Abstract
The class II chelatases associated with heme, siroheme, and cobalamin biosynthesis are structurally related enzymes that insert a specific metal ion (Fe2+ or Co2+) into the center of a modified tetrapyrrole (protoporphyrin or sirohydrochlorin). The structures of two related class II enzymes, CbiXS from Archaeoglobus fulgidus and CbiK from Salmonella enterica, that are responsible for the insertion of cobalt along the cobalamin biosynthesis pathway are presented in complex with their metallated product. A further structure of a CbiK from Desulfovibrio vulgaris Hildenborough reveals how cobalt is bound at the active site. The crystal structures show that the binding of sirohydrochlorin is distinctly different to porphyrin binding in the protoporphyrin ferrochelatases and provide a molecular overview of the mechanism of chelation. The structures also give insights into the evolution of chelatase form and function. Finally, the structure of a periplasmic form of Desulfovibrio vulgaris Hildenborough CbiK reveals a novel tetrameric arrangement of its subunits that are stabilized by the presence of a heme b cofactor. Whereas retaining colbaltochelatase activity, this protein has acquired a central cavity with the potential to chaperone or transport metals across the periplasmic space, thereby evolving a new use for an ancient protein subunit.
Keywords: enzyme mechanism, tetrapyrrole biosynthesis
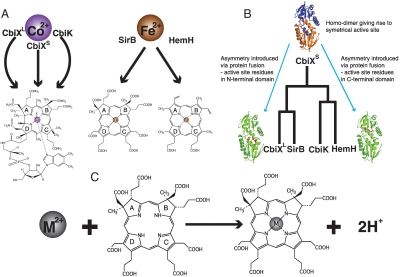
The class II chelatases are enzymes that insert divalent metal ions (Co2+, Fe2+) into a range of modified tetrapyrroles to generate heme, siroheme, and cobalamin (1–4). The simplest of these enzymes is CbiXS, which has a subunit composed of around 130 amino acids (2). The structure of the Archaeoglobus fulgidus CbiXS (Af–CbiXS) reveals that it forms a homodimer with a symmetrical active site and functional studies have shown that it is responsible for the chelation of Co2+ into sirohydrochlorin (SHC) in the biosynthesis of cobalamin (5). These small forms of the enzyme are generally found in the Archaea and are thought to represent a primordial form of the protein. In most other organisms the chelatases are about twice the size of CbiXS, resulting from a gene duplication and fusion event (2). Hence, structures of the Salmonella enterica CbiK (Se–CbiK) (6) and the Bacillus subtilis HemH (7), which are responsible for the insertion of Co2+ and Fe2+ into cobalamin and heme respectively, are bilobal enzymes consisting of two alpha/beta domains with a pseudo two-fold similarity, where the main catalytic groups are found in the C-terminal domain of the proteins. In contrast, the Bacillus megaterium CbiXL and SirB enzymes insert Co2+ and Fe2+ into SHC in the biosynthesis of cobalamin and siroheme, respectively, but in this case the active site residues are located in the N-terminal region of these proteins (2, 8, 9). Thus, these enzymes have evolved by a process of gene duplication and fusion followed by maintenance of the active site residues in either the N- or C-terminal region of the protein. The function and phylogenetic relationship between these proteins is summarized in Fig. 1.
Fig. 1.
(A) The enzymes CbiXL, CbiXS, and CbiK catalyse the insertion of Co(II) in cobalamin biosynthesis and SirB and HemH the insertion of Fe(II) in siroheme and heme biosynthesis, respectively. (B) The ancestral homodimer, has duplicated and fused to give the larger chelatases. (C) The reaction catalysed by cobalto-chelatase is presented, metal is inserted and two protons removed from the pyrrole rings.
Intriguingly, CbiK also appears to have a separate function in a number of Gram-negative bacteria. For instance, in Porphyromonas gingivalis, CbiK (Pg–CbiK) is found with an N-terminal leader sequence that directs it to the outer membrane. Indeed, Pg–CbiK was first identified as a 30-kDa major antigenic heme-binding protein (10, 11). Its gene is found in a small operon where it is transcribed with genes encoding a putative ABC transporter and a ferrichrome binding protein. Thus, it has been suggested that the Pg–CbiK can also function as part of a metal uptake mechanism, perhaps by helping to remove iron from exogenous heme (10). Notwithstanding the spatial location of CbiK in P. gingivalis, which is not located in the cytosolic site of tetrapyrrole biosynthesis, the enzyme is still functional as a cobaltochelatase and thus the new function for the protein has not involved the loss of any chelatase substrate-binding or functional residues (12). In Desulfovibrio vulgaris, there are two versions of CbiK encoded within the genome, which were named Dv–CbiKC and Dv–CbiKP, reflecting their respective localization in the cytoplasm (C) and periplasm (P) of the cell (13). As with the Pg–CbiK, the Dv–CbiKP is also isolated with a bound heme group, with a stoichiometry of one molecule per protein dimer (13). However, both Dv–CbiKC and Dv–CbiKP are active as cobaltochelatases, although given their cellular distribution it would seem more likely that Dv–CbiKC is the enzyme involved in the biosynthesis of cobalamin, whereas Dv–CbiKP presumably plays a similar role to Pg–CbiK in a transport process.
Mechanistically, it is thought that the tetrapyrrole-derived substrate binds to the type II chelatase in a distorted fashion, to allow pyrrole-deprotonation and metal ligation to the resulting pyrrolines (1). In support of this, N-methylmesoporphyrins, which are intrinsically distorted, are found to act as strong inhibitors of the protoporphyrin ferrochelatases (14). Indeed, structural studies on a bacterial ferrochelatase in complex with an N-methylmesoporphyrin found the tetrapyrrole bound in a distorted fashion consistent with such a model (15). Furthermore, antibodies raised against a similar compound found them to be catalytically active as a protoporphyrin ferrochelatase, suggesting that a strained porphyrin acts as a transition state in the chelation mechanism (16, 17). In the structures of the bacterial protoporphyrin ferrochelatase in complex with N-methylmesoporphyrin the deformation of the inhibitor is assisted by binding the macrocycle across an active site phenylalanine, which acts as a molecular anvil, providing the inertia to forge distortion into the substrate (15, 18). However, the mode of binding of the true substrate, protoporphyrin, to human ferrochelatase is different as the substrate is rotated around by 100° and is accommodated approximately 5 Å deeper within the active site. Moreover, only a slight warp is observed in the plane of the macrocycle indicating that just minor ruffling of the tetrapyrrole is required to enhance metal insertion (19, 20). The latter, therefore, represents a more accurate substrate/product binding site than that identified by the former inhibitor complex.
In the protoporphyrin ferrochelatase the metal binding site is thought to be formed around the highly conserved active site histidine and glutamate residues because crystallographic studies with the yeast ferrochelatase have shown that both cobalt and cadmium bind in this area (21), supporting previous data derived from the bacterial enzyme that the histidine plays an important role in defining the metal binding location (7). Moreover, site directed mutagenesis of the histidine residue is also consistent with this hypothesis (22). Nonetheless, evidence of metal binding to an area behind the active site in human protoporphyrin ferrochelatase has led to a controversial idea that the metal ion may travel to the active site via a channel spanning a distance of around 20 Å (20, 23). Thus, there is still some debate on not only how the metal ion binds to the chelatase but also how the metal ion is delivered to the enzyme.
Progress on the sirohydrochlorin cobaltochelatases has been slower due mainly to the difficulty in producing the tetrapyrrole-derived substrate, SHC, which is highly oxygen-sensitive. The active site of CbiK contains two conserved histidine residues, corresponding to the Glu and His found in protoporphyrin ferrochelatases (6). The site also contains the phenylalanine residue against which the macrocycle is likely to stack. With CbiXS the active site has two-fold symmetry and therefore possesses two pairs of histidines, and as a consequence of the symmetry lacks the active site phenylalanine (5). This arrangement promotes questions about the number of metal binding sites and the orientation of the incoming SHC. In this paper we explore how sirohydrochlorin binds to both Se–CbiK and Af–CbiXS, how cobalt binds to Dv–CbiKP and propose a mechanism for metal insertion, demonstrating how greater protein complexity results in enhanced catalysis. We also show how further oligomerization of CbiK leads to heme-binding in Dv–CbiKP, revealing a fascinating insight into a protein that is caught in evolutionary flux between two distinct processes.
Results
Binding of Sirohydrochlorin to CbiXS and CbiK—Evolution of Specificity.
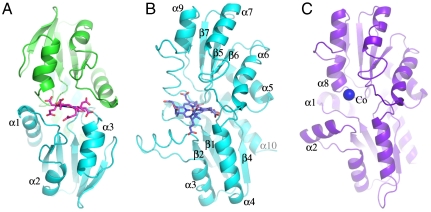
Crystals of Af–CbiXS were grown and diffraction data collected to 1.6 Å, improving on the previously reported resolution of the enzyme structure (5). When the protein was crystallized anaerobically in the presence of SHC, purple colored crystals were observed, which diffracted to 2.0 Å. The resulting Fourier synthesis revealed clear electron density within the active site of the enzyme, into which the C and D rings of SHC could be easily fitted (Figs. 2A and 3A). A similar approach of cocrystallization of SHC was undertaken with both Se–CbiK and Dv–CbiKP, although only purple crystals were observed with the Salmonella enzyme. Diffraction data from the Se–CbiK crystals were collected and a substrate-complex structure was determined to 1.9 Å (Figs. 2B and 3C). In both the Af–CbiXS and Se–CbiK SHC cocrystals the structure of the observed bound tetrapyrrole clearly shows the presence of a metal ion inserted into the core of the macrocycle. The identity of the metal ion was not determined but may possibly be as a result of some carryover nickel from the His-bind purification of the protein. Therefore, these structures represent predominantly product rather than substrate complexes. Nonetheless, in comparison to both apo Af–CbiXS and Se–CbiK, it can be seen that the binding of SHC is accompanied by changes around the active site (Fig 3 B and D), which are described in more detail below. The product complex binds in the same orientation in both Af–CbiXS and Se–CbiK (Fig. 2), with the two adjacent propionate side chains of rings C and D of the tetrapyrrole-derived macrocycle fastening deep into the substrate-binding cleft and making hydrogen bonds to main chain amides.
Fig. 2.
Cartoon representations of the crystal structures of (A) Archaeoglobus fulgidus CbiXS (homodimer) with the two subunits shown in blue and green, (B) Salmonella enterica CbiK, both with metallo-sirohydrochlorin bound, and (C) Desulfovibrio vulgaris CbiKP with cobalt (blue sphere) bound to the active center.
Fig. 3.
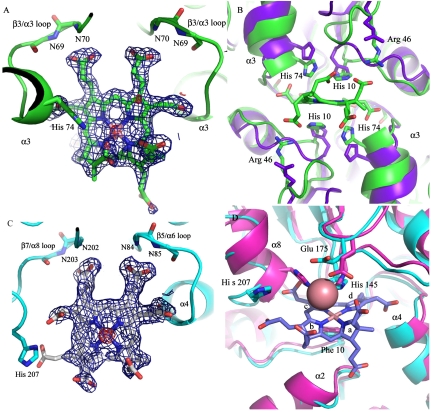
(A) σA-weighted difference Fourier synthesis of metallo-SHC bound to Af–CbiX (blue mesh), the anomalous map confirms the presence of a metal at the active center of the macrocycle. The propionates of rings C and D bind to the main-chain amides of the β3/α3 loops. (B) Superimposition of the free and bound forms of Af–CbiX showing the closing up of the active center around the bound macrocycle. (C) and (D) Similar images for Se–CbiK except that in D, the cobalt complex of Dv–CbiK is superimposed. In both complexes the product is bound and there is substantial closing of the active center around the bound sirohydrochlorin.
SHC Binding to Af–CbiXS.
An Af–CbiXS tetramer, a dimer of dimers, occupies the asymmetric unit of the crystal; there is therefore no crystallographic requirement for the two subunits forming the active center to have strict two-fold symmetry, but the rotation axis passing through the active centre is in fact very close to a strict two-fold axis. Remarkably, given the symmetry of the homodimer, SHC appears to have a unique binding mode in one of the two active centers with the C and D rings and the decorating side chains being particularly well defined and with reasonably good definition also for the A and B rings. The C and D ring acetates interact with the main chain amides of residues 73 and 74 at the N-terminal end of helix α3 and with ND1 of His74 of the two subunits. His74s are approximately perpendicular to the tetrapyrrole ring and His10 of both subunits lie roughly in the same plane as the SHC. The acetate from ring A and the propionate from ring B interact with the two-fold related Arg46s (Fig. 3B). The deeply bound C ring propionate hydrogen bonds with the main chain amides of residues 69 and 70 of the two subunits that make the catalytic homodimer but surprisingly the D ring hydrogen bonds only with the amide of residue 69 and significantly more weakly with the amide of residue 70; this asymmetry was unexpected given the otherwise close two-fold symmetry of the active center and SHC binding (Fig. 2A). When SHC binds to CbiXS the active center closes around the substrate (Fig. 3B); the loops bearing Arg46 and His10 are pulled closer to the center as are the helices bearing His74s and the adjacent amides that hydrogen bond the C and D ring acetates. The acetates on rings C and D appear to play a major role in this induced fit in contrast to the propionates on rings C and D that bind to a relatively static part of the structure.
SHC Binding to Se–CbiK.
In contrast to CbiXS, there is regio-specific binding of SHC in the active center of Se–CbiK such that the upper face of SHC is adjacent to the two active site histidines. The propionate side chain attached to ring C makes hydrogen bonds to the amides of residues 202 and 203 and that of ring D to the amides of residues 84 and 85 (Fig. 3D). The tetrapyrrole lies across one face of Phe10; in fact it looks as if the tetrapyrrole is bent over this phenylalanine residue. His145 is approximately in the same plane as the tetrapyrrole ring and is roughly positioned over the inserted metal ion. The major conformational change induced by the binding of the SHC is in the position of the His207 and the polypeptide neighboring this histidine (Fig. 3D). The beta carbon of His207 changes by approximately 5.5 Å as the histidine side chain goes out of the way of the tetrapyrrole to a position where it hydrogen bonds the acetate group on the C ring. Movement of the main chain away from the acetate group results in a distortion to the helical structure of this region. The region that moves, residues 204–209 (inclusive), is adjacent to the backbone amides binding the C ring propionates.
The Structure of Dv–CbiKP and the Binding of Cobalt.
Crystals of Dv–CbiKP, which are orange in color due to the presence of the associated heme b, diffracted to 1.9 Å resolution. Subsequently, full data were collected and the protein phases determined by multiple anomalous dispersion methods. Overall, the subunit structure of Dv–CbiKP is closely similar to the architecture of Se–CbiK (6) (Fig. 2 B and C). However, unlike the previously reported structure of Se–CbiK, Dv–CbiKP is tetrameric, the biologically authentic tetramer being generated when the crystallographic symmetry is applied to the subunit present in the asymmetric unit. The tetrameric structure in the crystal is consistent with previous biochemical studies in solution (13).
Cocrystallization of Dv–CbiKP in the presence of cobalt revealed that the protein binds cobalt via residues His154, Glu184, and His216 (Dv numbering, Fig. 3D and Fig. S1). Besides these residues, there is evidence for bound peroxide and water molecules. In comparison to apo Se–CbiK and Dv–CbiKP, the imidazole ring of His154 is rotated by 63° relative to its position in the cobalt-containing structure of Dv–CbiKP, indicating some minor rearrangement of the protein ligands upon cobalt binding. The interaction of the cobalt with His154 is remarkable, because rather than the cobalt binding to the lone pair of the NE2 atom, the cobalt appears to interact with histidine ring via the NE2-CE2 edge of the histidine side chain.
Structure of Dv–CbiKP—Protein Oligomerization Evolves Another Function.
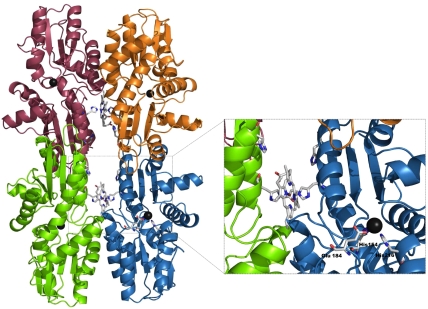
The tetrameric structure reveals how heme is bound in Dv–CbiK, which is well removed from the chelatase active site of the enzyme (Fig. 4). The heme b in Dv–CbiKP is found located in-between two monomers, consistent with previous biochemical studies (13) that determined a ratio of 0.5 heme/monomer. The heme is axially coordinated by a histidine residue from each monomer, His96, with a coordination distance of ca. 2.0 Å to the iron atom. In the tetramer, the two hemes are quite separate from each other, with a distance between the two iron atoms of ca. 32 Å (Fig. 4). The distance between the heme iron and the cobalt site is ca. 20 Å. The heme is located in a hydrophobic pocket formed by residues Pro91, Phe95, Leu99 and Pro159 from each vertical monomer. The heme propionate groups do not make protein contacts, but are directed toward the tetramer interface where they are exposed to the solvent.
Fig. 4.
Tetrameric assembly of D. vulgaris CbiKP. Each monomer is colored differently (A, red; B, green; C, blue; D, yellow) and the hemes b in-between subunits are represented as sticks together with the axial iron ligand His96; the cobalt sites are displayed as black spheres in each monomer. A detailed view of the central tetramer cavity is shown in the Inset.
In the tetramer, the two hemes are nearly coplanar, with the propionate groups pointing toward its center (Fig. 4) and the four monomers are arranged in such a way that the porphyrin binding clefts are facing outward, thus being easily accessible to the solvent. The cavity at the center of the tetramer has dimensions approximately 16 × 15 × 17 Å and can be partially blocked by movement of His103.
Discussion
These results provide unprecedented molecular detail on the mechanism of cobalt chelation and an insight into how protein complexity leads to the evolution of a different function (24). Although there is clear similarity in the topologies of the protoporphyrin ferrochelatases (HemH) (7, 20) and the sirohydrochlorin cobaltochelatases (CbiK and CbiX) (5, 6) the present work shows that the binding of the tetrapyrrole substrate is distinct. Specifically, the propionate groups of pyrrole rings C and D of SHC bind deep inside the substrate-binding pocket of CbiX and CbiK, in contrast to the protoporphyrin ferrochelatase substrate complex. Thus, superimposition of structures using DaliLite (25) reveals that although the human protoporphyrin ferrochelatase binds protoporphyrin in the same equivalent spatial location as SHC is found in CbiK/X, the porphyrin is rotated by about 90° in an orientation where the hydrophobic groups of the A and B rings are tightly bound (Fig. S2) (15, 19). In the inhibitor complex with the bacterial HemH, the N-mesomethylporphyrin is rotated by a further 90° and is not so buried within the active site cleft (Fig. S2) (15). This shows conservation of chelation function and preservation of a related mechanism involving conservation of the histidine that delivers the metal, but evolution of the substrate binding mode. The large differences in substrate orientation between the protoporphyrin and SHC chelatases would be consistent with these proteins diverging at an early stage from a common ancestor that had not evolved substrate discrimination.
By modeling the position of the bound cobalt found in Dv–CbiKP onto the structure of Se–CbiK a mechanism for chelation can be deduced. Theoretical calculations predict that metallation involves a number of steps including removal of water from the metal ion annulus and deprotonation of the modified tetrapyrrole nitrogens (26). The binding of cobalt to the Se–CbiK His145, His207, and Glu175 triad promotes the desolvation steps. However, the metal is held weakly as His145 interacts with the cobalt with its lone pair (NE2) apparently free,f whereas His207 has a more conventional nitrogen (NE2) lone pair–cobalt interaction. The nonideal geometry of the ligation of the cobalt ion means that the lone pair on His145 is available to abstract a proton from one of the tetrapyrrole nitrogen atoms. The C ring acetate group apparently plays a major role in inducing the conformational changes that accompany metal ion insertion by moving His207, weakening the grip of the enzyme on the metal and encouraging its insertion into the macrocycle by the exquisitely positioned His145. CbiK His145 is the equivalent to His263 in human ferrochelatase, and in both CbiK and human ferrochelatase this residue moves relatively modestly (27) compared to His207 that is substantially pushed away when SHC binds. The conserved nature of His145 (His263) suggests that this residue plays an important role in metal delivery and together with His207 may also contribute to substrate specificity, product entrapment and enhancement of catalytic rate. In CbiK, the D ring of SHC is locked in position by hydrogen bonding of the acetate group to the main chain amides of residues 88 and 89 at the N-terminal end of the helix (α4). The favorable N-terminal helix dipole interaction with the acetate will stabilize this interaction. This coupled with the distortion introduced into the isobacteriochlorin macrocycle, by binding across Phe10 to encourage pyrrole-deprotonation, sets the scene for a transition state. In this scenario the metal is displaced from its initial binding site by the introduction of the SHC acetic acid side chain, with either one or both of the histidines acting as general bases by deprotonating the pyrroles, thereby ensuring the formation of a nitrogen-cobalt bond and chelation.
With Af–CbiXS the process is slightly different. The metal binding site is likely to be composed of the two histidine residues found in each subunit (His10 and His74), because the equivalent group to Se–CbiK Glu175 is taken by an arginine (Arg44). On binding SHC, Af–CbiXS undergoes conformational rearrangement with the active center closing in around the bound tetrapyrrole, but less distortion is introduced into the tetrapyrrole because the comparable position of Se–CbiK Phe10 is taken by Af–CbiXS His10 and the binding cleft has pseudo two-fold symmetry that cannot by its nature exert distorting forces on the bound tetrapyrrole ring in the same way that the asymmetric active center of Se–CbiK can. The lack of distortion is presumably compensated by the fact that protons can be abstracted from either face of the bound SHC. Thus, if cobalt is bound to subunit A of the catalytic dimer, then His10 on subunit B is well positioned to deprotonate the pyrrole nitrogen, encouraging the formation of the first isobacteriochlorin-metal bond. Interestingly, metal insertion may also be stabilized through a metal-oxygen bond with the propionate side chain on either ring A or B of SHC, looping round underneath the macrocycle.
D. vulgaris contains two types of cobaltochelatase, Dv–CbiKP and Dv–CbiKC, and their alignment with Se–CbiK, shown in Fig. S3, reveals a high degree of similarity including conservation of the cobalt binding residues. However, the heme b ligand, His96, identified in the structure of Dv–CbiKP is absent from the sequences of both Dv–CbiKC and Se–CbiK and no heme cofactor has ever been detected in either of the latter (6, 13). Remarkably, a truncated form of Dv–CbiKP, lacking the N-terminal first twenty-eight amino acid residues (signal peptide) is unable to bind heme although the truncated protein exhibited the same tetrameric oligomerization state in solution as the full-length protein (13), showing that the heme is not necessary for the assembly of the Dv–CbiKP tetramer. The binding of heme to the cavity of Dv–CbiKP tetramer may provide the protein another function, as is implicated by its genomic organization. The Dv–CbiKP gene is part of a putative operon that includes genes with similarity to those encoding permeases, Fe(III) siderophores transporter systems and periplasmic binding proteins. Such an organization would be consistent with CbiKP-type proteins being involved in either heme or metal transport.
In summary, the work reported here provides a unique insight into the evolution of enzyme form and function. It demonstrates how a small protein can oligomerise to give a symmetrical active site and how asymmetry is subsequently afforded through gene duplication and fusion. Finally, new functions can be accommodated by further oligomerization, which generates new cavities for further binding activities.
Experimental Procedures
Protein Crystallization.
Dv–CbiKP was produced recombinantly and purified as previously described (13). Crystals were grown using the hanging drop vapour diffusion method at 20 °C, where the best crystals were obtained by mixing 1.0 μL Dv–CbiKP protein solution (10 mg/mL) with 2.0 μL reservoir solution and equilibrating the drop against a 0.5 mL reservoir, with a solution composed of 100 mM Tris-HCl. pH 8.5, containing 2.0 M ammonium sulphate (condition #30 of the Classic Screen, Nextal). The cocrystallization of Dv–CbiKP with cobalt was achieved by mixing 1.0 μL Dv–CbiKP with 1.8 μL reservoir solution and 0.2 μL 1.0 M cobalt chloride. Both the Se–CbiK and Af–CbiXS were purified and crystallized as previously described (5, 6). The SHC complexes of Se–CbiK and Af–CbiXS were produced by anaerobic cocrystallization of equimolar quantities of CbiK and SHC.
Data Collection, Processing, and Refinement.
Experimental phasing using MAD exploited the anomalous scattering from cobalt. CHOOCH (28) was used to determine the wavelengths for data collection and XDS (29) / SCALA (30) used for data processing. The structure was solved using SHELXE (31) and modeled using ARP/WARP (32). The other structures were solved by molecular replacement using PHASER (33) or MOLREP (34). All structures were refined and rebuilt with REFMAC (35) and COOT (36). Structures were superposed using SSM (37). Crystallographic statistics are given in Tables S1 and S2.
Supplementary Material
Acknowledgments.
We thank Prof. Michael James and Dr. Jiang Yin (University of Alberta) for the plasmid carrying the AfCbiXS gene. This work was funded by Fundação para a Ciência a Tecnologia Grants POCTI/BME/37406/2002 and PTDC/BIA-PRO/67107/2006. Susana Lobo is a recipient of Grant SFRH/BPD/63944/2009. We thank the European Synchrotron Radiation Facility (ESRF) for support with the ID14-EH1 data collection, as well as the European Molecular Biology Laboratory (EMBL) Grenoble Outstation for providing support for measurements at the ESRF EMBL-CRG BM14 beamline under the European Community—Access to Research Infrastructure Action FP6 Program “Structuring the European Research Area Specific Program,” with Contract RII3-CT-2004-506008. The help and advice given by Martin A. Walsh and Hassan Belrhali (EMBL, Grenoble) during the BM14 data collections is also gratefully acknowledged. We also acknowledge the support of the Biotechnology and Biological Research Council (BB/E002137, BB/D016568, and BB/E024203), Engineering and Physical Sciences Research Council (studentship to D.L.) and the use of the Diamond Light Source (Oxford).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014298108/-/DCSupplemental.
Data deposition: The atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank (www.pdb.org) [PDB ID codes 2xvx (native Dv–CbiKP) , 2xvz (Dv–CbiKP plus Co2+), 2xws (native Af–CbiXS), 2xwq (Af–CbiXS metallated SHC complex), and 2xwp (Se–CbiK metallated SHC complex].
References
- 1.Al-Karadaghi S, et al. Chelatases: Distort to select? Trends Biochem Sci. 2006;31:135–142. doi: 10.1016/j.tibs.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brindley AA, Raux E, Leech HK, Schubert HL, Warren MJ. A story of chelatase evolution: identification and characterization of a small 13–15-kDa “ancestral” cobaltochelatase (CbiXS) in the archaea. J Biol Chem. 2003;278:22388–22395. doi: 10.1074/jbc.M302468200. [DOI] [PubMed] [Google Scholar]
- 3.Dailey HA, et al. Ferrochelatase at the millennium: Structures, mechanisms and [2Fe-2S] clusters. Cell Mol Life Sci. 2000;57:1909–1926. doi: 10.1007/PL00000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. The biosynthesis of adenosylcobalamin (vitamin B12) Nat Prod Rep. 2002;19:390–412. doi: 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- 5.Yin J, et al. Crystal structure of the vitamin B12 biosynthetic cobaltochelatase, CbiXS, from Archaeoglobus fulgidus. J Struct Funct Genomics. 2006;7:37–50. doi: 10.1007/s10969-006-9008-x. [DOI] [PubMed] [Google Scholar]
- 6.Schubert HL, Raux E, Wilson KS, Warren MJ. Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis. Biochemistry. 1999;38:10660–10669. doi: 10.1021/bi9906773. [DOI] [PubMed] [Google Scholar]
- 7.Al-Karadaghi S, Hansson M, Nikonov S, Jonsson B, Hedersted L. Crystal structure of ferrochelatase: The terminal enzyme in heme biosynthesis. Structure. 1997;5:1501–1510. doi: 10.1016/s0969-2126(97)00299-2. [DOI] [PubMed] [Google Scholar]
- 8.Leech HK, et al. Characterization of the cobaltochelatase CbiXL: Evidence for a 4Fe-4S center housed within an MXCXXC motif. J Biol Chem. 2003;278:41900–41907. doi: 10.1074/jbc.M306112200. [DOI] [PubMed] [Google Scholar]
- 9.Raux E, et al. Identification and functional analysis of enzymes required for precorrin-2 dehydrogenation and metal ion insertion in the biosynthesis of sirohaem and cobalamin Bacillus megaterium. Biochem J . 2003;370:505–516. doi: 10.1042/BJ20021443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dashper SG, et al. Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J Bacteriol. 2000;182:6456–6462. doi: 10.1128/jb.182.22.6456-6462.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendtlass A, Dashper SG, Reynolds EC. Identification of an antigenic protein Pga30 from Porphyromonas gingivalis W50. Oral Microbiol Immun. 2000;15:383–387. doi: 10.1034/j.1399-302x.2000.150608.x. [DOI] [PubMed] [Google Scholar]
- 12.Roper JM, et al. The enigma of cobalamin (Vitamin B12) biosynthesis in Porphyromonas gingivalis. Identification and characterization of a functional corrin pathway. J Biol Chem. 2000;275:40316–40323. doi: 10.1074/jbc.M007146200. [DOI] [PubMed] [Google Scholar]
- 13.Lobo SA, et al. Two distinct roles for two functional cobaltochelatases (CbiK) in Desulfovibrio vulgaris hildenborough. Biochemistry. 2008;47:5851–5857. doi: 10.1021/bi800342c. [DOI] [PubMed] [Google Scholar]
- 14.Dailey HA, Fleming JE. Bovine ferrochelatase. Kinetic analysis of inhibition by N-methylprotoporphyrin, manganese, and heme. J Biol Chem. 1983;258:11453–11459. [PubMed] [Google Scholar]
- 15.Lecerof D, Fodje M, Hansson A, Hansson M, Al-Karadaghi S. Structural and mechanistic basis of porphyrin metallation by ferrochelatase. J Mol Biol. 2000;297:221–232. doi: 10.1006/jmbi.2000.3569. [DOI] [PubMed] [Google Scholar]
- 16.Lavallee DK. In: Mechanistic Principles of Enzyme Activity. Liebman JF, Greenberg A, editors. New York: VCH; 1988. pp. 279–314. [Google Scholar]
- 17.Romesberg FE, et al. Structural and kinetic evidence for strain in biological catalysis. Biochemistry. 1998;37:14404–14409. doi: 10.1021/bi981578c. [DOI] [PubMed] [Google Scholar]
- 18.Karlberg T, et al. Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: The role of active site residues. J Mol Biol. 2008;378:1074–1083. doi: 10.1016/j.jmb.2008.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medlock A, Swartz L, Dailey TA, Dailey HA, Lanzilotta WN. Substrate interactions with human ferrochelatase. Proc Natl Acad Sci USA. 2007;104:1789–1793. doi: 10.1073/pnas.0606144104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu CK, et al. The 2.0 Å structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nat Struct Biol. 2001;8:156–160. doi: 10.1038/84152. [DOI] [PubMed] [Google Scholar]
- 21.Karlberg T, et al. Metal binding to Saccharomyces cerevisiae ferrochelatase. Biochemistry. 2002;41:13499–13506. doi: 10.1021/bi0260785. [DOI] [PubMed] [Google Scholar]
- 22.Hansson MD, Karlberg T, Rahardja MA, Al-Karadaghi S, Hansson M. Amino acid residues His183 and Glu264 in Bacillus subtilis ferrochelatase direct and facilitate the insertion of metal ion into protoporphyrin IX. Biochemistry. 2007;46:87–94. doi: 10.1021/bi061760a. [DOI] [PubMed] [Google Scholar]
- 23.Sellers VM, Wu CK, Dailey TA, Dailey HA. Human ferrochelatase: Characterization of substrate-iron binding and proton-abstracting residues. Biochemistry. 2001;40:9821–9827. doi: 10.1021/bi010012c. [DOI] [PubMed] [Google Scholar]
- 24.Orengo CA, Thornton JM. Protein families and their evolution-a structural perspective. Annu Rev Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- 25.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 26.Shen Y, Ryde U. Reaction mechanism of porphyrin metallation studied by theoretical methods. Chemistry. 2005;11:1549–1564. doi: 10.1002/chem.200400298. [DOI] [PubMed] [Google Scholar]
- 27.Medlock AE, Dailey TA, Ross TA, Dailey HA, Lanzilotta WN. A pi-helix switch selective for porphyrin deprotonation and product release in human ferrochelatase. J Mol Biol. 2007;373:1006–1016. doi: 10.1016/j.jmb.2007.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans G, Pettifer RF. CHOOCH: A program for deriving anomalous-scattering factors from X-ray fluorescence spectra. J Appl Crystallogr. 2001;34:82–86. [Google Scholar]
- 29.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 30.Dodson EJ, Winn M, Ralph A. Collaborative Computational Project Number 4: Providing programs for protein crystallography. Methods Enzymol. 1997;277:620–633. doi: 10.1016/s0076-6879(97)77034-4. [DOI] [PubMed] [Google Scholar]
- 31.Sheldrick GM. Macromolecular phasing with SHELXE. Z Kristallogr. 2002;217:644–650. [Google Scholar]
- 32.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 33.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 34.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Applied Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. COOT: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.