Abstract
We have determined the 2.3-Å-resolution crystal structure of a myosin light chain domain, corresponding to one type found in sea scallop catch (“smooth”) muscle. This structure reveals hinges that may function in the “on” and “off” states of myosin. The molecule adopts two different conformations about the heavy chain “hook” and regulatory light chain (RLC) helix D. This conformational change results in extended and compressed forms of the lever arm whose lengths differ by 10 Å. The heavy chain hook and RLC helix D hinges could thus serve as a potential major and localized source of cross-bridge compliance during the contractile cycle. In addition, in one of the molecules of the crystal, part of the RLC N-terminal extension is seen in atomic detail and forms a one-turn alpha-helix that interacts with RLC helix D. This extension, whose sequence is highly variable in different myosins, may thus modulate the flexibility of the lever arm. Moreover, the relative proximity of the phosphorylation site to the helix D hinge suggests a potential role for conformational changes about this hinge in the transition between the on and off states of regulated myosins.
Keywords: protein crystal structure, light chain binding domain, glycine, regulation
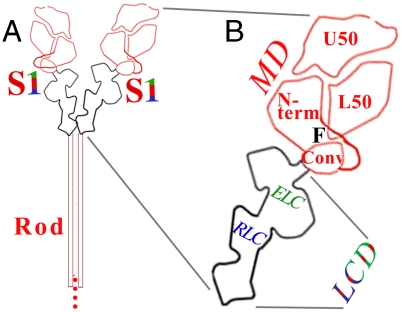
The function of the myosin head (S1) in muscle contraction and other cellular processes relies on remarkable molecular mechanical properties in which relatively rigid subdomains, joined by flexible linkers, are able to reorient relative to one another (1). Since the atomic structure of the myosin head was first determined in 1993 (2), a primary goal of crystallographic as well as other biophysical techniques has been to define the precise changes in structure that occur during the contractile cycle, as well as during the transition between the on and off states in regulated myosins. For both these functions, the light chain binding domain (LCD) [composed of the essential light chain (ELC), regulatory light chain (RLC), and a portion of the heavy chain (HC)] plays a central role.
In regulated myosins, the LCD (also referred to in the literature as “the regulatory domain”) is the location of the binding sites for regulatory ions. In certain invertebrate muscles, Ca2+ binds directly to the ELC portion of myosin (3). In vertebrate smooth muscles, Ca2+ activates myosin light chain kinase to phosphorylate the N-terminal region of the RLC (for review, see ref. 4). In both muscles, double-headed myosin is required for full regulation (5, 6). Sedimentation studies (7, 8) and electron microscope studies (8–10) show that the removal of the regulatory ions from the LCD of either myosin results in a conversion of the molecule to a relatively compact asymmetric conformation in the “off state.” Here, the two heads adopt different conformations and are bent back toward the alpha-helical coiled-coil rod. The detailed interactions in the off state are not yet established due to the lack of an atomic structure.
The LCD is the major element of myosin’s lever arm in the contractile cycle. The binding of nucleotide or actin to the motor domain (MD) induces small conformational changes between the upper 50 kDa subdomain, lower 50 kDa subdomain, and N-terminal subdomain (11). The region of the SH1–SH2 helix serves as a fulcrum about which these conformational changes are amplified by the converter and attached LCD (12, 13) (see Fig. 1). Hence, the LCD has often been called “the lever arm” in the literature. (Note that a lever arm, however, is mechanically defined as the region extending from the fulcrum, and in myosin would thus also include the converter.)
Fig. 1.
Schematic diagrams of (A) myosin and (B) an expanded view of the head (i.e., S1), showing locations of the LCD, converter (conv), and subdomains of the MD. F indicates the fulcrum.
The concept of a tilting cross-bridge (14, 15), or more specifically swinging lever arm (16), is critical. Here, the step size that myosin takes on actin is related to the length of the LCD (12, 17). Underlying this idea is the implicit assumption of a relatively rigid or semirigid lever arm that allows efficient transmission of force. Indeed, LCDs from bay scallop (Argopecten irradians) striated-muscle myosin (or from squid myosin) have shown only relatively small conformational changes in different crystal environments (see refs. 18 and 19 and below). Spectroscopic studies on vertebrate skeletal and gizzard myosins have also indicated a semirigid LCD (reviewed in ref. 20).
We have begun crystallographic studies on an LCD corresponding to one type found in the catch muscle of the sea scallop Placopecten magellanicus. The original goal of this study was to improve our understanding of regulation. No vertebrate smooth muscle myosin construct containing its phosphorylatable RLC has been crystallized, either in its phosphorylated or unphosphorylated state, perhaps due to the length of the N-terminal extension of this light chain. In the Placopecten catch muscle, there are two distinct RLCs present, SmoA and SmoB, formed by alternative splicing (21). (The SmoA RLC accounts for ∼40% of the catch muscle’s RLC population.) We chose the former, termed “Placo SmoA,” because this RLC contains a relatively short N-terminal extension, like that in the easily crystallizable LCDs from scallop striated muscles, and also because it contains the signature RxxS sequence necessary for phosphorylation, like that in vertebrate smooth muscle myosin RLC. We know that Ca2+ is essential for activating this Placopecten catch muscle isoform, but no information is yet available about the role of RLC phosphorylation. The Placo SmoA LCD construct does in fact crystallize, and we report here the atomic structure of this construct, in the unphosphorylated state and in the presence of Ca2+. The crystallographic results unexpectedly reveal major hinges in the RLC region that could contribute to cross-bridge compliance during the contractile cycle as well as to the attainment of the asymmetric structure of regulated myosins in the off state.
Results and Discussion
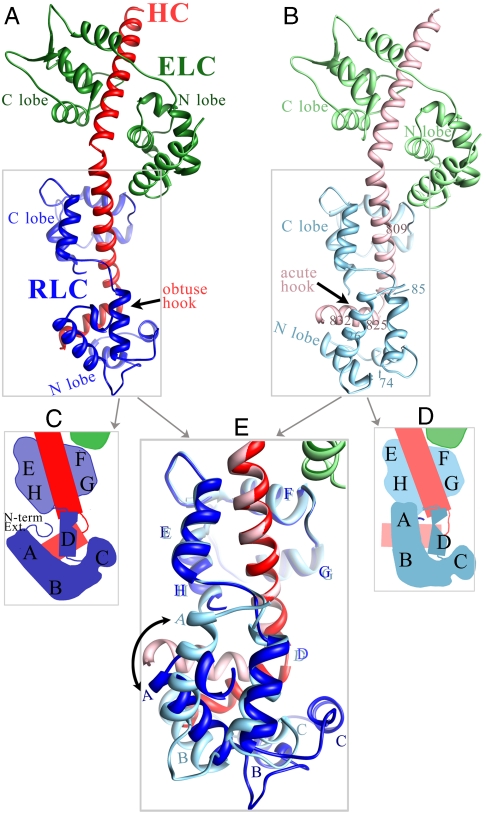
The crystal examined contains two molecules of Placo SmoA LCD in different packing environments in the unit cell, referred to as “molecule 1” (drawn in bright colors in Figs. 2 A and C, 3A, 4A, and 5A) and “molecule 2” (pale colors in Figs. 2 B and D, 3B, 4B, and 5B). These structures display major conformational differences in the RLC region (Fig. 2E and summarized in Fig. 2 legend), specifically about the heavy chain hook (Fig. 3) and RLC helix D (Fig. 4), as well as in the structure of the RLC N-terminal extension (Fig. 5). In addition to these structures in Placo SmoA, we also describe in this section their relevance to other myosin isoforms (Figs. 6 and 7), using Placo SmoA numbering unless otherwise noted.
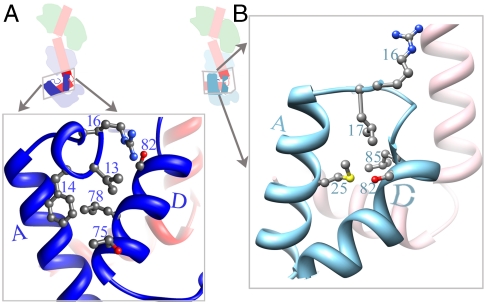
Fig. 2.
Crystal structure of a myosin LCD corresponding to that in Placo SmoA. As in other LCD structures, a portion of the HC (red) is bound to the ELC (green) and the RLC (blue). In this Placo SmoA crystal, however, there are two crystallographically independent LCD molecules whose conformations are similar in the ELC region and ELC–RLC interface but are highly divergent from each other in the RLC region (boxed). (A) In molecule 1 (bright colors), the HC segment bound to the RLC forms an obtuse-angled hook, the HC is relatively long, RLC helix D is straight, and its C-terminal portion is far from helix A. Also, the RLC N-terminal extension forms a short alpha-helix. (B) In molecule 2 (faded colors, and reoriented to align with molecule 1), the HC hook angle is acute, the HC is relatively short, RLC helix D is bent in the middle, and its C-terminal portion contacts helix A. Moreover, most of the RLC N-terminal extension is disordered. (Note that in the actual crystal lattice, molecule 2 is oriented differently than molecule 1, and that they contact each other using various parts of the ELC, RLC, and HC.) (C) Schematic of the RLC region of molecule 1. (D) Schematic of RLC region of molecule 2. (E) Overlay of the two Placo SmoA molecules based on fitting residues 811–818 of the HC (immediately N-terminal to the hook). Only the RLC region is shown. Note, close positional match of helix D, in particular its C terminus, as well as helices E–H, in the two molecules. Expanded views of the crystal structure are shown in Figs. 3, 4, and 5.
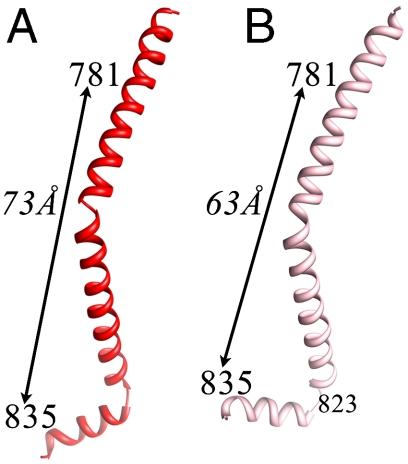
Fig. 3.
The HC adopts different angles about the hook in the two independent molecules of the Placo SmoA LCD crystal. As a result, the length of the LCD varies by 10 Å. (Residues shown define the endpoints of the HC bound by the light chains.) Relative to the obtuse-angled structure, formation of the acute-angled HC involves a partial shortening of the HC helix just prior to the hook, with a breaking of main-chain H bonds between residues 819–823 and 820–824. (A) HC of molecule 1. (B) HC of molecule 2.
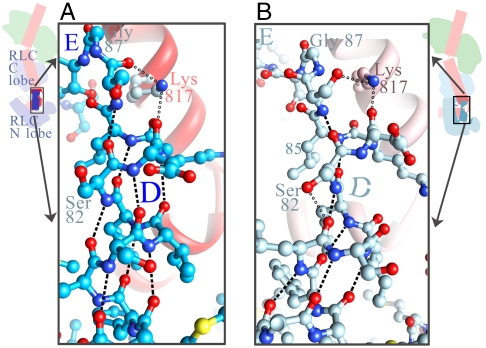
Fig. 4.
RLC helix D has different conformations about a hinge in its middle in the two independent Placo SmoA LCD molecules. (A) In molecule 1, helix D is an intact alpha-helix, where all main-chain H bonds are formed. (Serine 82 side chain forms an H bond with arginine 16, see Fig. 5). (B) In molecule 2, helix D is bent, and two of the main-chain H bonds at the location of the bend are not made; instead, an H bond is made between the side chain of serine 82 and the main-chain carbonyl of residue 78. (In both panels, dashes are H bonds between main-chain atoms; hollow circles are H bonds between a main-chain atom and a side-chain atom.)
Fig. 5.
A significant difference between the two crystallographically independent Placo SmoA LCD molecules also occurs for the RLC N-terminal extension. (A) In molecule 1, there is a short well-ordered alpha-helix in the RLC’s N-terminal extension between residues 12–16. (B) This alpha-helix is not formed in molecule 2, and only two residues N-terminal to RLC helix A are ordered. Note difference in contacts of this extension with the helix D hinge in the two molecules. The figure also shows the different angle between helix A and the C-terminal portion of helix D, largely as a result of the hinge in the middle of helix D. The conformation of molecule 2 involves the formation of apolar contacts of methionine 25 with leucine 85 and contacts of methionine 25 and leucine 17 with serine 82.
Fig. 6.
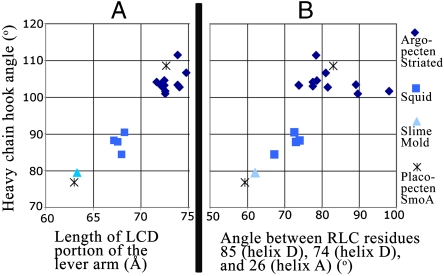
The large differences in the conformation of the LCD observed between the two independent Placo SmoA molecules has not yet been seen in any other isoform. (A) The length of the LCD (calculated between HC residues 781 and 835) and the magnitude of the angle of the HC about the hook (calculated between the alpha-carbons of residues 809, 825, and 832) are displayed. (B) Among the available S1 and LCD structures, the angle of the HC hook (y axis, as in A) correlates with the angle between residues (indicated) in RLC helices A and D, consistent with flexing about the hook and helix D being mechanically linked. For both panels, in addition to the current structure, the other isoforms examined for which multiple independent structures are available include bay scallop (Argopecten irradians) cross-striated-muscle S1 or LCD (Protein Data Bank ID code 3JTD, 3JVT, 1WDC, 1S5G, 1SR6, 1QVI, 1KQM, 1KWO, 1L2O, 1KK8, 1KK7) and squid S1 (3I5F, 3I5G, 3I5H, 3I5I). Slime mold (Physarum polycephalum) LCD (2BL0) is also shown.
Fig. 7.
Locations of glycines (blue, shaded) in diverse RLC sequences suggest that a hinge in helix D is a conserved feature of myosin RLC. See text. Alpha-helices (underlined) and residue numbers are indicated for Placo SmoA. Placo SmoA and human smooth-muscle RLCs, which include the phosphorylatable RxxS sequence (italicized), show identical sequences for the nearby nine-residue stretch in the N-terminal extension (lowercase red), despite being from such different organisms (see text). (Note N-terminal extensions prior to helix A are not aligned.) The databank identification numbers for the listed RLC sequences are: 1389849 (1. Placo SmoA), p13543 (2. Argo striated), Protein Data Bank ID code 3I5G (3. Squid), 18143650 (4. Physarum), P02609 (5. chicken skeletal), BAB88917.1 or GI:29568111 (6. human smooth), NP_000423.2 (7. human slow cardiac RLC2).
Flexing About the Heavy Chain Hook.
All myosin S1 and LCD structures show a sharp turn in the path of the HC approximately 11 residues before the head-rod boundary. The current structure of Placo SmoA LCD shows an angle of 109° about this hook in molecule 1 and 77° in molecule 2 (Figs. 2, 3, and 6). As a result, the length of the LCD portion of the lever arm (as measured by the distance between HC residues 781 and 835) is 72.7 Å for molecule 1 and 63.0 Å for molecule 2 (Fig. 3). Chicken skeletal S1 (2) and slime mold (Physarum polycephalum) LCD (22) structures have shown conformations similar to that of molecule 2; Argopecten striated-muscle S1 (13, 18, 23) and LCD (24, 25) structures are similar to that of molecule 1; squid S1 structures (19) are intermediate (Fig. 6). The current Placo SmoA structures are the first, however, to show two such different conformations within a single isoform.
Flexing About RLC Helix D.
In addition to the flexing of the HC helices about the hook, there is also a reorientation between the N and C lobes of the Placo SmoA RLC (Figs. 2 and 4). The center of the hinge in the RLC, however, is located in large part within helix D, near residue 82 (Fig. 4), rather than being located exclusively in the D–E linker that is normally considered the junction of the lobes. Hence, the C terminus of helix D together with the C lobe (helices E–H) adopt different orientations relative to helices A, B, and C of the N lobe in the two molecules (Figs. 2E and 4). In molecule 1, RLC helix D is straight (Fig. 4A), and its C-terminal portion is relatively far from helix A (Fig. 5A). In molecule 2, RLC helix D is bent about serine 82 (Fig. 4B), and its C-terminal portion forms apolar contacts with helix A (Fig. 5B).
The numerous contacts between the HC and RLC (26) coordinate the conformational changes of these two chains. RLC helices A, B, C, and the N-terminal part of helix D bind to the HC helix C-terminal to the hook (i.e., residues 824–837); the C terminus of RLC helix D and RLC helices E, F, G, and H bind to the HC helix N-terminal to the hook (residues 804–822). Moreover, among the current and previously determined structures, wherever the HC hook angle is relatively large (such as in molecule 1 of the current Placo SmoA structure and in all 11 of the Argopecten S1 and LCD structures) so also is the angle between RLC helix D and helix A. Wherever the HC hook angle is small or intermediate (as in molecule 2 of the current structure and in the slime mold LCD and squid S1 structures) so also is that between helices D and A (Fig. 6B). These observations show that the HC hook and RLC helix D hinges are mechanically linked to one another.
Certain residues of Placo SmoA appear to promote formation of the hinge in its RLC helix D and to link the C terminus of this helix with the RLC C lobe (Fig. 4). Although most of the HC helix immediately N-terminal to the hook is bound to the RLC C lobe, the positively charged side chain of HC lysine 817 caps the C terminus of RLC helix D in both molecule 1 and molecule 2 (Fig. 4) and helps strengthen the connection of the C terminus of helix D with the C lobe. (Such a connection has also been noted for Argopecten striated-muscle LCD; ref. 24.) In addition, the Placo SmoA RLC helix D contains two serines, including one at position 82, which is in the middle of the bend of this helix that occurs in molecule 2 (Fig. 4B). Here, the hydroxyl side chain of this serine is H-bonded to the main-chain carbonyl of residue 78, disrupting the optimal alpha-helical geometry of the main-chain H bond between residue 78 and 82 (see also ref. 27).
In addition to these observations, a comparison of sequences from different isoforms suggests that a hinge in helix D is likely to be a general feature of muscle myosin RLCs rather than a singularity of the current crystal. Although residue 817 of the HC is a lysine only in mollusks, residue 82 of the RLC is a serine in scallop sequences and is a glycine in most other myosins (Fig. 7). Due to the flexibility of its main chain, the frequency of glycine residues in the middle of intact alpha-helices is lower than all other (non-proline) amino acid residues (28, 29). In RLCs, nearly all of the loops between the alpha-helices contain glycines, and nearly all glycines are located in the loops—or in helix D (Fig. 7).
Confirmation of our prediction of significant flexibility about the HC hook and RLC helix D hinges in diverse myosins will require further experimental verification, which is currently limited. Although spectroscopic measurements (of skeletal and gizzard myosins) have indicated some flexibility about the ELC-RLC junction (albeit less than that about the MD-LCD junction, see review, ref. 20), there appear to be no similar experiments that have directly tested the flexibility between the N and C lobes of the RLC or about the HC hook. As indicated above, X-ray crystallographic studies of the 11 Argopecten S1 and LCD structures and of the four squid S1 structures have shown only a relatively small degree of flexibility about these hinges within each isoform (Fig. 6) (18). Note, however, that the presence of the complete RLC was confirmed only in the two Argopecten LCD structures obtained by reconstitution of the domain using RLC purified by a nonproteolytic method (25); the remaining Argopecten and squid structures were obtained by proteolysis, and the RLC N-terminal extension (shown to be nicked in these Argopecten structures) is not visualized. This extension, as we next explain, may influence the flexibility of the molecule.
The RLC N-Terminal Extension.
The Placo SmoA LCD construct that we have crystallized includes the complete RLC (see Methods). In addition to the conformational differences about the HC hook (Fig. 3) and RLC helix D (Fig. 4), there is also a change between molecule 1 and molecule 2 in the order and conformation of the extension immediately N-terminal to RLC helix A (Fig. 5). In molecule 2, this extension is poorly ordered: The first residue observed is arginine 16, whose side chain is oriented away from the helix-A–helix-D interface (Fig. 5B).
In molecule 1, the extension N-terminal to helix A is better ordered. Here the structure is visualized starting at residue 12 (immediately following the phosphorylation site at residue 11) and forms a well-ordered short alpha-helix between residues Asn 12 and Arg 16 (Fig. 5A). This helix is oriented roughly perpendicular to the main helix A, and places valine 13 and phenylalanine 14 in a position to form apolar contacts with residues 75 and 78 near the N-terminal part of helix D. Arginine 16 is also reoriented relative to that in molecule 2, and in molecule 1 forms an H bond with Ser 82 side chain, thus preventing it from disrupting the alpha-helical geometry of helix D. An alpha-helix N-terminal to RLC helix A occurs in certain other members of the calmodulin superfamily, such as in troponin C. The current observation of this alpha helix in an RLC confirms predictions from sequence (25).
In addition to the current Placo SmoA crystal structure, contacts are also observed between RLC helix D and the RLC N-terminal extension (albeit with higher B factors) in the two Argopecten LCD structures that contain a complete RLC (25). These contacts suggest a role for this extension in affecting the flexibility of the structure about helix D, and hence about the HC hook. High sequence variability of the N-terminal extension (Fig. 7) may then indicate that different isoforms have inherently different levels of flexibility about these hinges. Vertebrate smooth muscle RLC and Placo SmoA RLC do, however, share a strikingly identical sequence between residue 7–15, which includes the phosphorylation site and certain helix D contacting residues (Fig. 7). These isoforms may thus share a similar (high) level of flexibility about the helix D and HC hook hinges.
Implications for the On State.
The ability of actin and myosin to extend axially or to shorten in response to changes in force affects the contractile properties of muscle, and numerous studies have sought to define the precise molecular sources of such compliance. The presence of an elastic element in the myosin cross-bridge has long been theorized (30). X-ray fiber diffraction and mechanical studies of frog skeletal muscle suggest that the cross-bridge contributes about a quarter, i.e., 14 Å/isometric force, of the total compliance in a half sarcomere (31–33). Mechanical experiments on mutations of human slow skeletal muscle beta-myosin indicate that a region of the converter, located near its junction with the LCD, could be one source of cross-bridge compliance in this isoform (34). Polarized fluorescence studies of vertebrate skeletal muscle (35) and cryoelectron microscopy reconstructions of insect flight muscle (36) also indicate flexibility near the ELC-RLC interface. Compliance has also been modeled as a relatively uniform bending throughout the lever arm (37, 38).
The current atomic resolution crystal structure of Placo SmoA LCD reveals that the HC hook (Fig. 3) and RLC helix D (Fig. 4) might well contribute another significant, as well as localized, source of compliance in the cross-bridge (see also ref. 18). The two conformations that are observed about these hinges show a difference of 10 Å in the length of the lever arm (Figs. 3 and 6A). The precise component of this conformational change that can contribute to axially directed compliance of the cross-bridge would, of course, vary during the contractile cycle as the orientation of the lever arm, and perhaps also of the hook, changes relative to the axis of the actin filament. A preliminary model corresponding to the end of the power stroke indicates a change of ∼5–10 Å in the axial displacement of the end of the lever arm between the two conformations.
A variety of mutational and light chain dissociation studies have suggested that an increase in the compliance of the lever arm yields a decrease in actin filament velocity (34, 39, 40). The slower contractile velocity of vertebrate smooth muscle relative to skeletal muscle (41) is probably due to numerous factors. The current results, however, provide a specific atomic framework for experiments that could determine the magnitude and physiological consequences of flexibility in the RLC region in different myosin isoforms.
Implications for the Structure of the Off State.
The flexibility of the RLC may contribute to regulation. Upon dephosphorylation, actin binding of vertebrate smooth muscle heavy meromyosin is reduced, but by a small amount relative to the reduction in ATPase activity (42). This feature has been accounted for by the blocking of the actin binding site of one myosin head, but not the other, i.e., by an asymmetric conformation, as seen in electron microscopic reconstructions of the off state (9). This asymmetry has been modeled by different conformations about the MD-LCD junction in the two heads (see also ref. 10). It may turn out that the asymmetry in the off state also extends to the conformation about the HC hook and RLC helix D hinges. A role for conformational changes about these hinges in the off state and/or in the transition between the on and off states is also suggested by the proximity of the helix D hinge to the phosphorylation site in the current crystal structure.
Coda.
These crystallographic studies have revealed that the RLC appears to have a critical role in the mechanical and contractile properties of muscle, in addition to its well-known role in regulation. This high-resolution study provides an atomic model for a potential source of muscle compliance. Further studies, including mutational experiments on the HC hook, RLC helix D, and the N-terminal extension, will be required to assess the functional implications of their conformational variability for both the off and on states of myosin. Visualization of the atomic structures of double-headed myosins, as well as of the phosphorylated state of the current construct, will make decisive contributions to this goal.
Methods
Protein Purification.
We have formed an LCD which mimics that of Placopecten catch muscle with 100% RLC SmoA. The Placopecten striated muscle was used to obtain the ELC and the heavy chain fragment (HCF) for several reasons: it is much more plentiful than the catch muscle, gives a fourfold myosin yield over catch muscle, and the amino acid sequences of the HCF portion and the ELC of the striated muscle are identical to those of the catch muscle (21, 43).
The expressed Placo RLC SmoA (made by C. Perreault-Micale; ref. 21) was dried by speed vacuum and stored at -20 °C. The original DNA sequencing confirms that the complete N terminus of this RLC is present, and that there are five additional amino acids when compared to the Placo SmoB and striated RLCs’ sequences.
Placopecten striated-muscle myosin was prepared according to Stafford and coworkers (44). The ELC was obtained from a guanidine total light chain preparation of Placopecten striated-muscle myosin as described by ref. 45. Subsequent purification of the ELC from the RLC was accomplished by denaturation/reduction of the light chains followed by a DE-52 column where buffer A is 25 mM NaCl, 5 mM NaPi (pH 6.5), 3 mM NaN3, 0.1 mM DTT, 0.5 μg/mL leupeptin, 0.7 μg/mL pepstatin-A, and buffer B is the same except for 0.6 M NaCl. The remaining RLC impurity was removed by a SepPak C-18 column run. ELC collected at 60% CH3CN/0.1% TFA was checked for purity using urea gels, then dried by speed vacuum and stored at -20 °C.
The HCF was obtained from Placopecten striated-muscle myosin resuspended in 70 mM NaCl, 10 mM NaPi (pH 7), 3 mM NaN3, 2 mM MgCl2, 0.5 mM CaCl2, and digested at 20 °C for 100 min with affinity-purified papain at a wt/wt ratio of between 1∶500 and 1∶600 papain∶myosin. Digestion was stopped with 50 μg/mL leupeptin. All subsequent buffers contained at least 0.5 μg/mL leupeptin. The HCF portion of the papain LCD was purified away from the light chains as described in ref. 46. HCF was stored in 40% CH3CN/0.1% TFA at -20 °C until use.
The three separate components were reconstituted into an LCD as previously described (44) with the following change: All buffers contained 0.5 μg/mL leupeptin and 0.7 μg/mL pepstatin-A. The reconstituted material was then purified from free light chains with a Fast-Q column run where buffer A is 25 mM NaCl, 5 mM Hepes (pH 7), 3 mM MgCl2, 0.2 mM CaCl2, 0.1 mM EGTA, 0.5 mM DTT, 0.5 μg/mL leupeptin, 0.7 μg/mL pepstatin-A, and buffer B is the same except for 1 M NaCl. Peaks were checked on 4–20% Tris•HCl gradient gels (run in SDS) as well as 12.5% urea gels. The purified LCD (fractions with equimolar proportions of all three components) was concentrated using Amicon Ultra-4 spin units, exchanging buffer during concentration, and clarified prior to crystallization.
The final sample buffer contains 25 mM NaCl, 5 mM Hepes (pH 7), 3 mM MgCl2, 0.2 mM CaCl2, 0.1 mM EGTA, 0.5 mM DTT, 0.5 μg/mL leupeptin, and 0.7 μg/mL pepstatin-A.
Crystallization, Data Collection, and Structure Determination.
Placo SmoA LCD, in the unphosphorylated state, was crystallized in the presence of CaCl2 (0.5–1 mM) by vapor diffusion in hanging or sitting drops at 4 °C. Drops initially contained 1.1–1.6 mg/mL protein in 5 mM Hepes buffer (pH 7.0), 20 mM NaCl, 1.5 mM MgCl2, 0.5–1 mM CaCl2, 0.1 mM EGTA, 0.5 mM DTT, 2 mM NaN3, 0.5 μg/mL leupeptin, 0.7 μg/mL pepstatin-A, and 7.5% (wt/vol) of PEG 3350 or polyethylene glycol monomethyl ether 2000 (MME PEG 2 K). The reservoir solution consisted of 1 mL of 10 mM Hepes (pH 7.0), 40 mM NaCl, 3 mM MgCl2, 2 mM NaN3, and 15% PEG 3350 or MME PEG 2 K. Crystals appeared in 1–3 d and grew to sizes up to 0.1 × 0.1 × 0.2–0.3 mm. The crystals were gradually equilibrated with a cryoprotectant solution of 7.5 mM Hepes (pH 7), 30 mM NaCl, 2 mM MgCl2, 1 mM DTT, 0.75–1.5 mM CaCl2, and containing 20–25% PEG (3350 or MME 2 K) and 20% PEG 400, and then flash frozen in liquid nitrogen.
A 2.25-Å resolution dataset from a single crystal was collected at 100 K with synchrotron radiation at beamline x29 at Brookhaven National Laboratory. Diffraction data were processed using HKL2000 (47). Initial phase information was obtained by molecular replacement with the program PHASER (48) using the coordinates of Argopecten striated-muscle regulatory domain (Protein Data Bank ID code 1WDC). Iterative rounds of simulated annealing refinement with CNS (49) and model adjustment with COOT (50) were carried out to build the model. Final rounds of positional, ADP, and total least-squares refinement were carried out using the PHENIX package (51). See Table 1 for data processing and refinement statistics.
Table 1.
Crystallographic statistics
| Data collection | |
| Space group | P1 |
| a, b, c, Å | 50.7, 68.9, 79.4 |
| α, β, γ, ° | 77.3, 85.9, 73.6 |
| Resolution, Å | 50–2.25 (2.33–2.25) |
| Rmerge, redundancy | 0.074 (0.59), 3.6 (2.5) |
| I/σI, completeness, % | 16.9 (1.3), 96.2 (82.0) |
| Refinement | |
| Resolution, Å | 45.3–2.25 |
| No. unique reflections | 42,919 |
| Rwork, Rfree | 19.2, 24.1 |
| No. non-H protein atoms | 5,914 |
| No. water, metal atoms | 490; 4 |
| Average protein B factor | 46.3 |
| rmsd bond lengths , Å | 0.008 |
| rmsd angles, ° | 1.172 |
Values in parentheses are for highest-resolution shell.
Structural figures were prepared using the University of California, San Francisco Chimera package (52).
Acknowledgments.
We are grateful to Cynthia Perreault-Micale; without her excellent sequencing work, along with expression, purification, and long-term storage of large quantities of this most interesting RLC, this project would not have been possible. We also thank the staff of the Brookhaven National Laboratory for assistance with data collection, and Hugh Huxley, Kathleen Trybus, and Daniel Himmel for a critical reading of the manuscript. This work has been supported by National Institutes of Health Grant AR017346 (to C.C.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank, www.pdb.org (PDB ID code 3PN7).
See Commentary on page 5.
References
- 1.Houdusse A, Kalabokis VN, Himmel D, Szent-Györgyi AG, Cohen C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: A novel conformation of the myosin head. Cell. 1999;97:459–470. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
- 2.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 3.Kwon H, et al. Isolation of the regulatory domain of scallop myosin: Role of the essential light chain in calcium binding. Proc Natl Acad Sci USA. 1990;87:4771–4775. doi: 10.1073/pnas.87.12.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sellers JR. Regulation of cytoplasmic and smooth muscle myosin. Curr Opin Cell Biol. 1991;3:98–104. doi: 10.1016/0955-0674(91)90171-t. [DOI] [PubMed] [Google Scholar]
- 5.Trybus KM. Role of myosin light chains. J Muscle Res Cell Motil. 1994;15:587–594. doi: 10.1007/BF00121066. [DOI] [PubMed] [Google Scholar]
- 6.Trybus KM, Freyzon Y, Faust LZ, Sweeney HL. Spare the rod, spoil the regulation: Necessity for a myosin rod. Proc Natl Acad Sci USA. 1997;94:48–52. doi: 10.1073/pnas.94.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki H, Stafford WF, 3rd, Slayter HS, Seidel JC. A conformational transition in gizzard heavy meromyosin involving the head-tail junction, resulting in changes in sedimentation coefficient, ATPase activity, and orientation of heads. J Biol Chem. 1985;260:14810–14817. [PubMed] [Google Scholar]
- 8.Stafford WF, et al. Calcium-dependent structural changes in scallop heavy meromyosin. J Mol Biol. 2001;307:137–147. doi: 10.1006/jmbi.2000.4490. [DOI] [PubMed] [Google Scholar]
- 9.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci USA. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung HS, et al. Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc Natl Acad Sci USA. 2008;105:6022–6026. doi: 10.1073/pnas.0707846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houdusse A, Szent-Györgyi AG, Cohen C. Three Conformational States of Scallop S1. Proc Natl Acad Sci USA. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uyeda TQ, Abramson PD, Spudich JA. The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc Natl Acad Sci USA. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Himmel DM, et al. Crystallographic findings on the internally-uncoupled and near-rigor states of myosin: Further insights into the mechanics of the motor. Proc Natl Acad Sci USA. 2002;99:12645–12650. doi: 10.1073/pnas.202476799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huxley HE. The mechanism of muscular contraction. Science. 1969;164:1356–1365. doi: 10.1126/science.164.3886.1356. [DOI] [PubMed] [Google Scholar]
- 15.Huxley AF, Simmons RM. Proposed mechanism of force generation in striated muscle. Nature. 1971;233:533–538. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- 16.Holmes KC. The swinging lever-arm hypothesis of muscle contraction. Curr Biol. 1997;7:R112–R118. doi: 10.1016/s0960-9822(06)00051-0. [DOI] [PubMed] [Google Scholar]
- 17.Oke OA, et al. Influence of lever structure on myosin 5a walking. Proc Natl Acad Sci USA. 2010;107:2509–2514. doi: 10.1073/pnas.0906907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gourinath S, et al. Crystal structure of scallop myosin S1 in the pre-power stroke state to 2.6 angstrom resolution: Flexibility and function in the head. Structure. 2003;11:1621–1627. doi: 10.1016/j.str.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y, et al. Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor. Structure. 2007;15:553–564. doi: 10.1016/j.str.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Thomas DD, Kast D, Korman VL. Site-directed spectroscopic probes of actomyosin structural dynamics. Annu Rev Biophys. 2009;38:347–369. doi: 10.1146/annurev.biophys.35.040405.102118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perreault-Micale CL, Jancso A, Szent-Györgyi AG. Essential and regulatory light chains of Placopecten striated and catch muscle myosins. J Muscle Res Cell Motil. 1996;17:533–542. doi: 10.1007/BF00124353. [DOI] [PubMed] [Google Scholar]
- 22.Debreczeni JE, et al. Structural evidence for non-canonical binding of Ca2+ to a canonical EF-hand of a conventional myosin. J Biol Chem. 2005;280:41458–41464. doi: 10.1074/jbc.M506315200. [DOI] [PubMed] [Google Scholar]
- 23.Risal D, Gourinath S, Himmel DM, Szent-Györgyi AG, Cohen C. Myosin S1 structures reveal a novel nucleotide conformation and a complex salt bridge that helps couple nucleotide and actin binding. Proc Natl Acad Sci USA. 2004;101:8930–8935. doi: 10.1073/pnas.0403002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Houdusse A, Cohen C. Structure of the regulatory domain of scallop myosin at 2 Å resolution: Implications for regulation. Structure. 1996;4:21–32. doi: 10.1016/s0969-2126(96)00006-8. [DOI] [PubMed] [Google Scholar]
- 25.Himmel DM, Mui S, O’Neall-Hennessey E, Szent-Györgyi AG, Cohen C. The on-off switch in regulated myosins: Different triggers but related mechanisms. J Mol Biol. 2009;394:496–505. doi: 10.1016/j.jmb.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowey S, Trybus KM. Common structural motifs for the regulation of divergent class II myosins. J Biol Chem. 2010;285:16403–16407. doi: 10.1074/jbc.R109.025551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ballesteros JA, Deupi X, Olivella M, Haaksma EE, Pardo L. Serine and threonine residues bend alpha-helices in the chi(1) = g(-) conformation. Biophys J. 2000;79:2754–2760. doi: 10.1016/S0006-3495(00)76514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deville J, Rey J, Chabbert M. Comprehensive analysis of the helix-X-helix motif in soluble proteins. Proteins. 2008;72:115–135. doi: 10.1002/prot.21879. [DOI] [PubMed] [Google Scholar]
- 30.Huxley AF. Muscle structure and theories of contraction. Prog Biophys Biophys Chem. 1957;7:255–318. [PubMed] [Google Scholar]
- 31.Huxley HE, Stewart A, Sosa H, Irving T. X-ray diffraction measurements of the extensibility of actin and myosin filaments in contracting muscle. Biophys J. 1994;67:2411–2421. doi: 10.1016/S0006-3495(94)80728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lombardi V, et al. X-ray diffraction studies of the contractile mechanism in single muscle fibres. Philos Trans R Soc Lond B Biol Sci. 2004;359:1883–1893. doi: 10.1098/rstb.2004.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakabayashi K, et al. X-ray diffraction evidence for the extensibility of actin and myosin filaments during muscle contraction. Biophys J. 1994;67:2422–2435. doi: 10.1016/S0006-3495(94)80729-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seebohm B, et al. Cardiomyopathy mutations reveal variable region of myosin converter as major element of cross-bridge compliance. Biophys J. 2009;97:806–824. doi: 10.1016/j.bpj.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knowles AC, et al. Orientation of the essential light chain region of myosin in relaxed, active, and rigor muscle. Biophys J. 2008;95:3882–3891. doi: 10.1529/biophysj.108.131508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Littlefield KP, et al. Similarities and differences between frozen-hydrated, rigor acto-S1 complexes of insect flight and chicken skeletal muscles. J Mol Biol. 2008;381:519–528. doi: 10.1016/j.jmb.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howard J, Spudich JA. Is the lever arm of myosin a molecular elastic element? Proc Natl Acad Sci USA. 1996;93:4462–4464. [PubMed] [Google Scholar]
- 38.Dobbie I, et al. Elastic bending and active tilting of myosin heads during muscle contraction. Nature. 1998;396:383–387. doi: 10.1038/24647. [DOI] [PubMed] [Google Scholar]
- 39.Sherwood JJ, Waller GS, Warshaw DM, Lowey S. A point mutation in the regulatory light chain reduces the step size of skeletal muscle myosin. Proc Natl Acad Sci USA. 2004;101:10973–10978. doi: 10.1073/pnas.0401699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowey S, Waller GS, Trybus KM. Skeletal muscle myosin light chains are essential for physiological speeds of shortening. Nature. 1993;365:454–456. doi: 10.1038/365454a0. [DOI] [PubMed] [Google Scholar]
- 41.Guilford WH, et al. Smooth muscle and skeletal muscle myosins produce similar unitary forces and displacements in the laser trap. Biophys J. 1997;72:1006–1021. doi: 10.1016/S0006-3495(97)78753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sellers JR, Eisenberg E, Adelstein RS. The binding of smooth muscle heavy meromyosin to actin in the presence of ATP. Effect of phosphorylation. J Biol Chem. 1982;257:13880–13883. [PubMed] [Google Scholar]
- 43.Perreault-Micale CL, Kalabokis VN, Nyitray L, Szent-Györgyi AG. Sequence variations in the surface loop near the nucleotide binding site modulate the ATP turnover rates of molluscan myosins. J Muscle Res Cell Motil. 1996;17:543–553. doi: 10.1007/BF00124354. [DOI] [PubMed] [Google Scholar]
- 44.Stafford WF, 3rd, Szentkiralyi EM, Szent-Györgyi AG. Regulatory properties of single-headed fragments of scallop myosin. Biochemistry. 1979;18:5273–5280. doi: 10.1021/bi00591a002. [DOI] [PubMed] [Google Scholar]
- 45.Kendrick-Jones J, Szentkiralyi EM, Szent-Györgyi AG. Regulatory light chains in myosins. J Mol Biol. 1976;104:747–775. doi: 10.1016/0022-2836(76)90180-7. [DOI] [PubMed] [Google Scholar]
- 46.Kalabokis VN, O’Neall-Hennessey E, Szent-Györgyi AG. Regulatory domains of myosins: Influence of heavy chain on Ca2+-binding. J Muscle Res Cell Motil. 1994;15:547–553. doi: 10.1007/BF00121160. [DOI] [PubMed] [Google Scholar]
- 47.Otwinowski Z, Minor W. Methods Enzymol. Vol. 276. New York: Academic; 1997. Processing of X-ray Diffraction Data Collected in Oscillation Mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 48.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brunger AT. Version 1.2 of the crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 52.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]