Abstract
Cirrhosis is the end result of chronic liver disease. Hepatic stellate cells (HSC) are believed to be the major source of collagen-producing myofibroblasts in cirrhotic livers. Portal fibroblasts, bone marrow-derived cells, and epithelial to mesenchymal transition (EMT) might also contribute to the myofibroblast population in damaged livers. Fibroblast-specific protein 1 (FSP1, also called S100A4) is considered a marker of fibroblasts in different organs undergoing tissue remodeling and is used to identify fibroblasts derived from EMT in several organs including the liver. The aim of this study was to characterize FSP1-positive cells in human and experimental liver disease. FSP1-positive cells were increased in human and mouse experimental liver injury including liver cancer. However, FSP1 was not expressed by HSC or type I collagen-producing fibroblasts. Likewise, FSP1-positive cells did not express classical myofibroblast markers, including αSMA and desmin, and were not myofibroblast precursors in injured livers as evaluated by genetic lineage tracing experiments. Surprisingly, FSP1-positive cells expressed F4/80 and other markers of the myeloid-monocytic lineage as evaluated by double immunofluorescence staining, cell fate tracking, flow cytometry, and transcriptional profiling. Similar results were obtained for bone marrow-derived and peritoneal macrophages. FSP1-positive cells were characterized by increased expression of COX2, osteopontin, inflammatory cytokines, and chemokines but reduced expression of MMP3 and TIMP3 compared with Kupffer cells/macrophages. These findings suggest that FSP1 is a marker of a specific subset of inflammatory macrophages in liver injury, fibrosis, and cancer.
Keywords: tumor microenvironment
Cirrhosis is the end result of chronic liver injury and is characterized by the presence of fibrosis and nodular regeneration throughout the liver. The cellular source of collagen in fibrotic liver has long been a matter of debate. Indeed, hepatocytes and sinusoidal endothelial cells were initially suggested to synthesize collagen (1, 2). The successful establishment of isolation protocols for a pure hepatic stellate cell (HSC) population has been an important technical breakthrough to study these cells in vitro. It was shown that activated HSCs synthesize and secrete different types of collagen, and it was therefore proposed that they represent the principal source of collagen in fibrotic liver (3).
However, the paradigm that HSCs are the sole source of all collagen in cirrhotic liver has been challenged in recent years. It has become increasingly apparent that myofibroblasts may also be derived from other cells in addition to resident HSCs. Portal fibroblasts, for example, may be of particular importance in cholestatic liver disease and are morphologically and functionally distinct from HSCs (4). A significant proportion of cells expressing myofibroblast markers are derived from bone marrow in human liver disease (5). The functional significance of these cells in the pathogenesis of liver fibrosis remains unclear (6–8). It was recently proposed that myofibroblasts may originate from epithelial cells via epithelial to mesenchymal transition (EMT) during liver fibrogenesis. Hepatocytes were proposed as a source of myofibroblasts, because treatment of cultured hepatocytes with TGFβ1 induces phenotypic and functional changes indicative of EMT (9–11). Cholangiocytes might also be a source of myofibroblasts via EMT to contribute to portal tract fibrosis in human and experimental cholestatic liver injury (12–15).
Fibroblast-specific protein 1 (FSP1) was identified by subtractive and differential mRNA hybridization as a gene expressed in fibroblasts but not in epithelial cells (16). FSP1 is also known as S100A4 and belongs to the S100 superfamily of cytoplasmic calcium-binding proteins. S100 proteins form homo- or heterodimers do not possess enzymatic activity but can regulate the function of other proteins by binding to them. FSP1/S100A4 is also secreted and has diverse functions on various cell types through binding to an unknown receptor (17). FSP1 is expressed in fibroblasts in different organs that undergo tissue remodeling including kidney, lung, and heart (16, 18, 19). In addition, FSP1 is commonly used as a marker to identify epithelial cells undergoing EMT during tissue fibrogenesis (20) and was used as proof of EMT by hepatocytes and cholangiocytes (10, 13–15). However, it remains elusive whether FSP1-positive cells contribute to extracellular matrix (ECM) deposition in liver injury.
In this study, we demonstrate that FSP1-positive cells neither synthesize type I collagen nor express other mesenchymal cell markers. Using a panel of different methods, we provide evidence that FSP1 is expressed by a subset of inflammatory macrophages in injured livers.
Results
FSP1-Positive Cells Are Increased in Human and Experimental Liver Disease.
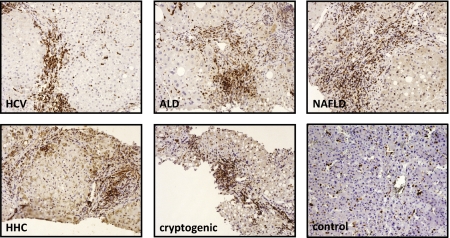
In normal human liver, a few FSP1-positive cells are found scattered throughout the parenchyma. The number of FSP1-positive cells is increased in all forms of chronic liver disease, including chronic hepatitis C virus infection, alcoholic liver disease, nonfatty liver disease, hereditary hemochromatosis, and cryptogenic cirrhosis (Fig. 1). FSP1-positive cells were small cells with scant cytoplasm typically located along fibrotic septa. To determine whether FSP1 is increased in experimental liver disease, C57Bl6 mice underwent two complementary models of liver fibrosis: bile duct ligation (BDL) or CCl4 treatment. The number of FSP1-positive cells increased over time in both liver injury models as evaluated by immunohistochemistry. Significant increases in FSP1 mRNA and protein levels paralleled increased cell number (Fig. S1). The specificity of the FSP1 antibody was validated by performing immunofluorescence staining for FSP1 using sections from FSP1-deficient mice receiving a single injection of CCl4 (Fig. S2).
Fig. 1.
FSP1-positive cells are increased in human liver disease. Paraffin-embedded liver sections from patients with chronic hepatitis C virus infection (HCV), alcoholic liver disease (ALD), nonalcoholic liver disease (NAFLD), hereditary hemochromatosis (HHC), cryptogenic liver disease, and controls without liver disease were evaluated for FSP1 expression by immunohistochemistry.
FSP1-Positive Cells Are Increased in Human and Experimental Liver Cancer.
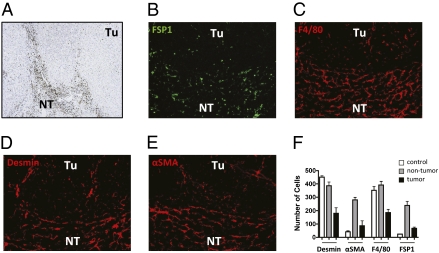
Liver cancer typically develops in the presence of cirrhosis, suggesting that the microenvironment of chronic liver disease favors initiation and/or progression of hepatocellular carcinoma (HCC). FSP1 was suggested to be a marker for tumor-associated fibroblasts (21). The number of FSP1-positive cells was increased in the cancer-free part of livers of patients with HCC, and individual FSP1-positive cells were scattered within tumors (Fig. 2A). To analyze whether FSP1-positive cells are also increased in the tumor microenvironment of a murine model of HCC, sections from mice challenged with diethylnitrosamine (DEN) were stained for FSP1. Parallel sections were also stained for desmin, αSMA, and F4/80 to identify quiescent HSCs, myofibroblasts, and macrophages, respectively (Fig. 2 B–E). The relative amount of each cell type in the tumor stroma of DEN-induced liver cancer was quantified. Mice without DEN treatment served as a control (staining not shown). F4/80- and desmin-positive cells were already abundant in livers of control mice, whereas only rare FSP1- and αSMA-positive cells were observed. In contrast, FSP1- and αSMA-positive cells were increased in the cancer-free part of mice with DEN-induced liver cancer (Fig. 2F).
Fig. 2.
FSP1-positive cells are increased in human and experimental liver cancer. Liver sections from patients with HCC were evaluated for FSP1 expression by immunohistochemistry (A). FSP1-positive cells were abundant in the nontumor part (NT) of the liver, and few FSP1-positive cells were scattered within the tumors (Tu). Liver sections prepared from DEN-induced HCC-bearing mice were also stained for FSP1 (B), F4/80 (C), desmin (D), and αSMA (E). The number of cells expressing each marker was quantified in untreated control mice (staining not shown, white bars) and in the tumor (black bars) and nontumor parts (gray bars) of livers of mice with DEN-induced cancer (F).
Hepatic Fibroblasts Do Not Express FSP1.
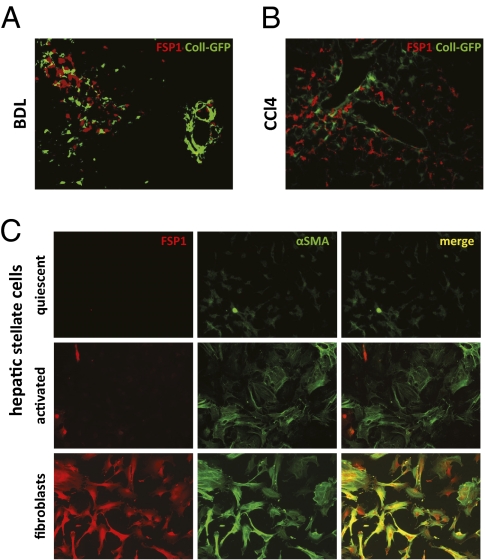
FSP1 is expressed by fibroblasts in different organs undergoing tissue remodeling (16, 18, 19). However, it remains unknown whether FSP1-positive cells are a source of extracellular matrix (ECM) in liver disease. To test whether FSP1-positive cells contribute to the population of collagen-producing cells, Coll-GFP reporter mice, in which the collagen α1(I) promoter/enhancer drives expression of GFP, were subjected to BDL or CCl4 treatment. A total of 6,185 GFP-positive cells were analyzed, but no colocalization of GFP and FSP1 was observed (Fig. 3 A and B).
Fig. 3.
FSP1 is not a marker of hepatic fibroblasts or hepatic stellate cells. Coll-GFP reporter mice were subjected to BDL (A) or CCl4 treatment (B). Sections were stained for FSP1. HSCs were isolated from mice and activated by adhesion to plastic. Cells were analyzed 1 d after plating (quiescent) and after 5 d (activated) in culture. Activated HSCs expressed αSMA but not FSP1. In contrast, dermal fibroblast isolated from mice expressed both markers (C).
HSCs are a major source of collagen-producing fibroblasts in liver disease. To evaluate whether HSCs express FSP1, HSCs were isolated from mice and activated by adhesion to plastic. After 1 d in culture, HSCs were small and round and did not express αSMA, indicating that they were still in a quiescent state. After culture on plastic for 5 d, HSCs acquired an activated phenotype indicative of myofibroblastic differentiation: increased cell size, a spread out stellate shape with numerous protrusions, and αSMA expression. However, activated HSCs still lacked FSP1 expression. In contrast, mouse skin fibroblasts expressed both FSP1 and αSMA (Fig. 3C).
FSP1 Is Not a Marker of Myofibroblasts.
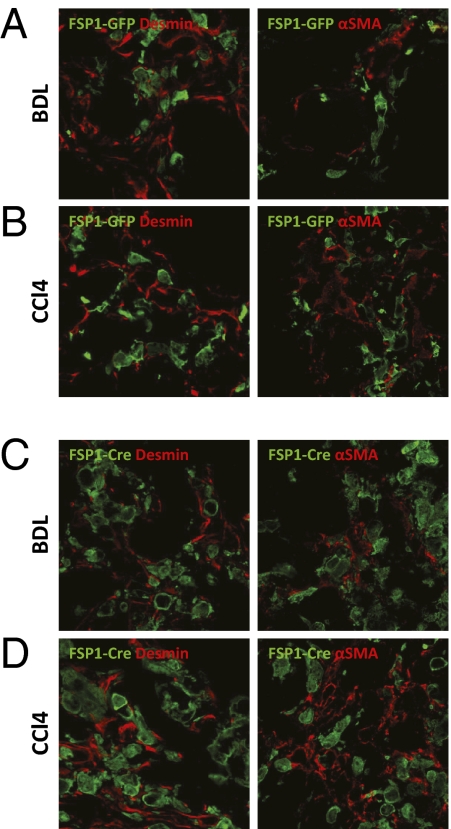
Recently, FSP1 was used as a marker to demonstrate EMT of hepatocytes and cholangiocytes (10, 13–15). Therefore, we tested whether FSP1 is a marker of myofibroblasts characterized by expression of mesenchymal markers including desmin and αSMA. For this purpose, FSP1-GFP reporter mice in which the FSP1 promoter drives GFP expression were subjected to BDL or CCl4 treatment. A total number of 3,148 GFP-positive cells were analyzed, but no colocalization of GFP and αSMA or desmin was observed (Fig. 4 A and B). As a control, sections from FSP1-GFP mice subjected to BDL or CCl4 treatment were also evaluated for FSP1 expression. Colocalization of FSP1 and GFP was observed, indicating that GFP expression parallels FSP1 expression in these mice (Fig. S3).
Fig. 4.
FSP1 is not a marker for myofibroblasts or their precursors. FSP-GFP reporter mice in which the FSP1 promoter drives expression of GFP were subjected to BDL or CCl4 treatment. Liver sections were stained for mesenchymal cell markers desmin and αSMA. No colocalization of GFP with desmin or αSMA was observed (A and B). Alternatively, FSP1-Cre mice were crossed to ROSA26-lox P-STOP-lox P-YFP mice. These mice were subjected to BDL or CCl4 treatment. No colocalization of GFP with desmin or αSMA was observed in liver sections (C and D).
Using two different kinds of reporter mice, these results provide evidence that FSP1-positive cells in liver do not synthesize collagen or express markers of myofibroblasts. To corroborate these findings, we performed double immunofluorescence staining for FSP1 and αSMA by using frozen liver sections from mice subjected to BDL or CCl4 treatment, but did not observe any colocalization (Fig. S4).
FSP1 Is Not a Marker for Precursors of Myofibroblasts.
Because FSP1-positive cells are present early in the course of experimental liver injury before the appearance of αSMA-positive myofibroblasts and deposition of collagen (Fig. S1), FSP1 might be a marker of uncommitted precursor cells that differentiate into myofibroblasts over the course of liver injury. To address whether FSP1-positive cells might lose expression of FSP1 and acquire a myofibroblast phenotype, we used genetic cell fate tracking. For this purpose, we crossed FSP1-Cre mice in which the FSP1 promoter drives expression of Cre recombinase to different reporter mice in which the ROSA26 promoter drives either expression of YFP or membrane-targeted GFP after Cre-mediated removal of a loxP-flanked STOP cassette (22, 23). This approach allows identification of cells that have expressed FSP1 at some point during differentiation although they may lack expression of FSP1 at the time of analysis. These mice were subjected to BDL or CCl4 treatment and expression of desmin and αSMA was evaluated by confocal microscopy. A total of 1,563 GFP-positive cells indicative of prior expression of Cre recombinase under the control of the FSP1 promoter were analyzed, but no colocalization with desmin or αSMA was observed (Fig. 4 C and D). We also isolated HSCs from these mice and evaluated expression of GFP over the course of culture on plastic. After 5 d, HSCs acquired an activated phenotype indicative of myofibroblast differentiation but did not express GFP, demonstrating that the FSP1 promoter is not functional in activated HSCs (Fig. S5).
FSP1 Is a Marker of Macrophages.
To identify the cellular lineage to which FSP1-positive cells in liver injury belong, we performed gene expression profiling on FSP1-positive cells isolated from CCl4-treated FSP1-GFP mice. These data were compared with publicly available datasets by using compatible platforms, and a cluster analysis was performed by using a “distance” function. FSP1-positive cells clustered with bone marrow-derived dendritic cells and peritoneal macrophages (Fig. S6A) and recalculation using datasets from only these three different cell types indicated that the gene expression profile of FSP1-positive cells is closest to that of peritoneal macrophages stimulated with zymosan (Fig. S6B). A heat map depicting the 44 genes that are most differentiating among these three cell types is presented (Fig. S6C).
FSP1-positive cells expressed markers and genes typical for macrophages and/or dendritic cells including CD68, Nramp1, Soat1, CD63, CD83, CD93, Clec4d, Clec4b1, Clec4n, Clec7a, and p22(phox). FSP1-positive cells also expressed several genes that play a pivotal role in innate immunity including the LPS receptors CD14 and Toll-like receptor 4 (TLR4) as well as TLR2, TLR7, and TLR8. Furthermore, FSP1-positive cells also expressed several cytokines and chemokines and genes involved in phagocytosis.
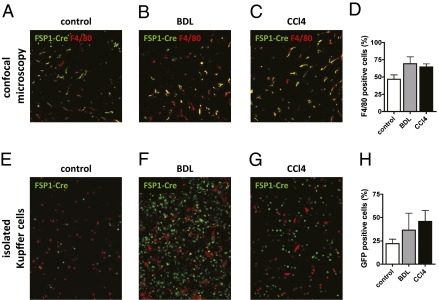
Gene expression profiling suggested that FSP1-positive cells in liver injury belong to myeloid-monocytic lineage. To corroborate this result, we isolated hepatocytes, Kupffer cells, endothelial cells, and HSCs from mice and analyzed FSP1 mRNA expression by quantitative PCR (qPCR). FSP1 mRNA was primarily detected in the Kupffer cell fraction. Similar results were obtained when cell fractions from mice undergoing BDL and CCl4 treatment were analyzed (Fig. S7). These observations also suggested that FSP1 might be a marker of macrophages or some other myeloid cell type. To test this hypothesis, we performed immunofluorescence staining with the macrophage marker F4/80 by using sections from mice genetically tagged for FSP1. Confocal microscopy revealed that in untreated mice, 46.7 ± 6.3% of cells which expressed FSP1 at some point during development also stain for F4/80. After BDL or CCl4 treatment, the percentage of FSP1-Cre–positive cells expressing F4/80 increased to 69.18 ± 9.99 and 64.45 ± 4.55, respectively (Fig. 5 A–D). We also isolated Kupffer cells/macrophages from FSP1-Cre mice crossed to ROSA26 reporter mice and quantified the percentage of GFP-positive cells. Consistent with results from confocal microscopy, the percentage of GFP-positive cells among Kupffer cells/macrophages increased following liver injury (Fig. 5 E–H).
Fig. 5.
FSP1 is expressed in macrophage/Kupffer cells in liver injury. FSP1-Cre mice were crossed to ROSA26-lox P-STOP-lox P-mGFP mice. These mice were subjected to BDL or CCl4 treatment, and livers sections were stained for macrophage marker F4/80 (A–C). The percentage of FSP1–Cre-positive cells (green) staining positive for F4/80 (red) was quantified (D). Alternatively, liver macrophages/Kupffer cells were isolated from these mice and evaluated for green fluorescence indicative of successful Cre-mediated recombination (E–H).
To confirm results obtained by confocal microscopy and further characterize FSP1-positive cells in liver injury, we performed flow cytometry. For this purpose, we isolated nonparenchymal cells from FSP1-GFP mice receiving two CCl4 injections and stained for cell surface markers characteristic of the monocytic-myeloid lineage. The majority of GFP-positive cells expressed CD45 (91.35 ± 3.08%), indicating that FSP1-positive cells are bone marrow derived (Fig. S8A). Furthermore, FSP1-positive cells also expressed CD11b (86.5 ± 4.22%), CD11c (80.94 ± 3.50%), and F4/80 (72.14 ± 1.45%), but lacked expression of the granulocyte maturation marker Gr1high (4.32 ± 1.82%) (Fig. S8 B–D, F, and G). A significant proportion of FSP1-positive cells also expressed CD103 (63.11 ± 0.66%), a marker for resident dendritic cells of the intestine and the skin (Fig. S8E).
To confirm that cells of the myeloid-monocytic lineage express FSP1, we generated bone marrow-derived macrophages from FSP1-Cre mice bred to ROSA26 reporter mice. As a control, we used LysM-Cre mice and compared the efficiency of deletion between these two different Cre mice. After 7 d in culture, 99% of macrophages generated from FSP1-reporter mice were green, indicative of successful removal of a loxP-flanked red fluorescent protein (Fig. S9 A, E, and F). This efficiency of deletion exceeded the percentage of recombination in cells cultured from LysM-reporter mice (≈50%; Fig. S9 A, D, and F). Cultured cells expressed CD11b and F4/80, confirming successful differentiation of macrophages (Fig. S9 B and C). We also analyzed peritoneal macrophages isolated from FSP1-GFP mice as well as FSP1- and LysM-reporter mice. After i.p. injection with thioglycollate, FSP1-positve cells expressed CD11b (≈99%) and F4/80 (≈95%) (Fig. S10 B and C). Furthermore, >95% of macrophages isolated from FSP1-reporter mice were green, indicative of successful removal of a loxP-flanked red fluorescent protein (Fig. S10 E and F). Similar to bone marrow-derived macrophages, the efficiency of deletion observed in peritoneal macrophages isolated from FSP1-Cre mice exceeded that of LysM-Cre mice. These results demonstrate that FSP1 is also expressed in macrophages generated by standard methods.
FSP1-Positive Cells Are an Inflammatory Subpopulation of Macrophages.
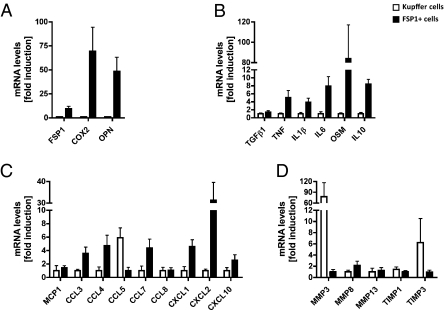
Our data indicates that FSP1 is a marker of a subpopulation of macrophage/Kupffer cells in livers undergoing tissue remodeling. To address whether FSP1 marks a functionally distinct subpopulation of macrophages, we performed qPCR for various genes including cytokines, chemokines, and genes involved in regulation of ECM turnover. FSP1 mRNA levels were ≈10-fold higher in FSP1-sorted cells, indicating successful enrichment of this subpopulation of macrophages. FSP1-positive cells are characterized by increased expression of COX2, ostepontin, cytokines including TNF, IL1β, IL6, IL10, oncostatin M, and chemokines including CCL3, CCL4, CCL7, CXCL1, CXCL2, CXCL10, but reduced expression of MMP3 and TIMP3 compared with macrophages (Fig. 6).
Fig. 6.
FSP1-positive cells are an inflammatory subpopulation of macrophages. mRNA was prepared from FSP1-positive cells (filled bars) and macrophage/Kupffer cells (open bars) isolated from CCl4-treated FSP1-GFP mice. FSP1-positive cells are characterized by increased expression of COX2 and ostepontin (A), cytokines including TNF, IL1β, IL6, IL10, and oncostatin M (B), and chemokines including CCL3, CCL4, CCL7, CXCL1, CXCL2, and CXCL10 (C), but reduced expression of MMP3 and TIMP3 compared with macrophages (D). Data are summarized as mean ± SE mean and are presented as fold induction compared with the cell population with lower expression levels of indicated genes.
Discussion
The liver is a complex organ consisting of parenchymal (hepatocytes) and nonparenchymal cells including biliary epithelial cells, HSCs, liver sinusoidal endothelial cells, resident macrophages (Kupffer cells), and other immune cells like B-, T-, and NK cells. Remarkably, most of these cells directly or indirectly contribute to liver fibrosis. Delineating the cellular and molecular mechanisms underlying liver fibrosis may lead to the development of more effective treatments.
FSP1 was identified as a transcript that is expressed in much higher amounts in fibroblasts than in epithelial cells and is commonly used to identify fibroblasts in organs undergoing tissue remodeling or as a marker of EMT (16, 18–20). Correspondingly, FSP1 was used as a marker for EMT of hepatocytes and cholangiocytes (10, 13–15). The increase of FSP1-positive cells and their evolution from genetically marked hepatocytes or costaining with cholangiocyte-specific markers led to the conclusion that EMT substantially contributes to the pool of ECM-producing cells in liver (10, 13, 24). However, our laboratory has challenged the notion that FSP1-positive cells are derived from hepatocytes or cholangiocytes and that epithelial cells contribute to the pool of collagen-producing cells in vivo (11, 25). Furthermore, because FSP1 has been suggested to be a marker of myofibroblasts, it was assumed—but never proven experimentally—that FSP1-positive cells are collagen-producing cells in liver injury. To test this hypothesis, we used Coll-GFP reporter mice. We analyzed >6,000 GFP-positive cells but failed to observe colocalization of FSP1 with GFP (Fig. 3). Similar results were reported in mice subjected to unilateral ureteral obstruction (26). Furthermore, microarray profiling failed to detect significant enrichment of any type of collagen in FSP1-positive cells of the livers. These data challenge the current assumption that FSP1—despite its auspicious name—is a marker of fibroblasts in experimental liver injury. Accordingly, HSCs, the primary cellular source of collagen in liver disease, did also not express FSP1 at any point of activation (Fig. 3 and Fig. S5).
We have demonstrated plasticity of myofibroblasts, which might lose or acquire expression of markers (27). We therefore evaluated coexpression of FSP1 with classical mesenchymal cell markers αSMA and desmin. Because FSP1 can also be secreted, immunofluorescence staining carries the risk of detecting secreted, extracellular FSP1 attached to some other cell type. Accordingly, immunofluorescence staining for FSP1 did not completely overlap with GFP fluorescence in FSP1-GFP reporter mice and mice with a GFP knockin at the endogenous FSP1 gene (Fig. S2 and Fig. S3). Careful analysis and interpretation of studies relying entirely on FSP1 staining is therefore warranted. To specifically identify FSP1-positive cells, we used FSP1-GFP reporter mice but failed to observe coexpression of GFP with mesenchymal markers desmin or αSMA (Fig. 4 A and B). In addition, similar results were obtained by double immunofluorescence staining for FSP1 and αSMA (Fig. S4). Using genetic lineage tracing experiments, we also failed to observe colocalization with αSMA or desmin, excluding the possibility that FSP1 might be a marker of uncommitted myofibroblasts (Fig. 4 C and D). Furthermore, gene expression profiling also failed to detect significant enrichment of mRNAs for αSMA or desmin in FSP1-positive liver cells. Conversely, we also isolated HSCs, liver sinusoidal endothelial cells, and hepatocytes from these mice, and FSP1 was not expressed at any time point by any of these cell types (Fig. 3 and Fig. S5). In summary, our current and previous studies (11, 25) indicate that FSP1 is not a marker of myofibroblastic cell populations in the liver and refute the utility of FSP1 as a marker for myofibroblasts in experimental liver fibrosis. Studies from other laboratories (12–15) have reported that cholangiocytes express FSP1 in human cholestatic fibrotic liver diseases, but our short-term experimental models do not address this issue.
Alternatively, five different approaches support the concept that macrophage/Kupffer cells express FSP1 in mouse liver undergoing tissue remodeling. First, the pattern of genes expressed by FSP1-positive cells in liver injury is characteristic for cells of the myeloid-monocytic lineage (Fig. S6). Second, Kupffer cells are the main source of FSP1 in the normal and injured liver (Fig. S7). Third, the majority of cells that expressed FSP1 at some point during differentiation are also stained for F4/80 (Fig. 5 A–D). Fourth, a significant percentage of isolated macrophages/Kupffer cells expressed FSP1 at some point during the course of liver injury (Fig. 5 E–H). Fifth, FSP1-positive cells isolated from injured mouse liver are of hematopoietic origin and belong to the myeloid-monocytic lineage. Accordingly, FSP1-positive cells express CD45 and surface markers typical of macrophages including CD11b, CD11c, and F4/80 (Fig. S8).
In accordance with our results, three independent groups reported coexpression of FSP1 with hematopoietic markers in rodent models of kidney injury (26, 28, 29). Flow cytometry showed that FSP1-positive cells express CD45, CD68, macrophage-1 antigen (Mac-1 also known as CD11b), Mac-2 (Lgals3), Mac-3 (LAMP2), and MHC class II (28). Consistent with these results, Lin et al. (26) observed coexpression of CD11b, CD68, and F4/80 with FSP1 in mice undergoing unilateral ureteral obstruction. Similar results were obtained by others who performed flow cytometry for myeloid dendritic cells showing that FSP1 is coexpressed with CD11b, CD11c, CD80, CD86, and MHC class II, but not with the T- and B-cell markers CD4, CD8, and B220 (30). In patients with rheumatoid arthritis and osteoarthritis, FSP1 colocalizes with CD68, providing further evidence that tissue macrophages express FSP1 (31). Similarly, FSP1 also colocalizes with CD68 in hearts of patients suffering from aortic stenosis and ischemic cardiomyopathy (19). In contrast, in primary cell cultures isolated from lung, fibroblasts, but not macrophages or type II alveolar epithelial cells, express FSP1 (18). Similarly, FSP1 is expressed by dermal fibroblasts (Fig. 3C) and stromal fibroblasts in mammary glands (32, 33). These observations suggest that there may exist considerable heterogeneity among cells expressing FSP1 in different organs.
It was recently shown that FSP1 is crucial for proper myosin-IIA assembly and colony-stimulating factor-1 receptor signaling in macrophages and that FSP1 mediates macrophage recruitment and chemotaxis in vivo (34). These data suggest that FSP1 apart from being a marker also has a biological function in macrophages.
In summary, FSP1-positive cells in livers are of hematopoietic origin and are recruited after liver injury to acquire a macrophage-like phenotype. FSP1-positve cells represent a subset of macrophages in livers undergoing tissue remodeling characterized by increased expression of proinflammatory genes. Whereas FSP1 is a marker of dermal fibroblasts and a subset of fibroblasts in some organs, cells of the myeloid-monocytic lineage also express this marker. Thus, its applicability to identify fibroblasts in organs undergoing tissue remodeling is limited and prone to interpretational pitfalls. Likewise, FSP1-Cre mediated gene deletion cannot be assumed to exclusively affect all mesenchymal cells.
Materials and Methods
Human Liver Samples.
Human liver biopsy specimens were obtained during routine diagnostic evaluations at the Department of Internal Medicine, General Hospital Oberndorf. Immunohistochemical staining of liver specimen was approved by the local ethical committee.
Mice.
Coll-GFP, FSP1-GFP, FSP1-Cre, FSP1 KO, and ROSA26 reporter mice were described (20, 22, 23, 27, 35, 36). All other experiments were conducted in C57BL6 mice purchased from the Jackson Laboratory or Taconic.
Mouse Models of Liver Disease.
Liver fibrosis and cancer were induced as described (37, 38).
Cell Isolation.
Cell fractions of mouse livers were isolated as described (39). Bone marrow cells were obtained from mouse tibias and femurs and cultured in DMEM (Invitrogen) with 10% FBS and 20% L929 supernatant containing macrophage-stimulating factor for 6 d. At day 7, media was changed to DMEM containing M-CSF (10 ng/mL), and cells were analyzed 24 h later. Peritoneal macrophages were isolated from untreated mice or mice 3 d after injection of 3% sterile thioglycollate medium by lavage with PBS.
Flow Cytometry and Fluorescent-Activated Cell Sorting.
Nonparenchymal cells were isolated from mouse liver by using standard protocols and analyzed by flow cytometry as described (40).
RNA Isolation and qPCR.
RNA was isolated by using TRIzol reagent (Invitrogen) and the RNAesy Kit (Qiagen) and reverse transcribed with the High Capacity cDNA Reverse Transcription Kit from Applied Biosystems. qPCR was carried out with commercially available primer-probe sets (Applied Biosystems) or using SYBR green (Bio-Rad). Catalog numbers of primer-probe sets and sequences of primers used for qPCR are available upon request.
Immunoblotting.
Protein samples were prepared by using a lysis buffer containing 1% Triton X-100, and protein concentration was determined by using a BCA assay (Pierce). Western blotting was performed by using standard methods.
Immunohistochemistry and Immunofluorescence.
Immunohistochemistry for FSP1 was performed by using standard methods. Immunofluorescence staining for αSMA (1:200; Abcam), desmin (1:500; LabVision), F4/80 (1:200; eBioscience) and FSP1 (1:1,000) was performed by using frozen sections as described (11). YFP expression in ROSA26 reporter mice and GFP expression in FSP1-GFP and FSP1 KO mice was visualized by immunofluorescence staining by using a goat-anti GFP antibody (1:1,000; Abcam). Detailed protocols are available upon request.
Fluorescence-Activated Cell Sorting.
Fluorescent-activated cell sorting was conducted as described (11).
Microarrays.
Total RNA was extracted from sorted cells by using the RNeasy kit (Qiagen). Biotinylated cRNA was prepared by using the Illumina RNA Amplification Kit (Ambion). cRNA was hybridized to MouseWG-6 v2 Expression BeadChip Arrays (Illumina) and scanned on the Illumina BeadArray reader. Data analysis was carried out by using the Illumina BeadStudio software. Raw data from the present experiment were combined with publicly available pro–B-cell expression data (41), bone marrow-derived dendritic cells (42), and peritoneal macrophages (datasets kindly provided by Amy Sullivan and Christopher K. Glass, University of California, San Diego, CA). The combined raw expression data were normalized by using a mloess algorithm (43). Based on the histogram of log2-transformed values, a gene is called “detected” when its expression value exceeds 64 in at least one sample, which results in 15,641 detected genes. For clustering of cell types, we characterized each cell type by the vector of log2-transformed expression values of all detected genes. The “distance” function was calculated as a weighted sum of squared differences between all detected genes, where the weight of each gene is its variance across all cell types being considered. Using this approach more weight is assigned to genes that discriminate between cell types and no gene is excluded from analysis.
Supplementary Material
Acknowledgments
We thank Dr. Eric G. Neilson from Vanderbilt University (Nashville, TN) for providing FSP1-GFP reporter, FSP1 KO, and FSP1-Cre mice as well as the FSP1-antibody. We express our gratitude to Drs. Taura (Kyoto) and Iwaisako (La Jolla, CA) for advice regarding cell isolations and providing mRNA of different liver cells and Amy Sullivan and Christopher K. Glass for sharing unpublished gene expression data. C.H.Ö. was supported by a Fellowship-to-Faculty Transition Award kindly provided by the American Gastroenterology Association and is a recipient of an Austrian Programme for Advanced Research and Technology fellowship from the Austrian Academy of Sciences at the Institute of Pharmacology, Center for Physiology and Pharmacology, Medical University Vienna. G.H. acknowledges support from National Institutes of Health Grants DK063491, CA023100, and DK080506. This work was supported by National Institutes of Health Grants (to M.K. and D.A.B.). Research in the laboratory of M.K. is supported by Superfund Basic Research Program Grant 5 P42 ES010337 and Wellcome Trust Grant 086755/Z/085/Z. M.K. is an American Cancer Society Research Professor.
Footnotes
The authors declare no conflict of interest.
1C.H.Ö. and M.P.-Ö. contributed equally to this work.
Data deposition: The microarray data reported in this paper have been deposited in the European Bioinformatics Institute ArrayExpress database (accession no. E-MTAB-287).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017547108/-/DCSupplemental.
References
- 1.Diegelmann RF, Guzelian PS, Gay R, Gay S. Collagen formation by the hepatocyte in primary monolayer culture and in vivo. Science. 1983;219:1343–1345. doi: 10.1126/science.6828863. [DOI] [PubMed] [Google Scholar]
- 2.Irving MG, Roll FJ, Huang S, Bissell DM. Characterization and culture of sinusoidal endothelium from normal rat liver: Lipoprotein uptake and collagen phenotype. Gastroenterology. 1984;87:1233–1247. [PubMed] [Google Scholar]
- 3.Friedman SL, Roll FJ, Boyles J, Bissell DM. Hepatic lipocytes: The principal collagen-producing cells of normal rat liver. Proc Natl Acad Sci USA. 1985;82:8681–8685. doi: 10.1073/pnas.82.24.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z, et al. Transforming growth factor-beta and substrate stiffness regulate portal fibroblast activation in culture. Hepatology. 2007;46:1246–1256. doi: 10.1002/hep.21792. [DOI] [PubMed] [Google Scholar]
- 5.Forbes SJ, et al. A significant proportion of myofibroblasts are of bone marrow origin in human liver fibrosis. Gastroenterology. 2004;126:955–963. doi: 10.1053/j.gastro.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 6.Kisseleva T, et al. Bone marrow-derived fibrocytes participate in pathogenesis of liver fibrosis. J Hepatol. 2006;45:429–438. doi: 10.1016/j.jhep.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 7.Russo FP, et al. The bone marrow functionally contributes to liver fibrosis. Gastroenterology. 2006;130:1807–1821. doi: 10.1053/j.gastro.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 8.Higashiyama R, et al. Negligible contribution of bone marrow-derived cells to collagen production during hepatic fibrogenesis in mice. Gastroenterology. 2009;137:1459–1466. doi: 10.1053/j.gastro.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Kaimori A, et al. Transforming growth factor-beta1 induces an epithelial-to-mesenchymal transition state in mouse hepatocytes in vitro. J Biol Chem. 2007;282:22089–22101. doi: 10.1074/jbc.M700998200. [DOI] [PubMed] [Google Scholar]
- 10.Zeisberg M, et al. Fibroblasts derive from hepatocytes in liver fibrosis via epithelial to mesenchymal transition. J Biol Chem. 2007;282:23337–23347. doi: 10.1074/jbc.M700194200. [DOI] [PubMed] [Google Scholar]
- 11.Taura K, et al. Hepatocytes do not undergo epithelial-mesenchymal transition in liver fibrosis in mice. Hepatology. 2009;51:1027–1036. doi: 10.1002/hep.23368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson H, Kirby JA, Yip WW, Jones DE, Burt AD. Biliary epithelial-mesenchymal transition in posttransplantation recurrence of primary biliary cirrhosis. Hepatology. 2007;45:977–981. doi: 10.1002/hep.21624. [DOI] [PubMed] [Google Scholar]
- 13.Omenetti A, et al. Hedgehog signaling regulates epithelial-mesenchymal transition during biliary fibrosis in rodents and humans. J Clin Invest. 2008;118:3331–3342. doi: 10.1172/JCI35875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Díaz R, et al. Evidence for the epithelial to mesenchymal transition in biliary atresia fibrosis. Hum Pathol. 2008;39:102–115. doi: 10.1016/j.humpath.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 15.Rygiel KA, et al. Epithelial-mesenchymal transition contributes to portal tract fibrogenesis during human chronic liver disease. Lab Invest. 2008;88:112–123. doi: 10.1038/labinvest.3700704. [DOI] [PubMed] [Google Scholar]
- 16.Strutz F, et al. Identification and characterization of a fibroblast marker: FSP1. J Cell Biol. 1995;130:393–405. doi: 10.1083/jcb.130.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boye K, Maelandsmo GM. S100A4 and metastasis: A small actor playing many roles. Am J Pathol. 2010;176:528–535. doi: 10.2353/ajpath.2010.090526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawson WE, et al. Characterization of fibroblast-specific protein 1 in pulmonary fibrosis. Am J Respir Crit Care Med. 2005;171:899–907. doi: 10.1164/rccm.200311-1535OC. [DOI] [PubMed] [Google Scholar]
- 19.Schneider M, et al. S100A4 is upregulated in injured myocardium and promotes growth and survival of cardiac myocytes. Cardiovasc Res. 2007;75:40–50. doi: 10.1016/j.cardiores.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 20.Iwano M, et al. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest. 2002;110:341–350. doi: 10.1172/JCI15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 22.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 24.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scholten D, et al. Genetic labeling does not detect epithelial-to-mesenchymal transition (EMT) of cholangiocytes in liver fibrosis in mice. Gastroenterology. 2010;139:987–998. doi: 10.1053/j.gastro.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin SL, Kisseleva T, Brenner DA, Duffield JS. Pericytes and perivascular fibroblasts are the primary source of collagen-producing cells in obstructive fibrosis of the kidney. Am J Pathol. 2008;173:1617–1627. doi: 10.2353/ajpath.2008.080433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magness ST, Bataller R, Yang L, Brenner DA. A dual reporter gene transgenic mouse demonstrates heterogeneity in hepatic fibrogenic cell populations. Hepatology. 2004;40:1151–1159. doi: 10.1002/hep.20427. [DOI] [PubMed] [Google Scholar]
- 28.Inoue T, Plieth D, Venkov CD, Xu C, Neilson EG. Antibodies against macrophages that overlap in specificity with fibroblasts. Kidney Int. 2005;67:2488–2493. doi: 10.1111/j.1523-1755.2005.00358.x. [DOI] [PubMed] [Google Scholar]
- 29.Le Hir M, Hegyi I, Cueni-Loffing D, Loffing J, Kaissling B. Characterization of renal interstitial fibroblast-specific protein 1/S100A4-positive cells in healthy and inflamed rodent kidneys. Histochem Cell Biol. 2005;123:335–346. doi: 10.1007/s00418-005-0788-z. [DOI] [PubMed] [Google Scholar]
- 30.Boomershine CS, et al. Autoimmune pancreatitis results from loss of TGFbeta signalling in S100A4-positive dendritic cells. Gut. 2009;58:1267–1274. doi: 10.1136/gut.2008.170779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senolt L, et al. S100A4 is expressed at site of invasion in rheumatoid arthritis synovium and modulates production of matrix metalloproteinases. Ann Rheum Dis. 2006;65:1645–1648. doi: 10.1136/ard.2005.047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng N, et al. Loss of TGF-beta type II receptor in fibroblasts promotes mammary carcinoma growth and invasion through upregulation of TGF-alpha-, MSP- and HGF-mediated signaling networks. Oncogene. 2005;24:5053–5068. doi: 10.1038/sj.onc.1208685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trimboli AJ, et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461:1084–1091. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li ZH, Dulyaninova NG, House RP, Almo SC, Bresnick AR. S100A4 regulates macrophage chemotaxis. Mol Biol Cell. 2010;21:2598–2610. doi: 10.1091/mbc.E09-07-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue C, Plieth D, Venkov C, Xu C, Neilson EG. The gatekeeper effect of epithelial-mesenchymal transition regulates the frequency of breast cancer metastasis. Cancer Res. 2003;63:3386–3394. [PubMed] [Google Scholar]
- 37.Osterreicher CH, et al. Angiotensin-converting-enzyme 2 inhibits liver fibrosis in mice. Hepatology. 2009;50:929–938. doi: 10.1002/hep.23104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park EJ, et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140:197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taura K, et al. Hepatic stellate cells secrete angiopoietin 1 that induces angiogenesis in liver fibrosis. Gastroenterology. 2008;135:1729–1738. doi: 10.1053/j.gastro.2008.07.065. [DOI] [PubMed] [Google Scholar]
- 40.Grivennikov SI, et al. Distinct and nonredundant in vivo functions of TNF produced by t cells and macrophages/neutrophils: protective and deleterious effects. Immunity. 2005;22:93–104. doi: 10.1016/j.immuni.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 41.Greig KT, et al. Critical roles for c-Myb in lymphoid priming and early B-cell development. Blood. 2010;115:2796–2805. doi: 10.1182/blood-2009-08-239210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klotz L, et al. Increased antigen cross-presentation but impaired cross-priming after activation of peroxisome proliferator-activated receptor gamma is mediated by up-regulation of B7H1. J Immunol. 2009;183:129–136. doi: 10.4049/jimmunol.0804260. [DOI] [PubMed] [Google Scholar]
- 43.Sásik R, Woelk CH, Corbeil J. Microarray truths and consequences. J Mol Endocrinol. 2004;33:1–9. doi: 10.1677/jme.0.0330001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.