Abstract
The centrosome is generally maintained at the center of the cell. In animal cells, centrosome centration is powered by the pulling force of microtubules, which is dependent on cytoplasmic dynein. However, it is unclear how dynein brings the centrosome to the cell center, i.e., which structure inside the cell functions as a substrate to anchor dynein. Here, we provide evidence that a population of dynein, which is located on intracellular organelles and is responsible for organelle transport toward the centrosome, generates the force required for centrosome centration in Caenorhabditis elegans embryos. By using the database of full-genome RNAi in C. elegans, we identified dyrb-1, a dynein light chain subunit, as a potential subunit involved in dynein anchoring for centrosome centration. DYRB-1 is required for organelle movement toward the minus end of the microtubules. The temporal correlation between centrosome centration and the net movement of organelle transport was found to be significant. Centrosome centration was impaired when Rab7 and RILP, which mediate the association between organelles and dynein in mammalian cells, were knocked down. These results indicate that minus end-directed transport of intracellular organelles along the microtubules is required for centrosome centration in C. elegans embryos. On the basis of this finding, we propose a model in which the reaction forces of organelle transport generated along microtubules act as a driving force that pulls the centrosomes toward the cell center. This is the first model, to our knowledge, providing a mechanical basis for cytoplasmic pulling force for centrosome centration.
Keywords: endosome, lysosome, pronuclear migration, the centrosome-organelle mutual pulling model, yolk granule
The centrosome is a major microtubule-organizing center in animal cells. It is generally positioned in the cell center. The central positioning of the centrosome is critical for proper microtubule-dependent cellular activities such as cell division and organelle distribution (1–3). Centrosome centration is an active process in which centrosomes move toward the cell center. For example, after fertilization in many species, the centrosomes associated with the male pronucleus migrate from the cell periphery toward the cell center (4–6). Centrosome centration is achieved by forces that act through the microtubules (6–8). Actin filaments are reported to modulate the speed of centration, but they are not essential for this process (9, 10). Two microtubule-dependent forces, namely, the pushing force and pulling force, are thought to position the centrosome to the cell center. The pushing force is dependent on microtubule polymerization. The plus ends of the growing microtubules can push the cell cortex to move the centrosome away from the cortex (11). Centration of microtubule-organizing centers in fission yeast is driven by microtubule pushing forces (12, 13). In contrast, the major driving force for centrosome centration in animal cells is microtubule pulling forces through the action of cytoplasmic dynein, a minus end-directed motor protein complex (14, 15). The pulling force, but not the pushing force, accounts for the behavior of the centrosomes in Caenorhabditis elegans embryos (16). However, it is still unclear where cytoplasmic dynein is anchored inside the cell to pull the centrosomes toward the center (6, 17–20). In the present report, we refer to the structure as the centrosome centration anchor.
The centrosome centration anchor may be located at the cell cortex (17). Cytoplasmic dynein has been reported to function at the cortex in many cells (21–24). However, these cytoplasmic dyneins at the cortex may not be required for centrosome centration. In sand dollar eggs, centrosome centration occurs in a cortex-independent manner (25). In large amphibian eggs, the plus ends of microtubules do not reach the cell cortex in the traveling direction during centrosome centration (20). In C. elegans, molecules involved in anchoring dynein at the cell cortex are identified, but they are dispensable for centrosome centration. Gα proteins (GOA-1, GPA-16) and their regulators GPR-1 and GPR-2 are involved in anchoring dynein at the cortex (23, 24), and are required for the asymmetric displacement or rocking movements of the mitotic spindle. For centrosome centration, Gα/GPR are dispensable and have an inhibitory effect because the centrosomes reach the cell center within a shorter time in Gα/GPR-suppressed embryos (26–28). It should be noted here that Gα/GPR-dependent forces can modulate centrosome centration as knockdown of Gα/GPR regulators, LET-99 or casein kinase 1 (CSNK-1), positions the centrosomes at a posterior or anterior position, respectively (29, 30). The observations mentioned earlier collectively indicate that dyneins anchored at locations other than the cortex can sufficiently drive centrosome centration.
Alternatively, the centrosome centration anchor may be located throughout the cytoplasm. In this case, the pulling force per microtubule increases as the length of the microtubule increases because the longer the microtubule, the more contact with cytoplasmic dynein (6). This length-dependent pulling mechanism was initially proposed by Hamaguchi and Hiramoto in sand dollar eggs (25). In a computer simulation assuming that the centrosome centration anchors are located throughout the cytoplasm, the in vivo profile of centrosome centration in C. elegans was reproduced (16). However, we are aware of no reports that have elucidated the actual structure in the cytoplasm that anchors cytoplasmic dynein to pull the centrosomes (19).
The purpose of this study was to characterize the centrosome centration anchor. By using C. elegans embryos, we found that DYRB-1, which belongs to the dynein light chain/roadblock (LC7) family, is selectively involved in centration. Interestingly, DYRB-1 was also required for minus end-directed movement of organelles. Knockdown of genes involved in organelle movement revealed that minus end-directed transport of organelles is tightly linked to centrosome centration. These results support the mutual pulling model in which organelles function as cytoplasmic anchors to generate pulling forces for centrosome centration.
Results
A Dynein Light Chain Subunit, DYRB-1, as a Molecule Selectively Involved in Centrosome Centration Anchor.
We investigated male pronuclear migration in the C. elegans embryo as a model for studying the centrosome centration mechanism. After fertilization, the centrosomes and associated male pronucleus migrate from the cell periphery (i.e., near sperm-entry site) to the cell center (Fig. S1A and Movie S1) (4, 5). We first searched for genes specifically involved in centrosome centration anchor by using the PhenoBank database, in which each phenotype during early embryogenesis in C. elegans under full-genome RNAi (19,075 genes) is recorded and categorized (31). In this database, the genes involved in the anchor should be categorized into the P0 pronuclear centration and P0 pronuclear migration (male) defect groups. A total of 56 genes were found in these groups.
To exclude genes required for the motor activity of dynein and the elongation of microtubules, we examined whether the male and female pronuclei met in embryos defective for these genes. This pronuclear meeting occurs during centration and is also dependent on long microtubules and the dynein heavy chain subunit in C. elegans (DHC-1) (32, 33). Therefore, if the genes specifically required for the centrosome centration anchor are inhibited, the meeting of the male and female pronuclei occurs, but the nucleus–centrosome complex does not reach the cell center. We viewed all of the videos (N = 726) in PhenoBank showing the phenotypes of RNAi of the 56 genes and calculated the frequency of the videos displaying the expected phenotype (Table S1). For nine genes, more than 50% of the RNAi embryos showed the expected phenotype. We then checked the phenotype of RNAi-treated cells for the nine genes ourselves and confirmed the phenotype for eight genes (Table 1).
Table 1.
Effect on pronuclear meeting and centration of knockdown of the top nine genes identified in PhenoBank analysis as involved in centration
| Under gpr-1/2 (RNAi)‖ |
|||||||
| Rank* | Genotype | Meeting defect† | Centration defect‡ | Score, %§ | Meeting defect† | Centration defect‡ | Score, %§ |
| 1 | dyrb-1(RNAi) | 0/9 | 9/9 | 100 | 2/13 | 13/13 | 85 |
| 2 | dnc-1(RNAi) | 2/7 | 7/7 | 71 | 8/12 | 12/12 | 33 |
| 2 | dnc-2(RNAi) | 2/6 | 6/6 | 67 | 4/9 | 7/9 | 33 |
| 3 | arp-1(RNAi) | 5/12 | 12/12 | 58 | 8/11 | 11/11 | 27 |
| 4 | gpb-1(RNAi) | 0/10 | 10/10 | 100 | 0/14 | 2/14 | 14 |
| 5 | let-754(RNAi) | 0/9 | 6/9 | 67 | 0/13 | 0/13 | 0 |
| 5 | gpc-2(RNAi) | 0/8 | 8/8 | 100 | 0/13 | 0/13 | 0 |
| 5 | lrg-1(RNAi) | 0/12 | 12/12 | 100 | 0/15 | 0/15 | 0 |
| — | csn-1(RNAi) | 0/14 | 4/14 | 29 | — | — | — |
| — | WT¶ | 0/17 | 0/17 | 0 | 0/7 | 0/7 | 0 |
| — | dhc-1(RNAi) | 6/7 | 6/7 | 0 | 8/8 | 8/8 | 0 |
*Ranking of genes based on score [under the gpr-1/2 (RNAi) condition].
†Number of embryos displaying pronuclear meeting defect divided by total number of embryos in each RNAi.
‡Number of embryos displaying centration defect divided by total number of embryos in each RNAi.
§Percentage of embryos displaying centration defect but not pronuclear meeting defect.
‖Knockdown of top eight genes performed under the gpr-1/2 (RNAi) condition.
¶CAL0092 strain was used as WT.
These eight genes included genes known to be dispensable for centration, such as gpb-1 and gpc-2. Disruption of these genes inhibits centration via hyperactivation of the cortical pulling force (26–28). To exclude such genes, we performed RNAi of the eight genes in embryos with reduced cortical pulling force. To eliminate the cortical pulling force, gpr-1 and gpr-2 were knocked down by RNAi (Movie S2) (34, 35). Consistent with a previous report (27), the centration defect in gpb-1 (RNAi) or gpc-2 (RNAi) embryos was lost in gpr-1/2 (RNAi) embryos, which indicates that gpb-1 and gpc-2 were not involved in centrosome centration anchor (Table 1). let-754 and lrg-1 were also eliminated as they showed similar phenotypes (Table 1). dnc-1, dnc-2, and arp-1 were also eliminated in this stage of the screening for a different reason. These genes were eliminated because knockdown of these genes in the gpr-1/2 (RNAi) embryos inhibited the meeting of pronuclei. We found that these embryos were defective in centrosome separation (Table 1 and Table S2). Without centrosome separation, elongation of astral microtubules is sterically hindered, and thus the pronuclear meeting was inhibited.
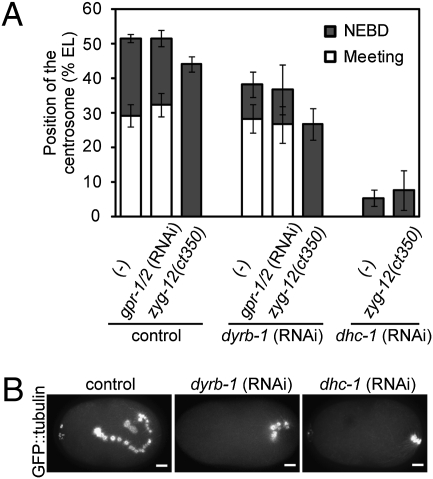
dyrb-1 was the only gene for which RNAi produced a centration defect but no meeting defect in more than 50% of the embryos, even under the gpr-1/2 RNAi condition (Table 1). DYRB-1 in the C. elegans embryo was characterized in past studies, and its involvement in centrosome centration was mentioned (23, 36). dyrb-1 (RNAi) embryos exhibited pronuclear meeting, but further centration was significantly impaired (Fig. 1A and Movie S3). The centrosomes in dyrb-1 (RNAi) embryos fail to reach the cell center [i.e., 50% of egg length (EL)], but still reach approximately 40% of EL (Fig. 1A). We suspected that this partial centration in dyrb-1 (RNAi) embryos was caused by the effect of pronuclear meeting. Pronuclear meeting is thought to be driven by pulling between male and female pronuclei through the function of dyneins anchored on the surface of pronuclei by a nuclear membrane protein ZYG-12 (32). When we eliminated the effect of pronuclear meeting by using a zyg-12 mutant, the centrosome in dyrb-1 (RNAi) embryos reach only 27% of EL (Fig. 1 A and B).
Fig. 1.
DYRB-1 is required for centrosome centration. (A) Mean position of the centrosomes at pronuclear meeting (white bars) and nuclear envelope breakdown (NEBD, gray bars) under the indicated conditions are shown. In dhc-1 (RNAi) embryos or the zyg-12(ct350) background at restrictive temperature, pronuclear meeting did not occur. Position is expressed as percentage of EL (posterior-most side of the egg, 0%). The average value ± SD is shown (n ≥ 5). (B) Time-lapse images of control (untreated), dyrb-1 (RNAi), and dhc-1 (RNAi) embryos expressing GFP-tubulin under the zyg-12(ct350) background at restrictive temperature were projected onto single images to visualize the trajectory of the centrosomes movement until NEBD. (Scale bars, 5 μm.)
dyrb-1 encodes a light chain subunit of cytoplasmic dynein and thus may reasonably function with(in) dynein (23, 36). One role of dynein accessory subunits such as DYRB-1 is to mediate specific associations between cytoplasmic dynein and various cellular structures, such as intracellular organelles (37–40). Therefore, we suspected that DYRB-1 had a direct role in mediating the association between dynein and the centrosome centration anchor.
Intracellular Localization of GFP::DYRB-1.
Previously, O'Rourke et al. and Couwenbergs et al. characterized DYRB-1 in C. elegans (23, 36). O'Rourke et al. focused on the repressive aspects of DYRB-1 in the regulation of DHC-1 activity (36). Because both DHC-1 and DYRB-1 positively contribute to centrosome centration, the role of DYRB-1 as a DHC-1 repressor should not function in the process of centrosome centration. Couwenbergs et al. focused on the function of the cortical fraction of DYRB-1, which acts with Gα and GPR proteins and regulates spindle positioning (23). Because incomplete centration was caused by dyrb-1 RNAi even in the gpr-1/2 loss-of-function background (23) (Fig. 1A), DYRB-1 has Gα/GPR-dependent and Gα/GPR-independent functions, and the latter is required for centrosome centration.
This Gα-independent function for centrosome centration is unlikely to be mediated by the cortical fraction of DYRB-1. If a cortical pulling force were responsible for centration, one would expect many microtubules to make contact with the anterior cortex. Visualization of microtubule plus ends during centrosome centration indicated that few microtubules reach the anterior cortex compared with those that reach the opposite direction (i.e., posterior) (Fig. S1 B–E). This observation suggests that pulling forces produced at the anterior cortex contributes little to centrosome centration.
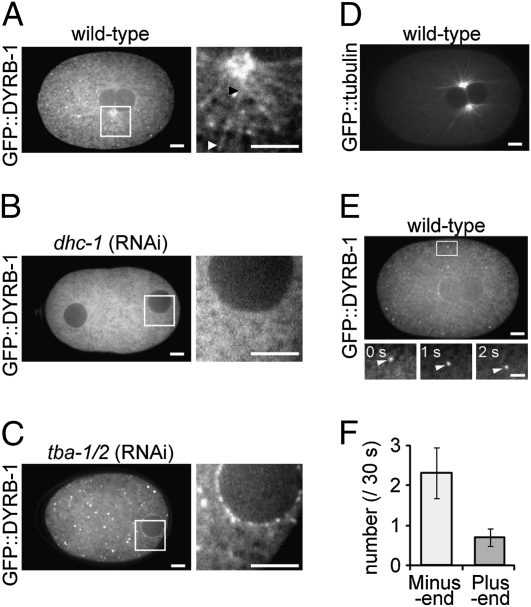
To reexamine the localization of DYRB-1, we generated transgenic worms expressing GFP fused to DYRB-1 (GFP::DYRB-1). This construct partially rescued the defect in organelle transport (DYRB-1 Is Required for the Minus End-Directed Movement of Organelles) caused by RNAi targeting 3′-UTR of dyrb-1 (Fig. S2). GFP::DYRB-1 was weakly localized throughout the cytoplasm, the cortex, and the spindle in early embryos (Fig. 2) (23, 36). We found filamentous signals of GFP::DYRB-1 that were similar to the pattern of astral microtubules (Fig. 2 A and D). Some GFP::DYRB-1 was observed in punctate form, which occasionally moved toward the centrosomes (Fig. 2 E and F and Table S3). The filamentous signal was not observed when DHC-1 or microtubules were impaired by RNAi-mediated knockdown of DHC-1 or α-tubulin (TBA-1/2), respectively (Fig. 2 B and C). The punctate signals were not observed in dhc-1 (RNAi) embryos (Fig. 2B), or did not show apparent centrosome-directed movement in tba-1/2 (RNAi) embryos (Table S3). These results indicate that cytoplasmic DYRB-1 moves along the astral microtubules in a dynein motor activity-dependent manner during centrosome centration.
Fig. 2.
Cytoplasmic localization of GFP::DYRB-1 during centrosome centration. Confocal fluorescence images of C. elegans embryos expressing GFP::DYRB-1 are shown (A–C). The boxed region in each panel is also shown magnified (Right). The arrowheads (black and white) in A indicate both ends of a filamentous signal of GFP::DYRB-1: (B) dhc-1 (RNAi) and (C) tba-1/2 (RNAi). (D) The pattern of astral microtubules revealed using a GFP::tubulin strain. (E) Tracking of DYRB-1 puncta. A fraction of DYRB-1 shows a punctate signal, a portion of which moves directionally as shown in the lower magnified images (bar, 2 μm). (F) The direction and frequency of fast (≥0.5 μm/s) and continuous (≥2 s) movements of DYRB-1 puncta within 30 s after pronuclear meeting (n = 10). The average number ± SEM is shown. (Scale bars, 5 μm except in E, Lower.)
DYRB-1 Is Required for the Minus End-Directed Movement of Organelles.
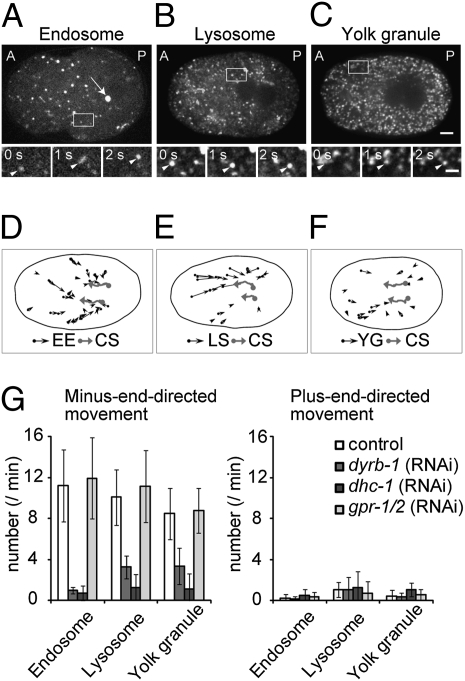
Because cytoplasmic DYRB-1 moves along microtubules, we investigated whether organelle movement along microtubules toward the minus ends occurs during centrosome centration and whether DYRB-1 is involved in the movement. We visualized the movements of early endosomes, yolk granules, or lysosomes by using strains expressing GFP::EEA-1 (FYVE*2) or GFP::VIT-2 or by exposing the worm to the lysosome marker reagent LysoTracker, respectively. In addition to the early endosomes as reported previously (41), yolk granules and lysosomes showed extensive movement toward the centrosomes during centrosome centration (Fig. 3).
Fig. 3.
Movements of intracellular organelles during centrosome centration. Confocal fluorescence images of (A) an embryo expressing both GFP::TBG-1 (arrow) and GFP::EEA-1(FYVE*2), (B) an embryo in which the lysosomes were stained with LysoTracker, and (C) an embryo expressing the yolk granule marker GFP::VIT-2. A, anterior; P, posterior. (Scale bar, 5 μm.) Three sequential fluorescence images of the boxed regions in A–C are magnified (bar, 2 μm) below each panel. The arrowhead marks a moving organelle. (D–F) A typical example of the movements of early endosomes (EE), lysosomes (LS), or yolk granules (YG; ≥0.5 μm/s, black arrows) for 2 min after pronuclear meeting is illustrated. Black ellipse represents the outline of the egg; gray arrows mark the movement of centrosomes (CS). (G) The number of each organelle that moved continuously for at least 2 s at a rate of at least 0.5 μm/s toward the minus end or plus end of the microtubules was counted for 2 min after pronuclear meeting in control, dyrb-1 (RNAi), dhc-1 (RNAi), and gpr-1/2 (RNAi) embryos (n ≥ 5). The average number ± SD of each organelle per 1 min is shown.
These minus end-directed movements of organelles were significantly reduced in dyrb-1 (RNAi) embryos (Fig. 3G). The result indicates a role of DYRB-1 in the cytoplasm during centrosome centration in mediating the interaction between DHC-1 and the organelles to move the organelles along microtubules toward the centrosomes. We did not detect colocalization of DYRB-1 with the organelles (e.g., lysosomes, RAB-5–positive vesicles). The number of motor proteins associated with intracellular vesicles is known to be small (42, 43) and thus hard to be detected by confocal microscopy.
We sought to determine whether the function of DYRB-1 in moving organelles is dependent on Gα/GPR. The minus end-directed movements of organelles were not impaired by RNAi-mediated knockdown of GPR-1/2 (Fig. 3G). Because centrosome centration and organelle movements were both mediated by a Gα/GPR-independent function of DYRB-1, we suspected causal relation between the two processes in a way that intracellular organelles associated with dynein serve as the centrosome centration anchor.
Working Hypothesis: Minus End-Directed Transport of Organelles Generates Cytoplasmic Pulling Forces For Centrosome Centration.
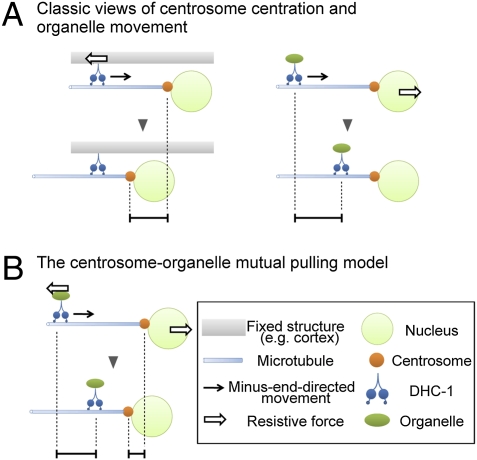
How can intracellular organelles be the centrosome centration anchors to produce forces for centrosome centration? Here, we propose a mutual pulling model in which the centrosomes and the organelles pull each other to appropriately position themselves (Fig. 4). A prevalent view of centrosome movement is that dynein is associated with a fixed structure, and upon dynein function, the centrosomes move toward the dynein (Fig. 4A, Left) (6). In contrast, for organelle movements, the centrosomes are fixed and thus the dynein and associated organelle move toward the centrosome (Fig. 4A, Right). Inside the cell, however, the centrosomes are not completely fixed and thus should somewhat move toward the dynein and organelle upon organelle movement. Although the movement of the centrosome aster should be small compared with that of a single organelle upon the mutual pulling, the rapid movement of many organelles toward centrosomes should generate significant amounts of force that can move the centrosomes (Discussion).
Fig. 4.
The centrosome–organelle mutual pulling model for centrosome centration. (A) Classic views of centrosome centration and organelle movement in which either dynein (Left) or the centrosomes (Right) are fixed and the other side (centrosomes or organelles, respectively) moves. (B) The centrosome–organelle mutual pulling model. When dyneins slide along the microtubule, both dynein–organelle complexes and the centrosomes move toward each other. This mutual movement positions the centrosomes at the cell center.
Correlation Between Movement of Early Endosomes and Centrosome Centration.
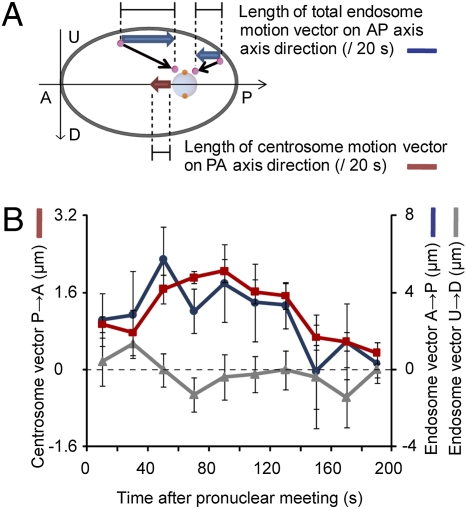
If the movements of the organelles toward the centrosomes generate the pulling force for centration, organelle movements should correlate with centrosome movement. To examine the temporal correlation, we quantified the total movement of the centrosomes and early endosomes every 20 s from the time just after pronuclear meeting (Fig. 5A). Because the centrosomes move mainly along the anterior-posterior (AP) axis of the embryo, we focused on the AP axis component of the movements in the confocal section containing the centrosomes. During the period studied, the speed of centrosome movement along the posterior-to-anterior direction was not constant. After the pronuclear meeting, the centrosomes initially moved toward the center at a relatively lower speed, and then the speed increased. As the centrosome neared the center, its speed slowed to near zero (Fig. 5B). When we summed the AP axis components of the endosome movements directed toward the posterior and subtracted the sum of those directed toward the anterior (Materials and Methods), the net movement gradually increased after the pronuclear meeting but then decreased to nearly zero as the centrosomes reached the cell center (Fig. 5B). This change in the net endosome movement is a result of the change in the direction of the movements as the centrosome moves. The correlation between centrosome movement and the net movements of early endosomes was statistically significant (r = 0.81; P < 0.005). As a negative control, we quantified the endosome movements along the axis perpendicular to the AP axis (“UD axis” in Fig. 5) and confirmed that the net movement along this axis was minimal and did not correlate with the centrosome movement along the AP axis (r = 0.22; P > 0.5). The strong temporal correlation between the net movement of endosomes and centrosome centration supports the idea that moving organelles drive centrosome centration.
Fig. 5.
The correlation between early endosome movement and centrosome centration. (A) Schematic of the measurement of the movement of early endosomes [GFP::EEA-1(FYVE*2), blue arrows] and centrosomes (GFP::TBG-1, red arrow) along the AP axis. The net movements of early endosomes in the anterior (A)-to-posterior (P) direction were calculated by subtracting the sum of P-to-A movements from the sum of A-to-P movements. Likewise, the net movements of early endosomes in the perpendicular direction to the AP axis [upside (U) – downside (D)] were calculated. (Gray ellipse, embryo; light blue circle, pronucleus; pink circle, early endosome; black arrow, movement of early endosome.) The length of movement of total early endosomes and the centrosomes was measured during a 200-s period just after pronuclear meeting. (B) The average length of movement of total early endosomes (blue line, AP axis component; gray line, perpendicular component) and centrosomes (red line, AP axis component) during each 20-s period on the equatorial plane is plotted (n = 9). Left y axis, the length of the centrosome movements (μm); right y axis, the length of total early endosome movements (μm); x axis, time of the measurement from after pronuclear meeting. Error bars represent SEM.
rilp-1 (C32A3.3), a C. elegans Counterpart of RILP, Is Required for Lysosome Transport and Timely Centrosome Centration.
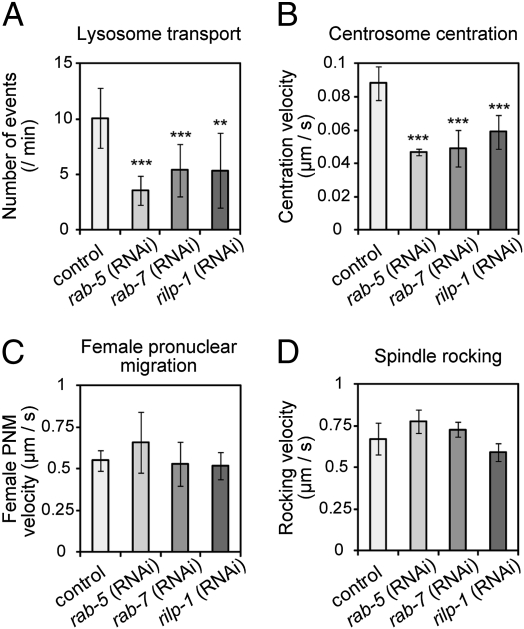
To test whether minus end-directed organelle movements are required for centrosome centration, we attempted to decrease organelle movements by knocking down genes involved in the movements. In mammalian culture cells, the dynein–dynactin complex interacts with RILP, which in turn interact with Rab7, which associates with the surface membrane of late endocytic compartments (44–47). We checked whether this pathway is conserved in C. elegans. RNAi knockdown of rab-7 and C32A3.3, the C. elegans putative homologue of RILP (48), significantly reduced the minus-end motility of lysosomes (P < 0.005; Fig. 6A). The result indicates that the molecular mechanism that anchors lysosomes to microtubules via dynein with RILP and Rab7 is conserved in C. elegans. Based on the primary sequence features and depletion phenotype, we named the C32A3.3 gene rilp-1, for encoding the C. elegans counterpart of RILP.
Fig. 6.
Centrosome centration was delayed when organelle movement was impaired. (A) The average number ± SD of lysosomes that moved continuously for at least 2 s at a rate of at least 0.5 μm/s during the 60 s after pronuclear meeting in WT, rab-5 (RNAi), rab-7 (RNAi), and rilp-1 (RNAi) embryos (n ≥ 7). (B) Average velocity ± SD (μm/s) of centrosome centration (n ≥ 7). (C) Average velocity ± SD (μm/s) of female pronuclear migration during the fast phase (n ≥ 8). (D) Average velocity ± SD (μm/s) of the maximum swing of the posterior spindle pole at metaphase (n ≥ 4). A statistically significant difference from control is indicated by asterisks (***P < 0.001, **P < 0.01).
When minus end-directed movement is impaired in rab-7 (RNAi) and rilp-1 (RNAi) embryos, the movement of the centrosomes toward the center was significantly delayed (P < 0.001; Fig. 6B). As negative controls, we confirmed that other events that are dependent on microtubules and dynein were not delayed under these RNAi conditions. These control events included the fast phase of female pronuclear migration (33) (P > 0.05; Fig. 6C), the rocking movement of the spindle (24) (P > 0.05; Fig. 6D), and centrosome separation (14) (Table S2). Selective inhibition of organelle movement and centrosome centration was also observed upon inhibition of rab-5 (Fig. 6). Rab5 regulates the early endocytic pathways and the motility of early endosomes on microtubules through the action of dynein (40, 49). These results indicate that rab-7, rilp-1, and rab-5 do not affect microtubule- or dynein-dependent processes in general. Importantly, RNAi of these genes did not affect the rocking movement of the spindle (Fig. 6D), which was affected by dyrb-1 RNAi through the regulation of Gα/GPR-dependent cortical pulling forces (Movie S3) (23). This finding suggests that the functions of rab-7, rilp-1, and rab-5 are independent of the function of DYRB-1 in the cortical Gα/GPR pathway and supports the notion that the cytoplasmic fraction of DYRB-1 contributes to centrosome centration.
Discussion
In this study, we propose a model in which the centrosome and organelles pull each other to position the centrosome at the cell center. For centrosome centration, the force exerted upon each microtubule is proposed to be proportional to its length (6, 16, 18, 25), but no mechanical basis for this idea exists. Because the organelles are distributed equally throughout the cell, the resultant pulling force per microtubule will be proportional to the microtubule length. Our model thus provides the first mechanical basis, to our knowledge, for the microtubule length-dependent force for centrosome centration.
In theory, movements of small organelles can produce sufficient force to move the nucleus-centrosome complex for centration. If we assume that the cytoplasm is a viscous fluid for which the Stokes law is valid, the drag force scales with the radius of the objects and its velocity (6, 50). In the C. elegans embryo, the pronuclear radius is approximately 5 μm, which is 10-fold larger than the radius of the organelles (e.g., approximately 0.5 μm for early endosomes). In contrast, the average velocity of organelle movement is approximately 1 μm/s, which is actually approximately 10 times faster than that of the nucleus–centrosome complex (0.1 μm/s) during centrosome centration. Therefore, if the Stokes law is valid for the movement of the nucleus–centrosome complex, fast movement of only one small organelle is sufficient to move the complex. The cytoplasm might not behave as an ideal viscous fluid, but this estimation suggests that organelle movements can exert sufficient forces to drive the centration of the nucleus-centrosome complex. Daniels et al. successfully estimated the viscosity in early C. elegans embryos (51), suggesting that the cytoplasm behaves as a viscous fluid. Further biophysical studies on the properties of cytoplasm are required for accurate estimation of the force balance under the centrosome centration.
In most cells, the centrosome is centrally located (8). Because cytoplasmic dyneins are required for the centration in many organisms (14, 15, 18), the mutual pulling model can be applied to these cells in general. Characterization of the relationships between organelle transport and the centration of the centrosomes and other structures in other cells is thus an interesting topic for future study. In the proposed mutual pulling model, we do not have to assume that a specific anchoring structure specialized for centrosome centration exists. Even if the centrosomes and organelles are randomly positioned, self-organization inside the cell will lead to the positioning of the centrosome at the center and the organelles around the centrosome. Centrosome centration is a fundamental feature of animal cells that is important for equal cell division. Our mutual pulling model provides a simple and robust mechanism for centrosome centration and thus the basis for organelle positioning inside the cell.
Materials and Methods
Strains and Manipulation of C. elegans.
C. elegans strains were maintained using standard techniques (52). RNAi was performed by injecting dsRNAs as described previously (53). Details of the strains and the procedure of RNAi experiments are described in SI Materials and Methods.
Microscopy, Quantification, and Statistical Analyses.
Image acquisition, quantification of the movements of organelles and centrosomes, and statistical analyses were performed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to the members of our laboratory for discussion and J. Ahringer (University of Cambridge, Cambridge, United Kingdom), H. Kagoshima, Y. Kohara, N. Kurata, K. Noguchi (National Institute of Genetics, Mishima, Japan), S. Onami (RIKEN, Yokohama, Japan), G. Seydoux (Johns Hopkins University, Baltimore), A. Sugimoto, M. Terasawa (RIKEN, Kobe, Japan), and T. Uemura (Kyoto University, Kyoto, Japan) for providing clones, strains, and technical advice. Strains were also provided by the Caenorhabditis Genetics Center, funded by the National Institutes of Health. We appreciate P. Gönczy (École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland), I. Tolic-Nørrelykke (Max-Planck-Institute, Dresden, Germany), M. Ginzberg and M. Wühr (Harvard Medical School, Boston) for reading the manuscript, and S. Bullock (Medical Research Council Laboratory of Molecular Biology, Cambridge, United Kingdom), T. Schüpbach, and E. Wieschaus (Princeton University, Princeton, NJ) for advice. K.K. is a National Institute of Genetics postdoctoral fellow. This study was supported by a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by the Transdisciplinary Research Integration Center of the Research Organization of Information and Systems, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013275108/-/DCSupplemental.
References
- 1.Howard J, Hyman AA. Dynamics and mechanics of the microtubule plus end. Nature. 2003;422:753–758. doi: 10.1038/nature01600. [DOI] [PubMed] [Google Scholar]
- 2.Rios RM, Bornens M. The Golgi apparatus at the cell centre. Curr Opin Cell Biol. 2003;15:60–66. doi: 10.1016/s0955-0674(02)00013-3. [DOI] [PubMed] [Google Scholar]
- 3.Doxsey S, McCollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- 4.Nigon V, Guerrier P, Monin H. L'architecture polaire de l'oeuf et les mouvements des constituants cellulaires au cours des premieres etapes du developpement chez quelques nematodes. Bull Biol Fr Belg. 1960;93:131–202. [Google Scholar]
- 5.Albertson DG. Formation of the first cleavage spindle in nematode embryos. Dev Biol. 1984;101:61–72. doi: 10.1016/0012-1606(84)90117-9. [DOI] [PubMed] [Google Scholar]
- 6.Reinsch S, Gönczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;111:2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- 7.Euteneuer U, Schliwa M. Mechanism of centrosome positioning during the wound response in BSC-1 cells. J Cell Biol. 1992;116:1157–1166. doi: 10.1083/jcb.116.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kellogg DR, Moritz M, Alberts BM. The centrosome and cellular organization. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- 9.Strome S, Wood WB. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- 10.Goulding MB, Canman JC, Senning EN, Marcus AH, Bowerman B. Control of nuclear centration in the C. elegans zygote by receptor-independent Galpha signaling and myosin II. J Cell Biol. 2007;178:1177–1191. doi: 10.1083/jcb.200703159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holy TE, Dogterom M, Yurke B, Leibler S. Assembly and positioning of microtubule asters in microfabricated chambers. Proc Natl Acad Sci USA. 1997;94:6228–6231. doi: 10.1073/pnas.94.12.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran PT, Marsh L, Doye V, Inoué S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tolić-Nørrelykke IM, Sacconi L, Thon G, Pavone FS. Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Curr Biol. 2004;14:1181–1186. doi: 10.1016/j.cub.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 14.Gönczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burakov A, Nadezhdina E, Slepchenko B, Rodionov V. Centrosome positioning in interphase cells. J Cell Biol. 2003;162:963–969. doi: 10.1083/jcb.200305082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura A, Onami S. Computer simulations and image processing reveal length-dependent pulling force as the primary mechanism for C. elegans male pronuclear migration. Dev Cell. 2005;8:765–775. doi: 10.1016/j.devcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Grill SW, Hyman AA. Spindle positioning by cortical pulling forces. Dev Cell. 2005;8:461–465. doi: 10.1016/j.devcel.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 18.Vallee RB, Stehman SA. How dynein helps the cell find its center: a servomechanical model. Trends Cell Biol. 2005;15:288–294. doi: 10.1016/j.tcb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 20.Wühr M, Dumont S, Groen AC, Needleman DJ, Mitchison TJ. How does a millimeter-sized cell find its center? Cell Cycle. 2009;8:1115–1121. doi: 10.4161/cc.8.8.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heil-Chapdelaine RA, Oberle JR, Cooper JA. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J Cell Biol. 2000;151:1337–1344. doi: 10.1083/jcb.151.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dujardin DL, et al. A role for cytoplasmic dynein and LIS1 in directed cell movement. J Cell Biol. 2003;163:1205–1211. doi: 10.1083/jcb.200310097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couwenbergs C, et al. Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J Cell Biol. 2007;179:15–22. doi: 10.1083/jcb.200707085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen-Ngoc T, Afshar K, Gönczy P. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol. 2007;9:1294–1302. doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- 25.Hamaguchi MS, Hiramoto Y. Analysis of the role of astral rays in pronuclear migration in Sand dollar eggs by the colcemid-UV method. Dev Growth Differ. 1986;28:143–156. doi: 10.1111/j.1440-169X.1986.00143.x. [DOI] [PubMed] [Google Scholar]
- 26.Gotta M, Ahringer J. Distinct roles for Galpha and Gbetagamma in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat Cell Biol. 2001;3:297–300. doi: 10.1038/35060092. [DOI] [PubMed] [Google Scholar]
- 27.Tsou MF, Hayashi A, Rose LS. LET-99 opposes Galpha/GPR signaling to generate asymmetry for spindle positioning in response to PAR and MES-1/SRC-1 signaling. Development. 2003;130:5717–5730. doi: 10.1242/dev.00790. [DOI] [PubMed] [Google Scholar]
- 28.Kimura A, Onami S. Local cortical pulling-force repression switches centrosomal centration and posterior displacement in C. elegans. J Cell Biol. 2007;179:1347–1354. doi: 10.1083/jcb.200706005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsou MF, Hayashi A, DeBella LR, McGrath G, Rose LS. LET-99 determines spindle position and is asymmetrically enriched in response to PAR polarity cues in C. elegans embryos. Development. 2002;129:4469–4481. doi: 10.1242/dev.129.19.4469. [DOI] [PubMed] [Google Scholar]
- 30.Panbianco C, et al. A casein kinase 1 and PAR proteins regulate asymmetry of a PIP(2) synthesis enzyme for asymmetric spindle positioning. Dev Cell. 2008;15:198–208. doi: 10.1016/j.devcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sönnichsen B, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 32.Malone CJ, et al. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- 33.Oegema K, Hyman AA. Cell division. WormBook. 2006;Jan 19:1–40. doi: 10.1895/wormbook.1.72.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colombo K, et al. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 2003;300:1957–1961. doi: 10.1126/science.1084146. [DOI] [PubMed] [Google Scholar]
- 35.Gotta M, Dong Y, Peterson YK, Lanier SM, Ahringer J. Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr Biol. 2003;13:1029–1037. doi: 10.1016/s0960-9822(03)00371-3. [DOI] [PubMed] [Google Scholar]
- 36.O'Rourke SM, Dorfman MD, Carter JC, Bowerman B. Dynein modifiers in C. elegans: Light chains suppress conditional heavy chain mutants. PLoS Genet. 2007;3:e128. doi: 10.1371/journal.pgen.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 38.Mallik R, Gross SP. Molecular motors: strategies to get along. Curr Biol. 2004;14:R971–R982. doi: 10.1016/j.cub.2004.10.046. [DOI] [PubMed] [Google Scholar]
- 39.Pfister KK, et al. Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet. 2006;2:e1. doi: 10.1371/journal.pgen.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satoh D, et al. Spatial control of branching within dendritic arbors by dynein-dependent transport of Rab5-endosomes. Nat Cell Biol. 2008;10:1164–1171. doi: 10.1038/ncb1776. [DOI] [PubMed] [Google Scholar]
- 41.Andrews R, Ahringer J. Asymmetry of early endosome distribution in C. elegans embryos. PLoS ONE. 2007;2:e493. doi: 10.1371/journal.pone.0000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shubeita GT, et al. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 2008;135:1098–1107. doi: 10.1016/j.cell.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hendricks AG, et al. Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr Biol. 2010;20:697–702. doi: 10.1016/j.cub.2010.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng Y, Press B, Wandinger-Ness A. Rab 7: An important regulator of late endocytic membrane traffic. J Cell Biol. 1995;131:1435–1452. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Méresse S, Gorvel JP, Chavrier P. The rab7 GTPase resides on a vesicular compartment connected to lysosomes. J Cell Sci. 1995;108:3349–3358. doi: 10.1242/jcs.108.11.3349. [DOI] [PubMed] [Google Scholar]
- 46.Jordens I, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- 47.Johansson M, et al. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): The Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bucci C, et al. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 50.Berg HC. Random Walks in Biology. Princeton: Princeton University Press; 1993. [Google Scholar]
- 51.Daniels BR, Masi BC, Wirtz D. Probing single-cell micromechanics in vivo: the microrheology of C. elegans developing embryos. Biophys J. 2006;90:4712–4719. doi: 10.1529/biophysj.105.080606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hara Y, Kimura A. Cell-size-dependent spindle elongation in the Caenorhabditis elegans early embryo. Curr Biol. 2009;19:1549–1554. doi: 10.1016/j.cub.2009.07.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.