA, which encodes A-type lamins, result in disparate diseases, known collectively as laminopathies, that affect distinct tissues, including striated muscle and adipose tissue. Lamins provide structural support for the nucleus and sites of attachment for chromatin, and defects in these functions may contribute to disease pathogenesis. Recent studies suggest that A-type lamins may facilitate connections between the nucleus and the cytoskeleton mediated by nuclear envelope nesprin and SUN proteins. In mammalian cells, however, interfering with A-type lamins does not affect the localization of these proteins. Here, we used centrosome orientation in fibroblasts, which requires separate nuclear and centrosome positioning pathways, as a model system to understand how LMNA mutations affect nucleus-cytoskeletal connections. We find that LMNA mutations causing striated muscle diseases block actin-dependent nuclear movement, whereas most that affect adipose tissue inhibit microtubule-dependent centrosome positioning. Genetic deletion or transient depletion of A-type lamins also blocked nuclear movement, showing that mutations affecting muscle exhibit the null phenotype. Lack of A-type lamins, or expression of variants that cause striated muscle disease, did not affect assembly of nesprin-2G and SUN2 into transmembrane actin-associated nuclear (TAN) lines that attach the nucleus to retrogradely moving actin cables. Nesprin-2G TAN lines were less stable, however, and slipped over the nucleus rather than moving with it, indicating that they were not anchored. Nesprin-2G TAN lines also slipped in SUN2-depleted cells. Our results establish A-type lamins as anchors for nesprin-2G–SUN2 TAN lines to allow productive movement and proper positioning of the nucleus by actin.
Keywords: nesprin SUN, linker of nucleoskeleton and cytoskeleton complex, muscular dystrophy
Lamin A and lamin C, the predominant A-type lamins, are expressed in most differentiated somatic cells. Yet, different mutations in the LMNA gene encoding these proteins result in a variety of diseases that affect specific tissues. LMNA mutations cause autosomal dominant Emery–Dreifuss muscular dystrophy (EDMD) and related diseases with dilated cardiomyopathy (DCM) that affect cardiac muscle and skeletal muscle to variable degrees (1). Other LMNA mutations cause Dunnigan-type familial partial lipodystrophy (FPLD) that affects adipose tissue (2), Charcot–Marie–Tooth type 2B1 disease that affects peripheral neurons (3), and progeroid syndromes with features of accelerated aging (4, 5). The mechanism whereby mutations in a single gene that is widely expressed cause such diverse diseases remains a puzzle.
There are two prevailing hypotheses to explain the pathogenesis of laminopathies. The mechanical stress hypothesis proposes that alterations in A-type lamins compromise nuclear integrity in tissues susceptible to stress, such as striated muscle. This model is supported by findings that A-type lamin deficiency disrupts nuclear integrity in model systems (6, 7). The second hypothesis postulates that LMNA mutations alter interactions of the nuclear lamina with chromatin, resulting in aberrant gene expression. This model is supported by findings that lamin A is necessary to repress inhibitors of MyoD expression (8).
A third possibility is that alterations in lamin A/C might cause disease by compromising interactions between the nucleus and cytoskeleton that are necessary for positioning the nucleus. Interactions between the nucleus and cytoskeleton are mediated by the linker of nucleoskeleton and cytoskeleton (LINC) complex, which spans the nuclear envelope via nesprin proteins in the outer nuclear membrane and SUN proteins in the inner nuclear membrane (9). Nesprins interact with cytoskeletal elements, and both nesprins and SUNs contribute to nuclear movement and positioning during development and in migrating cells (10–13). Recent work has shown that multiple LINC complexes assemble into transmembrane actin-associated nuclear (TAN) lines that attach nuclei to actin filaments during nuclear movement as fibroblasts polarize to migrate (13).
The nuclear lamina may contribute to LINC complex function during nuclear positioning. Disruption of the nuclear lamina affects nuclear positioning and cell polarity in Drosophila photoreceptor cells (14) and oocytes (15) and is required for nuclear localization of Unc84, a SUN protein involved in nuclear positioning in Caenorhabditis elegans (16). Mammalian LMNA mutations also seem to affect linkage of the nucleus to the cytoskeleton (17, 18), but nesprins and SUNs localize to the nucleus without A-type lamins (19); thus, the function of A-type lamins in nuclear-cytoplasmic connections is unclear.
The hypotheses discussed above have yet to be supported by data that might explain how distinct LMNA mutations cause tissue-specific diseases. We examined nuclear movement during centrosome orientation in migrating fibroblasts, which requires A-type lamins (18). We screened 16 different disease-causing lamin A variants and found that nearly all affected centrosome orientation but only those that cause muscle disease affected nuclear movement and positioning. A-type lamins were not required for assembly of LINC complexes into TAN lines but were required to anchor TAN lines so that actin cable movement produced nuclear movement. These data identify a unique function for A-type lamins in anchoring nuclear-cytoplasmic connections and reveal a subcellular process affected by LMNA mutations that segregates with disease.
Results
We used wounded NIH 3T3 fibroblast monolayers to explore whether disease-associated lamin A variants affected nuclear or centrosome positioning events that orient the centrosome toward the wound edge (20). In this system, the serum factor lysophosphatidic acid (LPA) activates Cdc42-dependent pathways that trigger actin and myosin II-dependent rearward nuclear movement (away from the leading edge) and microtubule- and dynein-dependent maintenance of the centrosome at the cell center (20–22). The combined effect of these two processes is the reorientation of the centrosome to a position between the nucleus and the leading edge.
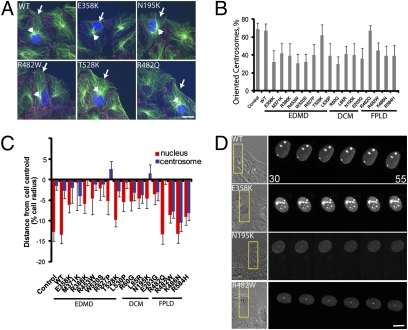
Mutant prelamin A cDNA constructs were expressed by microinjection into starved NIH 3T3 fibroblasts at the wound edge, and centrosome orientation was then triggered with LPA. Prelamin A constructs are processed to mature lamin A, and all mutant versions localized to the nucleus (23) (Fig. 1 and Fig. S1). Nearly all (14 of 16) disease-causing lamin A variants blocked LPA-stimulated centrosome orientation, whereas WT lamin A did not (Fig. 1 A and B). To determine whether nuclear or centrosome positioning was affected by expression of the lamin A variants, we measured the positions of the nucleus and centrosome relative to the cell centroid in fixed cells (20). Expression of lamin A variants that cause EDMD or DCM prevented rearward positioning of the nucleus compared with nonexpressing or WT lamin A-expressing controls (Fig. 1 A and C). Conversely, expression of variants that cause FPLD did not affect nuclear positioning but caused the centrosome to be positioned rearward with the nucleus rather than near the cell center as in control cells (Fig. 1 A and C). Direct time-lapse imaging showed that nuclei failed to move rearward in cells expressing lamin A variants that cause EDMD (E385K) or DCM (N195K), whereas nuclei moved rearward in cells that expressed WT lamin A or a variant that causes FPLD (R482W) (Fig. 1D). These results show that the lamin A variants inhibiting centrosome orientation did so by inducing specific phenotypes that segregated with respect to disease.
Fig. 1.
Disease-causing variants of lamin A block centrosome orientation. (A) LPA-stimulated NIH 3T3 fibroblasts expressing FLAG-tagged lamin A variants and stained for FLAG tag (blue), pericentrin for centrosomes and β-catenin for cell edges (red), and microtubules (green). Arrows denote expressing cells, and arrowheads denote the centrosome. (Scale bar, 10 μm.) (B) Centrosome orientation in NIH 3T3 fibroblasts expressing lamin A variants. Centrosomes were considered oriented when they were in a pie-shaped section facing the wound edge; random orientation is 33% by this measure (21). Error bars are SEM for >45 cells from three independent experiments, and centrosome orientation was significantly different from that of WT control (P < 0.001) for all lamin A variants except T528K and R482Q. (C) Nuclear (red) and centrosome (blue) positions along the axis perpendicular to the wound for cells expressing lamin A variants. The cell centroid is zero; positive values are located toward the leading edge, and negative values are located toward the cell rear. Values are expressed as a percentage of the cell radius to account for variations in cell size (20, 40). Error bars are SEM for >30 total cells from at least three independent experiments for each variant. The nuclear position in all EDMD and DCM variants (except T528K) was significantly different from that of WT (P < 0.001), and the centrosome position in all FPLD variants (except R482Q) was significantly different from that of WT (P < 0.001). (D) Montages from movies of NIH 3T3 fibroblasts expressing red fluorescent protein-tagged versions of the indicated lamin A variants. The leftmost panel shows a phase image. The box (Left) indicates the region shown at higher magnification in the fluorescence images (Right). Images were acquired every 5 min (time is in minutes after addition of LPA). (Scale bar, 5 μm.)
Loss of lamin A results in impaired cell migration (18, 24). We tested whether disease-causing lamin A variants affected cell migration by expressing the variants in wound edge cells and stimulating migration with serum. Cells expressing lamin A variants that cause muscle disease fell behind the wound edge during migration, whereas control cells that expressed GFP, WT lamin A, or variants that cause FPLD remained at the wound edge (Fig. S1A). These results indicate that defective nuclear movement in cells expressing lamin A variants that cause muscle disease interferes with cell migration, consistent with earlier studies (18, 24).
We tested whether the differential incorporation of the lamin A variants into the lamina contributed to the different effects on nuclear and centrosome positioning. Resistance to detergent extraction was used as a measure of incorporation into the lamina, and we found that with one exception, T528K, all the variants were resistant to extraction to about the same degree as WT (Fig. S1B). Furthermore, confocal microscopy of cells expressing the lamin A variants showed that the ratio of fluorescence in the nuclear rim to the nuclear interior was similar for WT and each disease-causing lamin A variant except T528K (Fig. S1 C and D). These results show that differential incorporation does not explain the disease-based segregation of polarity defects. That the T528K variant does not affect centrosome orientation may reflect its lack of incorporation into the nuclear envelope and may also explain why T528K was an outlier in a study on lamin A regulation of ERK signaling (25).
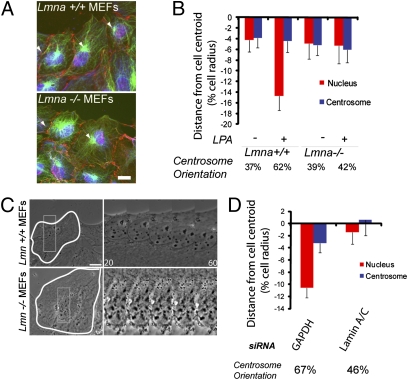
Defective nuclear or centrosome positioning caused by lamin A variants may represent a gain or loss of function, so we examined these processes in Lmna−/− mouse embryonic fibroblasts (MEFs) (26). As observed earlier (18), Lmna−/− MEFs failed to orient their centrosome in response to LPA compared with Lmna+/+ MEFs (Fig. 2 A and B). Further analysis showed that LPA-stimulated rearward positioning of the nucleus was defective in Lmna−/− MEFs (Fig. 2B). Time-lapse microscopy showed that the nucleus remained stationary in Lmna−/− MEFs in response to LPA, whereas it moved rearward in the Lmna+/+ MEFs (Fig. 2C). Acute depletion of lamin A/C by siRNA in NIH 3T3 fibroblasts yielded similar results on centrosome orientation, nuclear positioning, and nuclear movement (Fig. 2 D and Fig. S2A). Depletion of lamin A/C from NIH 3T3 fibroblasts also reduced cell migration velocity compared with control NIH 3T3 fibroblasts (22, 27) (Fig. S2B).
Fig. 2.
Lamin A/C is necessary for nuclear movement. (A) Images of LPA-stimulated Lmna+/+ and Lmna−/− MEFs in a wounded monolayer stained for pericentrin and β-catenin (red), nuclei (blue), and microtubules (green). Arrowheads indicate the centrosome, and the wound is located at the top. (Scale bar, 10 μm.) (B) Nucleus (red) and centrosome (blue) positions in LPA-stimulated Lmna+/+ and Lmna−/− MEFs. Centrosome orientation (percentage of cells) is located below the histogram. Error bars are SEM for >100 cells from at least three independent experiments. The nuclear position is significantly different between stimulated Lmna+/+ and Lmna−/− cells (P < 0.001). (C) Phase contrast images from movies of Lmna+/+ and Lmna−/− MEFs. The boxed region (Left) is shown at higher magnification (Right). Images were acquired every 5 min (time is in minutes). (Scale bar, 10 μm.) (D) Nucleus (red) and centrosome (blue) positions in NIH 3T3 fibroblasts depleted of lamin A/C and GAPDH. Centrosome orientation (percentage of cells) is shown below the histogram. Error bars are SEM for >100 cells from three independent experiments. The nuclear position is significantly different between GAPDH- and lamin A/C-depleted NIH 3T3 fibroblasts (P < 0.001).
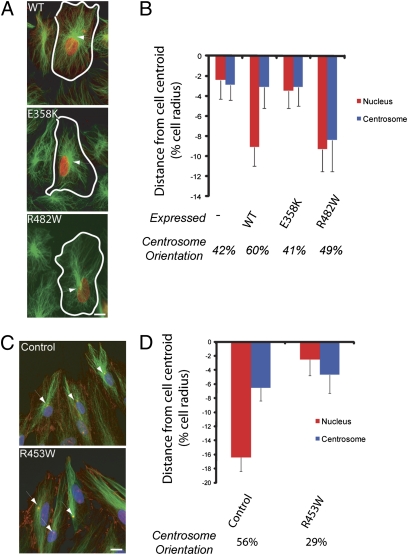
To test whether the effects of the disease-causing lamin A variants on centrosome orientation were intrinsic to the variant proteins, we expressed lamin A proteins in Lmna−/− MEFs. Expression of WT lamin A rescued centrosome orientation and nuclear positioning in Lmna−/− MEFs (Fig. 3 A and B). In contrast, expression of a variant that causes EDMD failed to rescue either centrosome orientation or nuclear movement, whereas expression of a variant that causes FPLD rescued nuclear movement but not centrosome orientation (Fig. 3 A and B). These results confirmed that lamin A was specifically required for nuclear movement and that the effects on centrosome orientation are intrinsic to the expressed lamin A proteins. Finally, we tested whether centrosome orientation was affected in cells from a patient with EDMD. Centrosome orientation and rearward nuclear positioning were significantly impaired in fibroblasts derived from an individual with EDMD (R453W mutation) (28) compared with fibroblasts derived from an unaffected individual (Fig. 3 C and D). Thus, A-type lamins contribute to nuclear movement in both human and mouse cells, and this activity is disrupted by LMNA mutations that cause EDMD.
Fig. 3.
Lamin A variants interfere with centrosome orientation and nuclear movement in Lmna−/− MEFs and in human fibroblasts. (A) Images of LPA-stimulated Lmna−/− MEFs expressing lamin A variants and stained for the expressed lamin A (red) and microtubules (green). The wound edge is located toward the top, and arrows indicate the centrosome. (Scale bar, 10 μm.) (B) Nucleus (red) and centrosome (blue) positions in the cells described in A. The centrosome orientation (percentage of cells) is located below the histogram. Error bars are SEM for >50 cells from at least three independent experiments. Nuclear position in E358K-expressing cells is significantly different from that in WT-expressing cells (P < 0.01) but not from that in nonexpressing controls. Nuclear position in R482W0-expressing cells is significantly different from that in nonexpressing controls (P < 0.01) but not from that in WT-expressing cells. The position of the centrosome in R482W-expressing cells is significantly different from that in each of the other conditions (P < 0.05). (C) Images of human fibroblasts from an unaffected control and from an individual with EDMD caused by the R453W LMNA mutation stimulated with LPA and stained for nuclei (blue), β-catenin and pericentrin (red), and microtubules (green). Arrows indicate the centrosome, and the wound edge is located at the top. (Scale bar, 10 μm.) (D) Nucleus (red) and centrosome (blue) positions for the cells described in C. Centrosome orientation (percentage of cells) is located below the histogram. Error bars are SEM for >100 cells from at least three independent experiments. Nucleus position and centrosome orientation in R453W are significantly different from those in control (P < 0.01).
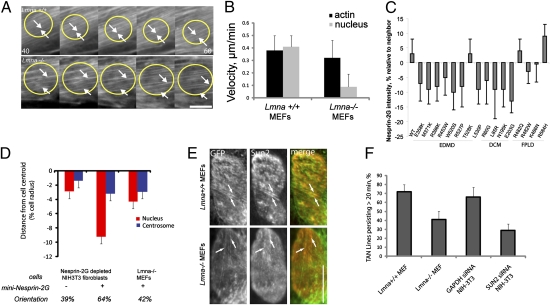
The effects of the FPLD variants on centrosome position are likely indirect because centrosome positioning involves dynein-dependent microtubule tethering at the cell cortex (22). We therefore focused on the immotile nuclear phenotype caused by EDMD and DCM LMNA variants. Nuclear movement in fibroblasts is driven by the coupling of retrogradely moving actin cables to the nucleus through nesprin-2G and SUN2 TAN lines (13); thus, nuclear movement could be disrupted by alterations in actin or nesprin-2G/SUN2. To examine actin retrograde flow, we expressed Lifeact-mCherry, which labels all F-actin detected by phalloidin and does not alter actin distribution in fibroblasts (29, 13). Movies of Lifeact-mCherry revealed that actin cables formed normally in response to LPA and that dorsal actin cables moved retrogradely at comparable rates in both Lmna+/+ and Lmna−/− MEFs (Fig. 4 A and B). Yet, the nucleus moved with dorsal actin cables in Lmna+/+ MEFs but remained stationary as actin cables flowed past in Lmna−/− MEFs (Fig. 4 A and B). These results indicate that defective nuclear movement in Lmna−/− MEFs is not caused by defects in actin retrograde flow.
Fig. 4.
Effect of Lamin A/C on actin cables and TAN lines (A) Panels from movies of Lmna+/+ (Upper) and Lmna−/− (Lower) MEFs expressing Lifeact-mCherry showing dorsal actin cables near the nucleus. The wound edge is located at the top, yellow circles denote nuclei, and arrows denote actin cables that move either with (Upper) or over (Lower) the nucleus. Time is in minutes after LPA stimulation. (Scale bar, 5 μm.) (B) Velocities of nuclei and actin cables from the cells described in A. Error bars are SEM from three experiments and 45 total cells. Nuclear velocity in LMNA−/− MEFs was significantly different from that in Lmna+/+ MEFs (P < 0.01; there was no significant difference in the velocity of actin cables). (C) Relative intensity of nesprin-2G immunofluorescence compared with nonexpressing neighboring cells. Values are the percentage difference from nonexpressing neighboring cells. Error is SEM for >30 cells from three separate experiments. Nesprin-2G intensity was significantly different (P < 0.05) for all variants that cause EDMD or DCM, except T528K, compared with WT- or FPLD-causing variants. (D) Nucleus (red) and centrosome (blue) positions in nesprin 2G-depleted NIH 3T3 fibroblasts and Lmna−/− MEFs expressing GFP-mininesprin-2G. Centrosome orientation (percentage of cells) is located below the histogram. Error bars are SEM for >45 cells from at least three independent experiments. Centrosome orientation and nuclear position in nesprin 2G-depleted cells expressing GFP-mininesprin-2G were significantly different from those in nonexpressing cells (P < 0.01). (E) Immunofluorescence images of nuclei from a Lmna+/+ MEF (Upper) and Lmna−/− MEF (Lower) expressing GFP-mininesprin-2G (green in merge) and stained for endogenous SUN2 (red in merge). Arrows indicate colocalized nesprin-2G and SUN2. (Scale bar, 5 μm.). (F) Percentage of GFP-mininesprin-2G TAN lines that persist >20 min. Error bars are SEM from >40 cells and three independent experiments. The differences between Lmna+/+ and Lmna−/− MEFs and between GAPDH-depleted and SUN2-depleted NIH 3T3 fibroblasts were significant (P < 0.05).
Because TAN lines provide the direct connection between the nucleus and actin, we examined the assembly and function of TAN lines in fibroblasts lacking lamin A/C or expressing lamin A variants. The levels of SUN2 are not affected in Lmna−/− cells or NIH 3T3 fibroblasts depleted of lamin A/C (19, 30); however, nesprin-2G has been reported to be present at reduced levels in nuclei from Lmna−/− MEFs (31). We observed a small reduction (5–15%) in nesprin-2G levels in nuclei of cells expressing 11 of 12 lamin A variants that cause EDMD or DCM but not in nuclei of cells expressing WT lamin A or variants that cause FPLD (Fig. 4 C and Fig. S3A). We tested whether increasing the levels of nesprin-2G would rescue the defective nuclear movement in Lmna−/− MEFs by expressing a chimeric construct of nesprin-2G (GFP-mininesprin-2G) that contains the N-terminal actin-binding domain and the C-terminal transmembrane and KASH domains and rescues nuclear movement in nesprin 2G-depleted NIH 3T3 fibroblasts (13) (Fig. 4D and Fig. S3B). GFP-mininesprin-2G localized to the nucleus but did not rescue centrosome orientation or rearward nuclear positioning in Lmna−/− MEFs (Fig. 4D and Fig. S3B), suggesting that defective nuclear movement in Lmna−/− MEFs does not result from decreased levels of nesprin-2G.
We next explored whether TAN lines formed in cells lacking lamin A/C. GFP-mininesprin-2G is a probe for TAN lines (13), and when expressed in Lmna−/− MEFs, TAN lines containing both GFP-mininesprin-2G and endogenous SUN2 formed, although slightly less efficiently than in Lmna+/+ MEFs (Fig. 4E and Fig. S3C). Additionally, the TAN lines were less persistent in Lmna−/− MEFs (Fig. 4F).
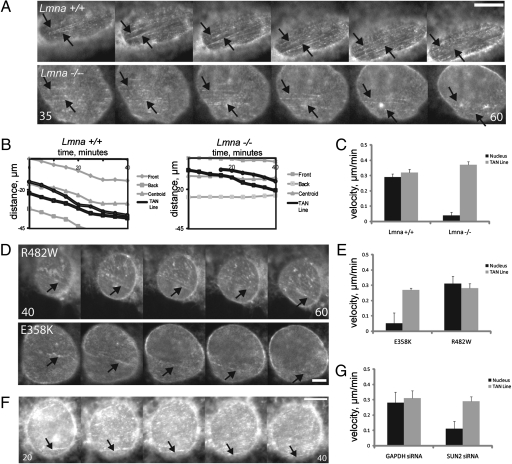
To test whether the decreased persistence of the TAN lines in Lmna−/− MEFs reflected defective interaction between TAN lines and actin cables, we imaged TAN lines and nuclear movements in Lmna+/+ and Lmna−/− MEFs. TAN lines and the nucleus moved at the same rate in Lmna+/+ MEFs, consistent with their role in linking actin cables to the nucleus for movement (13) (Fig. 5 A–C). TAN lines moved at a similar velocity in Lmna−/− MEFs compared with Lmna+/+ MEFs; however, they moved over a stationary nucleus rather than moving with the nucleus as in Lmna+/+ MEFs (Fig. 5 A–C). TAN lines moved with the nucleus in NIH 3T3 fibroblasts expressing an FPLD-causing lamin A variant, whereas they moved over a stationary nucleus in fibroblasts expressing an EDMD-causing lamin A variant (Fig. 5 D and E). These results indicate that lamin A/C anchors TAN lines and that lamin A variants causing striated muscle disease but not adipose disease disrupt this anchoring.
Fig. 5.
Lamin A/C provides a nucleoplasmic anchor for TAN lines. (A) Panels from movies of GFP-mininesprin-2G in nuclei of LPA-stimulated Lmna+/+ (Upper) and Lmna−/− (Lower) MEFs. The wound edge is located toward the top. Images were acquired every 5 min (time is minutes after LPA stimulation). Arrows indicate TAN lines that move with (Upper) or over (Lower) the nucleus. (Scale bar, 5 μm.) (B) Traces of the movement of the front, back, and centroid of the nucleus (gray) and two TAN lines (black) in a Lmna+/+ MEF (Left) and Lmna−/− MEF (Right). (C) Velocity of the nucleus and TAN lines in the cells described in A. Error bars are SEM for 30 nuclei/TAN Lines from three experiments. Nuclear velocity in Lmna−/− MEFs is significantly different from that in Lmna+/+ MEFs (P < 0.001); there is no significant difference between TAN line velocities. (D) Panels from movies of GFP-mininesprin-2G in nuclei of LPA-stimulated NIH 3T3 fibroblasts expressing the indicated red fluorescent protein-tagged lamin A variants. The wound edge is located toward the top. Images were acquired every 5 min (time is minutes after LPA stimulation). Arrows indicate TAN lines that move with the nucleus (Upper) or over the nucleus (Lower). (Scale bar, 2 μm.) (E) Velocities of nuclei and TAN lines in the cells described in D. Error bars are SEM for 30 nuclei and TAN lines from three experiments. Nuclear velocity in E358K was significantly different from that in R482W (P < 0.001); there is no significant difference between TAN line velocities. (F) Panels from a movie of GFP-mininesprin-2G in the nucleus of LPA-stimulated NIH 3T3 fibroblasts depleted of SUN2. The wound edge is located toward the top. Images were acquired every 5 min (time is minutes after LPA stimulation). Arrows indicate a TAN line that moves over the nucleus. (Scale bar, 5 μm.) (G) Velocity of nuclear and TAN line movements in SUN2- or control (GAPDH)-depleted NIH 3T3 fibroblasts stimulated with LPA. Error bars are SEM for 17 nuclei from four experiments. Nuclear velocity in SUN2-depleted cells was significantly different from that in GAPDH-depleted cells (P < 0.05); there was no significant difference in TAN line velocities.
Nesprin-2G resides in the outer nuclear envelope, whereas lamin A resides within the nucleoplasm, so one would expect that lamin A/C anchors nesprin-2G TAN lines through an inner nuclear membrane protein such as SUN2, which is also found in TAN lines. To test this possibility, we expressed GFP-mininesprin-2G in SUN2-depleted NIH 3T3 fibroblasts. GFP-mininesprin-2G TAN lines formed in SUN2-depleted cells, but they were less persistent than in controls (Fig. 4F and Fig. S3C) and moved over a stationary nucleus just as in the Lmna−/− MEFs (Fig. 5 F and G). Thus, deficiencies in lamin A/C or SUN2 lead to the same phenotype of slipping TAN lines on immobile nuclei.
Discussion
We have shown that lamin A/C plays an essential role in nuclear movement and positioning by contributing to the stability and anchoring of TAN lines. Lamin A/C interacts with components of the LINC complex (17, 19, 31–33), and it is likely through these interactions that it stabilizes and anchors TAN lines. Given that depletion of SUN2 yielded a similar phenotype on TAN line stability and anchoring, it is likely that lamin A/C interaction with SUN2 is responsible for anchoring the nesprin-2G TAN lines. Lack of lamin A/C had relatively little effect on the formation of the TAN lines despite the fact that nuclear nesprin-2G is reduced in cells lacking lamin A/C. This suggests that the assembly of the individual LINC complexes is not strongly affected by lamin A/C. The decreased persistence of TAN lines in cells lacking lamin A/C may reflect an inability of LINC complexes to resist forces exerted by the moving actin cables. Consistent with these interpretations, the overall mobility of mininesprin-2G and SUN2 in the nuclear envelope is increased in cells lacking lamin A/C (33).
TAN line anchoring by lamin A/C allows the force exerted on the nucleus by moving actin cables to be transmitted to the nucleus for productive movement. This anchoring function of lamin A/C is distinct from that involved in localizing some inner nuclear membrane proteins, because, for example, SUN2 localization is not altered in lamin A/C-deficient cells (19, 34). Also, whereas nesprin-2G levels are somewhat reduced in lamin A/C-deficient cells, increasing nesprin-2G levels did not rescue nuclear movement, showing that lamin A/C plays a role beyond simply localizing nesprin-2G to the nucleus. There is precedent for anchoring of nesprin and SUN proteins to resist force: In Schizosaccharomyces pombe, orthologs of nesprins and SUN proteins are anchored by Ima1, a heterochromatin-binding protein (35). Given that lamin A/C interacts with heterochromatin, it will be interesting to see whether heterochromatin-binding proteins are also involved in anchoring TAN lines in mammalian cells.
A key finding of our study is that only mutations in LMNA that cause muscle disease affect the LINC complex anchoring function of lamin A/C. The single (1 of 12) muscle disease-causing lamin A variant that failed to affect nuclear movement (T528K) also failed to incorporate into the nuclear lamina, suggesting that lamin A variant incorporation into the lamina may be necessary to disrupt nuclear-cytoskeletal interactions. Mechanistically, the variants that cause muscle disease appear to disrupt the ability of the lamina to anchor TAN lines, and thus resist force. Because this appears to occur through a dominant-negative effect, it may reflect a requirement for multiple points of anchorage and/or for lamina integrity that is disrupted by the lamin A variants.
Nuclei are precisely positioned in skeletal muscle, with a small number clustered under the neuromuscular junction and the remainder equally spaced at the periphery of the fiber. In individuals afflicted with muscular dystrophies, including EDMD, skeletal muscle nuclei are mispositioned, further suggesting that the role of A-type lamins in regulating nuclear movement and positioning may be physiologically relevant. Mouse studies have shown that nesprins and SUNs contribute to proper nuclear positioning in skeletal muscle (36, 37) and that mutations in nesprin genes are associated with EDMD (38). Coupled with our findings that lamin A variants causing muscle disease affect nuclear movement and positioning, these studies suggest that aberrant nuclear movement and positioning may underlie the pathogenesis of muscle disorders and provide a previously undescribed avenue for future studies of muscle disease.
Materials and Methods
Reagents.
Reagents are described in SI Materials and Methods.
Cell Culture.
NIH 3T3 fibroblasts were cultured and serum-starved as described previously (39). Lmna+/+ and Lmna−/− MEFs (26) were cultured in DMEM plus 10% (vol/vol) FBS and were starved similar to NIH 3T3 fibroblasts. Human dermal fibroblasts were obtained from a patient with EDMD and a normal individual and were cultured as described (28). Centrosome orientation was stimulated with 10 μM LPA (Avanti) dissolved in serum-free DMEM as previously described (31).
DNA Constructs.
FLAG-tagged lamin A (23), Lifeact-mCherry (29), and GFP-mininesprin-2G (12) constructs were previously described. Modified red fluorescent protein (mRFP) constructs were made by digesting FLAG-tagged constructs in pGAD424 with EcoN1 and BglII and inserting the digestion products into mRFP-C1(32) that had been digested with EcoN1 and BamH1. All constructs were verified by sequencing.
Immunofluorescence.
Cells were fixed in −20 °C methanol or 4% (wt/vol) paraformaldehyde and stained with antibodies as previously described (20).
Analysis of Centrosome Orientation and Position of the Nucleus and Centrosome.
Centrosome orientation and nuclear and centrosome positioning in fixed and stained cells at the wound edge were determined as previously described (20, 21, 40). For nucleus and centrosome position, only the distance along the axis perpendicular to the wound edge is reported because there was little movement along the axis parallel to the wound edge (19).
siRNA.
Duplex siRNA (21-mers) was purchased from Shanghai GenePharma and transfected using RNAiMAX (Invitrogen). Further details are presented in SI Materials and Methods.
Time-Lapse Microscopy.
Live cell imaging was as previously described (13, 20) and is detailed in SI Materials and Methods.
Additional Methods.
DNA Microinjection, Lamin A/C Incorporation, Quantification of Nesprin-2G Nuclear Levels, Analysis of TAN Lines, and Statistical Procedures are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Sue Shackleton (University of Leicester, Leicester, United Kingdom) and Glenn Morris (Wolfson Center for Inherited Neuromuscular Disease, Wrexham, United Kingdom) for antibodies and Colin Stewart (Institute of Medical Biology, Singapore) for MEFs. This study was supported by National Institutes of Health Grant GM062939 (to G.G.G.) and Grant NS059352 (to H.J.W.) and by Postdoctoral Fellowship 0625833T from the American Heart Association (to E.S.F.) and a postdoctoral fellowship from the Dystonia Medical Research Foundation (to G.W.G.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000824108/-/DCSupplemental.
References
- 1.Bonne G, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 2.Shackleton S, et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat Genet. 2000;24:153–156. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- 3.De Sandre-Giovannoli A, et al. Homozygous defects in LMNA, encoding lamin A/C nuclear-envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot-Marie-Tooth disorder type 2) and mouse. Am J Hum Genet. 2002;70:726–736. doi: 10.1086/339274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Sandre-Giovannoli A, et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 5.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broers JL, et al. Decreased mechanical stiffness in LMNA−/− cells is caused by defective nucleo-cytoskeletal integrity: Implications for the development of laminopathies. Hum Mol Genet. 2004;13:2567–2580. doi: 10.1093/hmg/ddh295. [DOI] [PubMed] [Google Scholar]
- 7.Lammerding J, et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakay M, et al. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain. 2006;129:996–1013. doi: 10.1093/brain/awl023. [DOI] [PubMed] [Google Scholar]
- 9.Crisp M, et al. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worman HJ, Gundersen GG. Here come the SUNs: A nucleocytoskeletal missing link. Trends Cell Biol. 2006;16:67–69. doi: 10.1016/j.tcb.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Starr DA, Han M. ANChors away: An actin based mechanism of nuclear positioning. J Cell Sci. 2003;116:211–216. doi: 10.1242/jcs.00248. [DOI] [PubMed] [Google Scholar]
- 12.Starr DA. A nuclear-envelope bridge positions nuclei and moves chromosomes. J Cell Sci. 2009;122:577–586. doi: 10.1242/jcs.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patterson K, et al. The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol Biol Cell. 2004;15:600–610. doi: 10.1091/mbc.E03-06-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillemin K, Williams T, Krasnow MA. A nuclear lamin is required for cytoplasmic organization and egg polarity in Drosophila. Nat Cell Biol. 2001;3:848–851. doi: 10.1038/ncb0901-848. [DOI] [PubMed] [Google Scholar]
- 16.Lee KK, et al. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol Biol Cell. 2002;13:892–901. doi: 10.1091/mbc.01-06-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale CM, et al. Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys J. 2008;95:5462–5475. doi: 10.1529/biophysj.108.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JS, et al. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J. 2007;93:2542–2552. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haque F, et al. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 21.Palazzo AF, et al. Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr Biol. 2001;11:1536–1541. doi: 10.1016/s0960-9822(01)00475-4. [DOI] [PubMed] [Google Scholar]
- 22.Schmoranzer J, et al. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr Biol. 2009;19:1065–1074. doi: 10.1016/j.cub.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Östlund C, Bonne G, Schwartz K, Worman HJ. Properties of lamin A mutants found in Emery-Dreifuss muscular dystrophy, cardiomyopathy and Dunnigan-type partial lipodystrophy. J Cell Sci. 2001;114:4435–4445. doi: 10.1242/jcs.114.24.4435. [DOI] [PubMed] [Google Scholar]
- 24.Houben F, et al. Disturbed nuclear orientation and cellular migration in A-type lamin deficient cells. Biochim Biophys Acta. 2009;1793:312–324. doi: 10.1016/j.bbamcr.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Muchir A, Shan J, Bonne G, Lehnart SE, Worman HJ. Inhibition of extracellular signal-regulated kinase signaling to prevent cardiomyopathy caused by mutation in the gene encoding A-type lamins. Hum Mol Genet. 2009;18:241–247. doi: 10.1093/hmg/ddn343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan T, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ezratty EJ, Bertaux C, Marcantonio EE, Gundersen GG. Clathrin mediates integrin endocytosis for focal adhesion disassembly in migrating cells. J Cell Biol. 2009;187:733–747. doi: 10.1083/jcb.200904054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muchir A, et al. Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve. 2004;30:444–450. doi: 10.1002/mus.20122. [DOI] [PubMed] [Google Scholar]
- 29.Riedl J, et al. Lifeact: A versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasan S, et al. Nuclear envelope localization of human UNC84A does not require nuclear lamins. FEBS Lett. 2006;580:1263–1268. doi: 10.1016/j.febslet.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Q, et al. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J Cell Sci. 2005;118:673–687. doi: 10.1242/jcs.01642. [DOI] [PubMed] [Google Scholar]
- 32.Ostlund C, Sullivan T, Stewart CL, Worman HJ. Dependence of diffusional mobility of integral inner nuclear membrane proteins on A-type lamins. Biochemistry. 2006;45:1374–1382. doi: 10.1021/bi052156n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Östlund C, et al. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J Cell Sci. 2009;122:4099–4108. doi: 10.1242/jcs.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Libotte T, et al. Lamin A/C-dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol Biol Cell. 2005;16:3411–3424. doi: 10.1091/mbc.E04-11-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.King MC, Drivas TG, Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–438. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schenk J, Wilsch-Bräuninger M, Calegari F, Huttner WB. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc Natl Acad Sci USA. 2009;106:16487–16492. doi: 10.1073/pnas.0908928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, et al. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet. 2007;16:2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- 39.Cook TA, Nagasaki T, Gundersen GG. Rho guanosine triphosphatase mediates the selective stabilization of microtubules induced by lysophosphatidic acid. J Cell Biol. 1998;141:175–185. doi: 10.1083/jcb.141.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomes ER, Gundersen GG. Real-time centrosome reorientation during fibroblast migration. Methods Enzymol. 2006;406:579–592. doi: 10.1016/S0076-6879(06)06045-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.