Abstract
N,N-Dodecyl,methyl-polyethylenimine coatings applied to solid surfaces have been shown by us to disinfect aqueous solutions of influenza viruses. Herein we elucidate the mechanism of this phenomenon. Infectivity-, protein-, RNA-, and scanning electron microscopy-based experiments reveal that, upon contact with the hydrophobic polycationic coating, influenza viruses (including pathogenic human and avian, both wild-type and drug-resistant, strains) irreversibly adhere to it, followed by structural damage and inactivation; subsequently, viral RNA is released into solution, while proteins remain adsorbed.
Keywords: antiviral, biocide, virucidal coating
Infectious diseases, including influenza, kill millions of people a year and sicken hundreds of millions (1, 2). In the United States alone, there are tens of thousands of influenza-related deaths annually (3). The influenza virus’s propensity for genomic recombination and mutation (antigenic shift and drift, respectively) can generate new strains to which humans have no previously developed immunity—a common factor in influenza pandemics (4, 5). A jarring example in modern history is the Spanish Flu of 1918–19, which killed up to 100 million people worldwide (6); the H1N1 (“Swine Flu”) virus of 2009 has shown that such pandemics remain an acute threat. Although vaccines and such antivirals as zanamivir (Relenza®) and oseltamivir (Tamiflu®) are somewhat effective, they have serious limitations: The former must predict the next seasonal flu strain months in advance for current egg-based vaccines (7), while the latter are unreliable because of rapid mutations in the influenza virus epitopes (8).
A complementary approach to vaccines and drugs is to inactivate the virus (or any pathogenic microbe) with biocides during its transmission through inanimate objects, which is a major route for nosocomial infections (9, 10). However, biocidal formulations commonly applied as solutions can evaporate and be used up or wiped away, making the efficacy of this approach dependent on the frequency of its reapplication.
We have demonstrated (11) that coating (“painting”) with certain hydrophobic polycations renders surfaces permanently antibacterial and antifungal, retaining their disinfectant properties even after multiple washes (12, 13). Recently, this antimicrobial activity has been expanded to influenza viruses (14, 15). Herein we mechanistically elucidate this phenomenon. Specifically, we have found that upon contact with N,N-dodecyl,methyl-PEI coatings, aqueous solutions of influenza A viruses (including human and avian, both wild-type and mutant zanamivir-resistant, strains) are completely disinfected; this correlates with a disappearance of viral proteins, although significant quantities of viral RNA are still in solution. Based on this and other evidence, we conclude that these solutions are disinfected by the removal of viral particles that irreversibly adhere to the hydrophobic polycationic coatings; the latter then cause disintegration (including RNA release) and inactivation of the adhered viruses.
Results and Discussion
Prior to embarking on a mechanistic investigation of virucidal properties of surfaces coated (“painted”) with N,N-dodecyl,methyl-PEI, we explored whether the nature of the underlying solid object plays a role. Because the virucidal activity of these hydrophobic polycations was previously discovered with coated glass slides (14, 15), we added to these studies chemically unrelated polyethylene and polypropylene. As seen in Table 1, bare (i.e., uncoated) slides of each material exhibited only partial or no virucidal activity against a waterborne influenza virus. In contrast, when coated with N,N-dodecyl,methyl-PEI, all three types of surfaces completely disinfected the aqueous solutions of the virus, indicating that this property is independent of the surface treated.
Table 1.
Effect of glass, polypropylene, and polyethylene slides coated with N - N-dodecyl,methyl-PEI on WSN influenza strain’s viral infectivity and concentration of viral particles in solution
| Infectivity (by plaque assay)* | Viral particles (by ELISA)*, † | |||||
| Surface | Initially | After contact with bare slides | After contact with polycation-coated slides | Initially | After contact with bare slides | After contact with polycation-coated slides |
| Glass | 20.4 ± 2.1 | 10.9 ± 0.5 | 0.0 ± 0.0 | 21.0 ± 2.2 | 1.4 ± 2.1 | 0.0 ± 1.6 |
| Polypropylene | 20.4 ± 2.1 | 18.1 ± 1.9 | 0.0 ± 0.0 | 21.0 ± 2.2 | 17.8 ± 1.1 | −1.0 ± 2.2 |
| Polyethylene | 20.4 ± 2.1 | 15.1 ± 3.2 | 0.0 ± 0.0 | 21.0 ± 2.2 | 17.1 ± 1.2 | 0.1 ± 1.2 |
*Values represent titers: (mean ± std dev) × 104 pfu/mL
†Assessed by measuring the concentration of viral nucleoprotein (NP); the high loss of viral particles for bare glass slides is likely due to nonspecific adsorption of exogenous viral NP
While quantifying the influenza virus by the plaque assay (16) allowed to titer the infectivity of solution, the fate of the viral particles heretofore remained obscure. In particular, we could not discriminate between the following possibilities: (i) The viruses collide with the coated surface, undergo irreversible inactivation, and bounce off back into solution; or (ii) the viruses collide with the coated surface and irreversibly adhere to it in either an infectious or noninfectious form. To distinguish between these alternative scenarios, we employed the viral nucleoprotein (NP) (a prevalent influenza protein, some 1,000 copies/viron) (4) as a marker for the viral particles. Note that although NP is an internal viral protein, it can be easily quantified following lysis of viral particles.
The ELISA data for NP—i.e., the viral particles—in aqueous solutions of an influenza virus (WSN strain) incubated between pairs of bare glass, polypropylene, or polyethylene slides revealed marked loss (presumably due to nonspecific adsorption) of virons for glass but little for the two polymers (Table 1). When the slides were coated with N,N-dodecyl,methyl-PEI, however, the viral particles completely disappeared from solution regardless of the material coated (Table 1). All subsequent studies reported herein were performed with polyethylene slides because they displayed far less background adsorption than glass and were easier to work with than polypropylene slides.
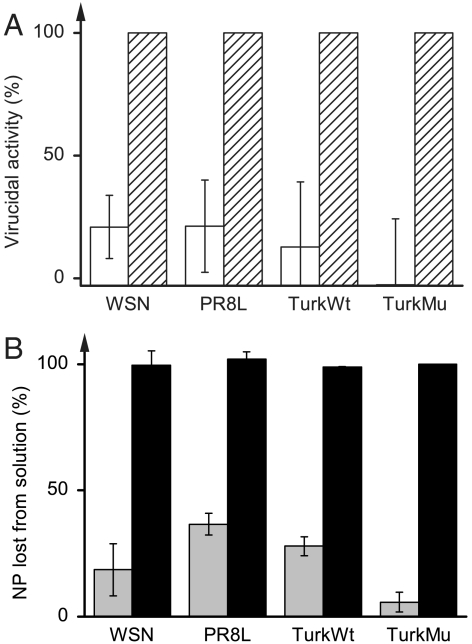
To test the generality of the foregoing observations, we expanded our investigation to other influenza viruses, namely a human pathogenic strain (PR8), as well as avian pathogenic wild-type (TurkWt) and mutant zanamivir-resistant (TurkMu) strains (Table 2). As seen in Fig. 1A, virucidal activities of bare polyethylene slides are negligible against all of these strains. In contrast, the N,N-dodecyl,methyl-PEI-coated slides are completely virucidal (Fig. 1A), thus indicating that the hydrophobic polycationic coating is able to disinfect aqueous solutions of a variety of diverse influenza strains. Separately, the independent ELISA results depicted in Fig. 1B show that while the bare slides had minor effect on viral particle concentrations, the viral particles disappeared from solution after incubation with the polycation-coated slides. Thus using influenza’s NP as a marker reveals a strong correlation between the disinfection of the viral solution and the disappearance of the viral particles from it, suggesting that the influenza viral particles adhere to the hydrophobic polycationic coatings. Note that this adherence takes place regardless of variables arising from the source of culture, suspension media, and antigenicity associated with the different strains used (Table 2).
Table 2.
Strains of influenza A viruses used in this study and their properties
| Short name | Influenza A viral strain | Initial titer (pfu/mL)* | Culture conditions; suspension conditions | Notes |
| WSN | WSN/33 (H1N1) | 4.5( ± 0.6) × 106 | Tissue; DME-HEPES/10% FBS | Laboratory strain, nonpathogenic |
| PR8H | PR/8/34 (H1N1) | 1.3( ± 0.5) × 108 | Egg; HEPES/saline | Human pathogenic; sucrose-gradient purified |
| PR8L† | PR/8/34 (H1N1) | 0.9( ± 0.2) × 106 | Egg; HEPES/PBS/saline | Human pathogenic; sucrose-gradient purified |
| TurkWt | turkey/MN/833/80 (H4N2) | 1.0( ± 0.1) × 106 | Egg; allantoic fluid | Avian pathogenic |
| TurkMu | turkey/MN/833/80 (H4N2) | 4.3( ± 0.3) × 106 | Egg; allantoic fluid | Avian pathogenic; zanamivir-resistant |
*Determined via plaque assay after ribonuclease pretreatment; the titer is that of a 10-μL droplet incubated with slides prior to a 100-fold dilution upon washing. See Materials and Methods for details.
†An approximately 100-fold dilution of PR8H with PBS
Fig. 1.
The effect of polyethylene slides coated with N,N-dodecyl,methyl-PEI challenged with aqueous solutions of various influenza A viruses. See Table 2 for viral strain abbreviations and details (note that “H” and “L” after PR8 stand for high and low titer of the virus, respectively). (A) Virucidal activity of bare (white bars) and polycation-coated polyethylene slides (hatch-marked bars) against various influenza virus strains. (B) Relative quantities of viral nucleoprotein (NP) disappeared from solution after incubation with bare (gray bars) and with polycation-coated polyethylene slides (black bars).
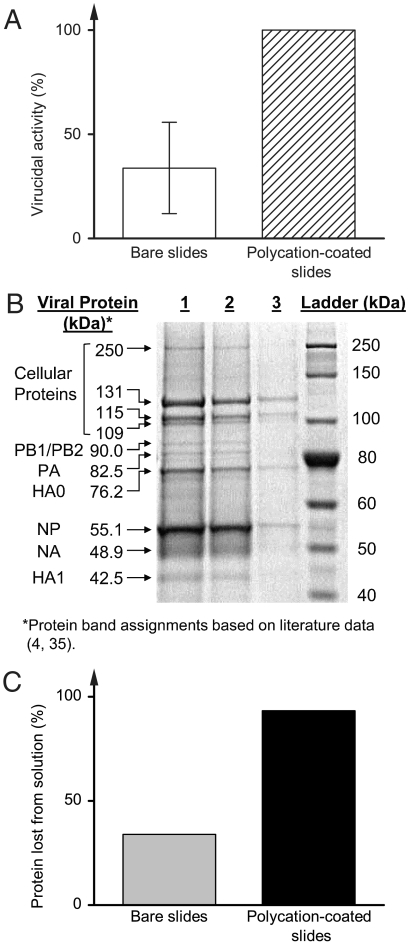
To independently verify these results and to test for other influenza viral proteins in solution, we examined N,N-dodecyl,methyl-PEI-coated slides against a solution of the purified PR8H virus (which differs from PR8L only by its much higher titer; Table 2) and confirmed its complete disinfection (Fig. 2A). Additionally, we analyzed the initial viral solution, as well as that after exposure to either bare or polycation-coated slides, by means of SDS/PAGE. Even a visual inspection of protein band profiles in lanes 1, 2, and 3 in Fig. 2B reveals some loss for each protein after the exposure, with by far the most dramatic one occurring with the virus incubated with the coated slides (lane 3 in Fig. 2B). Quantification of these observations by gel-scanning densitometry indicates a loss of 94% of viral protein after incubation with the coated slides versus only a 34% loss after incubation with control, bare slides (Fig. 2C). [That the bare slides demonstrated the same losses in virucidal activity (Fig. 2A) as in viral protein point to a nonspecific removal of the virus from solution.] The observed complete loss of virucidal activity and a concurrent nearly complete loss of viral protein suggest that a small fraction of inactivated influenza virus and/or protein-containing fragments thereof remains in solution even following incubation with coated slides, likely due to the much higher viral titer required for SDS/PAGE used in these experiments.
Fig. 2.
The effect of polyethylene slides coated with N,N-dodecyl,methyl-PEI on high-titer human pathogenic PR8 influenza virus strain (PR8H, Table 2). (A) Virucidal activity of PR8H by bare (white bars) and N,N-dodecyl,methyl-PEI-coated polyethylene slides (hatch-marked bars). (B) SDS/PAGE of the concentrated PR8H strain run on a 7% polyacrylamide gel and stained with the commassie blue dye. Lanes 1, 2, and 3 represent virus samples prior to assay, after contact with bare slides and after contact with polycation-coated slides, respectively. (C) Protein disappeared from solution by bare slides (gray bars) and polycation-coated slides (black bars), as determined by gel-scanning densitometry.
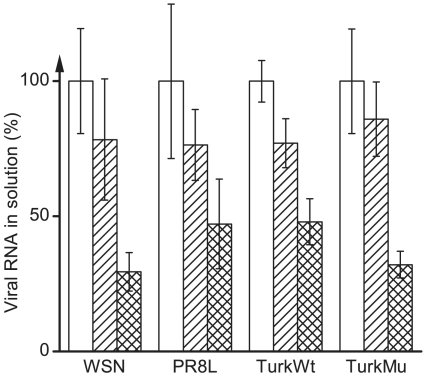
These findings support the second aforementioned scenario, namely that the viral particles irreversibly adhere to the N,N-dodecyl,methyl-PEI-coated surface, thereby vanishing from solution. To mechanistically explore this process further, we endeavored to assess the infectivity of the influenza viruses adhered to the coated surface. Because such viruses are not amenable to cell-based infectivity assays, we examined instead how much viral RNA could be detected in solution. Its substantial presence there would evidence the inactivation of the surface-bound viruses because they obviously cannot be infective without their RNA. To this end, we used real-time reverse-transcriptase PCR (qRT–PCR) with the probe and primers detecting influenza virus’s seventh RNA segment; i.e., the genomic region encoding the matrix (M1) protein (4, 17). Using the same influenza strains (Table 2), negligible losses of viral RNA from solution were observed in the case of bare polyethylene slides (Fig. 3). Importantly, and in sharp contrast to the results from our protein-based assays (Fig. 2), however, all viral solutions incubated with the N,N-dodecyl,methyl-PEI-coated slides exhibited significant quantities of viral RNA. This leakage of viral RNA from the surface-adhered viral particles points to a profound damage to the virus and its inevitable demise.
Fig. 3.
Relative quantities of influenza A viral RNA in solution prior to assay (white bars), after incubation with bare polyethylene slides (hatch-marked bars), and after incubation with N,N-dodecyl,methyl-PEI-coated polyethylene slides (cross-hatched bars).
To ensure that the RNA detected in solution was indeed leaked from the interior of the viral particle, we pretreated all viral samples with the ribonuclease enzymes to digest any exogenous single- or double-stranded RNA that may be present (18–20). Control experiments confirmed that this enzymatic pretreatment completely digests RNA and that addition of ribonuclease’s inhibitor prevents subsequent RNA digestion (Fig. S1). There was also no nonspecific amplification from N,N-dodecyl,methyl-PEI putatively leached into solution (Fig. S2). A comparison of genomic sequences between influenza viral samples before and after contact with the coating revealed at least a 95% sequence identity (less than a 100% identity is likely due to inherent errors in the sequencing technology) (21). All these results support the conclusion that the detected viral RNA is, in fact, endogenous viral genomic material leaked into solution after contact with the hydrophobic polycationic coating.
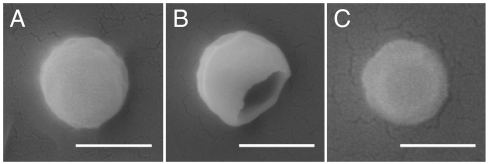
The recovery of RNA from the immobilized-N,N-dodecyl,methyl-PEI-contacted viral solutions was about half of that from the uncoated slides. This contrasts to our protein studies that have thus far suggested an all-or-none virus inactivation scenario. Because the root of this partial recovery of genomic material may point to an additional mechanism in the polycation’s virucidal activity, we directly visualized the viral particles on the surfaces using scanning electron microscopy (SEM). Micrographs of the WSN strain of influenza virus on a plain silicon wafer showed no visible damage to the viral structure (Fig. 4A). In contrast, exposure of the viruses to the coated surface revealed a mixture of two extremes: either a substantial structural damage leaving a gaping hole (Fig. 4B) or no noticeable effect (Fig. 4C). Specifically, out of 132 exposed viral particles surveyed, 54% exhibited the type of structural damage seen in Fig. 4B. Interestingly, this value agrees with the fraction of RNA recovered from the coated slides as compared to the bare slides (Fig. 3). As to the observed (Fig. 4 B and C) bimodal distribution, note that even viral particles that appear intact may be actually damaged on the side facing the N,N-dodecyl,methyl-PEI coating hidden from view; this type of damage would also likely hinder escape of viral RNA into solution.
Fig. 4.
Scanning electron microscopy (SEM) images of the WSN strain of influenza virus after exposure to plain (A) and N,N-dodecyl,methyl-PEI-coated (B and C) silicon wafers. Of the latter, a larger fraction of viral particles showed substantial structural damage (B), while a smaller fraction showed no visible damage (C); see text for details. (Scale bars, 100 nm.)
One might have expected that no RNA should be recovered in solution at all due to nonspecific interactions between the negatively charged nucleic acid and the surface-situated hydrophobic polycation molecules if not for the fact that the derivatization of PEI with long alkyl moieties sterically interferes with such interactions (22–24). Note in this regard that elemental analysis of N,N-dodecyl,methyl-PEI used herein shows approximately 1.6 dodecyl moieties per monomer unit, corresponding to, on average, 60% of monomers in PEI doubly dodecylated and 40% singularly dodecylated, which is considerably greater than in the previous reports (22–24).
Finally, we tested whether a monomeric structural analog of N,N-dodecyl,methyl-PEI, namely dodecyltrimethylammonium bromide (DTAB), was also capable of inactivating the influenza virus. To this end, we challenged the influenza virus with serial dilutions of DTAB for the latter’s concentration required for 50% inhibition (IC50) (25) (and also its toxic concentration TC50—i.e., that reducing MDCK cell viability by 50%). The DTAB’s IC50 value was found to be 21 ± 5 μg/mL [which was fourfold lower than its 84 ± 26 μg/mL TC50 value, in agreement with literature (26)]. Therefore, the antiviral activity observed with our surface-immobilized hydrophobic polycations appears to be inherent to the quaternary ammonium monomeric unit. Indeed, a similar quaternary ammonium salt, benzalkonium chloride, is also capable of inactivating influenza virus (27–29).
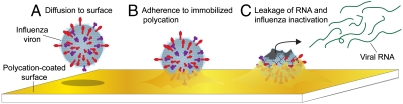
Based on the experimental evidence obtained herein, we propose a framework for influenza virus inactivation depicted in Fig. 5. When viral particles strike the N,N-dodecyl,methyl-PEI-coated surface due to thermal motion (Fig. 5A), they can adhere to it (Fig. 5B) through hydrophobic and electrostatic interactions (11). As a result of such interactions, virus’s disintegration ensues manifesting itself in a leakage of RNA into solution and a loss of infectivity (Fig. 5C). In this regard it is instructive to note that, upon interactions with polycations, model lipid vesicles can undergo lateral segregation and flip-flopping (30) of phospholipids, ultimately fluidizing their membrane (31). These types of effects would disrupt the ordered lipid rafts (32, 33) and leaflet asymmetry (34) in influenza viral envelopes.
Fig. 5.
Proposed mechanism of influenza virus inactivation by N,N-dodecyl,methyl-PEI coatings. (A) An influenza viral particle diffuses to the polycation-coated surface from solution and (B) adheres to it. Significant damage imparted by the immobilized hydrophobic polycations is then incurred by the adhered viral particle such that (C) its genomic material (RNA) leaks into solution leaving the viral particle inactivated on the surface.
Conclusions
Through investigation into how aqueous solutions of various human and avian influenza viruses are disinfected by N,N-dodecyl,methyl-PEI coatings, we found that this phenomenon was dependent on neither the nature of the surface that was coated nor the strain of influenza A virus. A direct correlation was found between the exposed solution’s losses of infectivity and of viral proteins, thereby indicating that the viral particles adhere to the surface-immobilized hydrophobic polycation. Additional analysis of the disinfected solution revealed significant quantities of RNA therein that stemmed from the viral particles, thereby evidencing that the integrity of the latter was compromised allowing the release of their genomic material. This conclusion was independently verified by direct SEM observations. A monomeric analog of N,N-dodecyl,methyl-PEI, DTAB, was also found to inactivate the influenza virus, suggesting that the antiviral activity is inherent to the hydrophobic quaternary ammonium salt moiety, which is retained in even a polymeric and surface-immobilized form.
Materials and Methods
Polymer Synthesis and Immobilization.
Unless otherwise noted, all chemicals were from Sigma-Aldrich Chemical Co. N,N-Dodecyl,methyl-PEI was synthesized as described previously (14, 16, 23). Briefly, linear 217-kDa PEI prepared by hydrolysis of 500-kDa poly(2-ethyl-2-oxazoline) in concentrated HCl at 125 °C (23) was dodecylated, followed either by methylation with CH3I to give the final quaternary ammonium polycation or by dialysis within a cellulose membrane (3.5-kDa cutoff) against hexane. Elemental analysis of N-dodecyl-PEI performed by Columbia Analytical Labs yielded a C∶H∶N of 72.3∶12.8∶4.0 by weight (or 21.1∶44.5∶1.0 by mol).
N,N-Dodecyl,methyl-PEI was applied from its 50 mg/mL solution in CHCl3 by painting slides with a 3/8 inch nylon-bristled paint brush (Loew–Cornell) and dried in a chemical hood; this painting procedure was repeated twice more. The square 2.5-cm slides were cleaned by sonication in isopropyl alcohol for 5 min and autoclaved prior to use. Glass slides were cut from standard microscope slides (VWR International), while low-density polyethylene and polypropylene slides were cut from 12“ × 12“ × 1/16“ sheets (McMaster–Carr Supply Co.).
Virus Culture and Ribonuclease Pretreatment.
The influenza A/WSN/33 (H1N1) virus was cultured from MDCK cells (14). Sucrose-gradient purified influenza A/PR/8/34 (H1N1) virus suspended in HEPES-saline buffer was used as obtained from Charles River Labs. Wild-type and zanamivir-resistant influenza A/MN/833/80 (H4N2) viruses were prepared from egg allantoic fluid (15).
Exogenous viral RNA (single- and double-stranded) was digested in all viral samples by incubating 100 μL of an influenza virus solution with 15 μL of ribonuclease III buffer, 50 μL of ribonuclease III enzyme (1 U/μL, Ambion), and 5 μL of 1,000-fold-diluted ribonuclease A (3,500 U/μL, Invitrogen) in PBS for 1 h at 37 °C (18–20). Subsequently, 5 μL of Antiribonuclease (20 U/μL, Ambion) was added, and the solution was incubated at RT for 15 min before being placed on ice.
A 10-μL droplet of a viral aqueous solution was sandwiched between two slides (glass, polyethylene, or polypropylene) in a 6 × 1.5 cm polystyrene Petri dish. After a 5-min incubation at RT, the slides were separated and washed with 990 μL of PBS. Viral samples were collected prior to contact with slides, after contact with bare slides, or after contact with polycation-coated slides (16).
Plaque assays were used to quantify the infectious viral particles (in pfu/mL) in solution by infecting a monolayer of MDCK cells in six-well plates with 200 μL of a viral solution (16).
ELISA.
An ELISA of influenza virus NP was performed with a 96-well maxisorp immunoplate (Nunc Intl.): the wells were incubated with 100 μL of a 1∶1,000 dilution of goat anti-influenza A polyclonal antibody (AB1074, Millipore) in PBS at 4 °C overnight and then washed thrice with 100 μL of a solution of 0.2% (w/v) I-block (Applied Biosystems) and 0.05% Tween 20 (v/v) in PBS (PBST), followed by a further incubation with an additional 100 μL of PBST for 3 h at RT. Then 100-μL volumes of viral samples, prepared by combining equal volumes of virus solution and 2X PBST, were added. After incubation overnight at 4 °C, the wells were washed thrice with 100 μL of PBST and incubated with 100 μL of a 3∶10,000 dilution of mouse anti-influenza A biotinylated monoclonal antibody (Millipore) in PBST for 3 h at RT. This was followed by washing thrice with 100 μL of PBST, incubation with 100 μL of a 1∶200 dilution of a streptavidin-horseradish peroxidase conjugate (Millipore) in PBS for 2 h at RT and washing thrice with 100 μL of PBST. Colorimetric development of 50 μL of 1-Step Ultra TMB (Thermo Scientific) at RT was stopped with 50 μL of 0.2 M H2SO4 after incubation for 12, 6, 9, or 4 min with WSN, PR8L, TurkWt, or TurkMu influenza viruses, respectively. Light absorbances measured at 450 nm for virus samples prior to assay, as well as after contact with bare or N,N-dodecyl,methyl-PEI-coated slides, were referenced to standard curves constructed from serially diluted stock samples and normalized to their plaque-assay determined titers.
SDS/PAGE.
Sucrose-gradient purified PR8H viral samples were disrupted with 1% SDS in PBS such that the final concentration was 0.1% SDS. Proteins from viral samples were concentrated and washed using Amicon Ultra 0.5-mL microcentrifuge filters (10-kDa cutoff, Millipore), according to the manufacturer’s instructions. Briefly, proteins were concentrated at RT by centrifugation at 14,000 × g and then washed thrice with 400 μL of 0.1% SDS in PBS by additional centrifugation. The final 14.0 μL of washed and concentrated protein solution was run on a SDS/PAGE using a 7% Tris-acetate NuPAGE gel (Invitrogen) and Tris-acetate buffer kit (Invitrogen) according to manufacturer’s instructions. Briefly, 14.0 μL of concentrated protein was combined with 5.4 μL of 4X lithium dodecylsulfate sample buffer, and 2.2 μL of 10X reducing agent prior to incubation at 70 °C for 10 min. The prepared samples and a boiled protein ladder (10–250 kDa, New England BioLabs) were added to the gel and subjected to electrophoresis for 60 min at 150 V. The resultant gel was stained with SimplyBlue comassie blue stain (Invitrogen) and imaged on an Alpha Imager 2200 (Cell Biosciences). Gel-scanning densitometry analysis was performed using ImageJ software (NIST) by calibrating the measured pixel density to optical density and then developing a standard curve with a power-law fit using the protein bands of serially diluted viral samples to which assay samples were referenced. Assignment of protein bands was based on the literature data (4, 35).
Viral RNA Extraction and Quantification with qRT-PCR.
RNA was extracted from 200 μL of a viral solution using the PureLink viral RNA/DNA kit (Invitrogen) according to manufacturer’s instructions giving a final 110 μL of viral RNA, stored at -80 °C. Quantification was performed with the RNA Ultrasense qRT-PCR mix (Invitrogen) according to manufacturer’s instructions, with the primers and probe (IDT) sequences used identical to those previously developed (17). Real-time reverse-transcriptase PCR was performed on an Applied Biosystems 7500 real-time PCR system as follows: 50 °C for 15 min, 95 °C for 2 min, and then 50 cycles of 95 °C for 15 sec and 60 °C for 32 sec. RNA samples from virus prior to assay, after contact with bare slides, and after contact with polycation-coated slides were referenced to standard curves constructed from serially diluted stock samples and normalized to plaque-assay titers (Applied Biosystems 7500 Software v2.0.1). All samples and standard curves were included on the same reaction plate.
RT-PCR and Sequencing.
Aliquots of 200 μL of WSN virus strain samples prior to assay and after contact with N,N-dodecyl,methyl-PEI-coated slides underwent RNA extraction as described above. Reverse transcription was performed using the Omniscript RT Kit (Qiagen) according to manufacturer’s instructions with Antiribonuclease (20 U/μL, Ambion AM2690) as the ribonuclease inhibitor with the aforementioned primers (IDT) (17). For amplification of the newly transcribed viral DNA, the TopTaq DNA Polymerase (Qiagen) with a dNTP mix (Promega) and primers were used according to manufacturer’s instructions. Sequencing was performed by the MIT Biopolymers Facility.
Detergent Studies.
The viability of MDCK cells after a 1-h incubation at RT with 100 μL of serially diluted DTAB in PBS was determined using the CellTiter96 Aqueous Nonradioactive Cell Proliferation Assay (Promega), an MTS-based assay for mitochondrial activity (36), and was performed according to manufacturer’s instructions. The measured 50% toxicity concentration (TC50) of DTAB was calculated using replicates of dose-response fitted curves (Origin Labs OriginPro v8.1).
The 50% inhibitory concentration (IC50) of DTAB against PR8 was measured by incubating serial dilutions of detergent with PR8 for 30 min at RT prior to determination of their infectivity by plaque assay (16, 25). Plaques were easily identifiable at and below 43 μg/mL DTAB, while above this level (e.g., at 85 μg/mL) cells were opaque, thus making potential plaques indistinguishable. Hence the IC50 value was calculated using replicates of dose-response fitted curves (Origin Labs OriginPro v8.1) to DTAB concentrations not exceeding 43 μg/mL.
Scanning Electron Microscopy.
Plain and N,N-dodecyl,methyl-PEI-coated 1-cm square silicon wafers were placed in a Petri dish, and a 10-μl droplet of an influenza virus solution was placed in the center before sandwiching with a plain silicon wafer to spread the droplet. This system was incubated at RT for 30 min before fixing the samples with Karnovsky’s fixative kit (Polysciences). To this end, the samples were incubated in the fixing solution (2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M Na phosphate buffer) for 2 h and rinsed for 10 min in the fresh buffer. The samples were then incubated in the dark in a 1% osmium tetroxide solution for 1 h before sequentially rinsing in 35%, 50%, 70%, 95%, and 100% aqueous ethanol for 10 min each, followed by dehydration thrice in 100% ethanol. The samples were then freeze-dried in liquid nitrogen and sputter-coated before imaging with a JEOL JSM-6700F SEM instrument at a 100,000X magnification.
Supplementary Material
Acknowledgments.
We thank Jayanta Haldar for providing the avian influenza A virus strains used herein; Peter Tieu for experimental assistance; and Liguo Wu, Koushik Mukherjee, Mishtu Dey, Peter Goldman, Eugene Antipov, Laura Jennings, Alyssa Larson, and Ching-Hung Shen for insightful discussions. This work was supported by the U.S. Army through the Institute of Soldier Nanotechnologies at the Massachusetts Institute of Technology under contract DAAD-19-02-D0002 with the Army Research Office.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017012108/-/DCSupplemental.
References
- 1.Beaglehole R, Irwin A, Prentice T. The World Health Report: Changing History. Geneva: World Health Organization; 2004. [Google Scholar]
- 2.Garibaldi RA. Epidemiology of community-acquired respiratory tract infections in adults: Incidence, etiology, and impact. Am J Med. 1985;78:32–37. doi: 10.1016/0002-9343(85)90361-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dushoff J, Plotkin JB, Viboud C, Earn DJD, Simonsen L. Mortality due to influenza in the United States—an annualized regression approach using multiple-cause mortality data. Am J Epidemiol. 2005;163:181–187. doi: 10.1093/aje/kwj024. [DOI] [PubMed] [Google Scholar]
- 4.Lamb RA, Krug RM. In: Fields Virology. Knipe DM, Howley PM, editors. Philadelphia: Lippincott; 2006. pp. 1488–1531. [Google Scholar]
- 5.Scholtissek C. Source for influenza pandemics. Eur J Epidemiol. 1994;10:455–458. doi: 10.1007/BF01719674. [DOI] [PubMed] [Google Scholar]
- 6.Osterholm MT. Preparing for the next pandemic. New Engl J Med. 2005;352:1839–1842. doi: 10.1056/NEJMp058068. [DOI] [PubMed] [Google Scholar]
- 7.Wright PF. Vaccine preparedness—are we ready for the next influenza pandemic? New Engl J Med. 2008;358:2540–2543. doi: 10.1056/NEJMp0803650. [DOI] [PubMed] [Google Scholar]
- 8.Weinstock DM, Zuccotti G. The evolution of influenza resistance and treatment. J Am Med Assoc. 2009;301:1066–1069. doi: 10.1001/jama.2009.324. [DOI] [PubMed] [Google Scholar]
- 9.Boyce JM, Potter-Bynoe G, Chenevert C, King T. Environmental contamination due to methicillin-resistant Staphylococcus aureus: Possible infection control implications. Infect Cont Hosp Ep. 1997;18:622–627. [PubMed] [Google Scholar]
- 10.Rosenthal VD, et al. Device-associated nosocomial infections in 55 intensive care units of 8 developing countries. Ann Intern Med. 2006;145:582–591. doi: 10.7326/0003-4819-145-8-200610170-00007. [DOI] [PubMed] [Google Scholar]
- 11.Klibanov AM. Permanently microbicidal materials coatings. J Mater Chem. 2007;17:2479–2482. [Google Scholar]
- 12.Lin J, Qiu SY, Lewis K, Klibanov AM. Mechanism of bactericidal and fungicidal activities of textiles covalently modified with alkylated polyethylenimine. Biotechnol Bioeng. 2003;83:168–172. doi: 10.1002/bit.10651. [DOI] [PubMed] [Google Scholar]
- 13.Mukherjee K, Rivera JJ, Klibanov AM. Practical aspects of hydrophobic polycationic bactericidal “paints”. Appl Biochem Biotechnol. 2008;151:61–70. doi: 10.1007/s12010-008-8151-1. [DOI] [PubMed] [Google Scholar]
- 14.Haldar J, An D, de Cienfuegos LA, Chen J, Klibanov AM. Polymeric coatings that inactivate both influenza virus and pathogenic bacteria. Proc Natl Acad Sci USA. 2006;103:17667–17671. doi: 10.1073/pnas.0608803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haldar J, Chen J, Tumpey TM, Gubareva LV, Klibanov AM. Hydrophobic polycationic coatings inactivate wild-type and zanamivir- and/or oseltamivir-resistant human and avian influenza viruses. Biotechnol Lett. 2008;30:475–479. doi: 10.1007/s10529-007-9565-5. [DOI] [PubMed] [Google Scholar]
- 16.Haldar J, Weight AK, Klibanov AM. Preparation, application and testing of permanent antibacterial and antiviral coatings. Nat Protoc. 2007;2:2412–2417. doi: 10.1038/nprot.2007.353. [DOI] [PubMed] [Google Scholar]
- 17.van Elden LJR, Nijhuis M, Schipper P, Schuurman R, van Loon AM. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J Clin Microbiol. 2001;39:196–200. doi: 10.1128/JCM.39.1.196-200.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majde JA, Guha-Thakurta N, Chen Z, Bredow S, Krueger JM. Spontaneous release of stable viral double-stranded RNA into the extracellular medium by influenza virus-infected MDCK epithelial cells: implications for the viral acute phase response. Arch Virol. 1998;143:2371–2380. doi: 10.1007/s007050050467. [DOI] [PubMed] [Google Scholar]
- 19.Nuanualsuwan S, Cliver DO. Pretreatment to avoid positive RT-PCR results with inactivated viruses. J Virol Methods. 2002;104:217–225. doi: 10.1016/s0166-0934(02)00089-7. [DOI] [PubMed] [Google Scholar]
- 20.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 22.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 23.Thomas M, et al. Full deacylation of polyethylenimine dramatically boosts its gene delivery efficiency and specificity to mouse lung. Proc Natl Acad Sci USA. 2005;102:5679–5684. doi: 10.1073/pnas.0502067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neamnark A, et al. Aliphatic lipid substitution on 2 kDa polyethylenimine improves plasmid delivery and transgene expression. Mol Pharm. 2009;6:1798–1815. doi: 10.1021/mp900074d. [DOI] [PubMed] [Google Scholar]
- 25.Hayden FG, Cote KM, Douglas RG. Plaque inhibition assay for drug susceptibility testing of influenza-viruses. Antimicrob Agents Chemother. 1980;17:865–870. doi: 10.1128/aac.17.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vieira OV, et al. Surfactants as microbicides and contraceptive agents: a systematic in vitro study. PLoS ONE. 2008;3:e2913. doi: 10.1371/journal.pone.0002913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abe M, et al. Effects of several virucidal agents on inactivation of influenza, newcastle disease, and avian infectious bronchitis viruses in the allantoic fluid of chicken eggs. Jpn J Infect Dis. 2007;60:342–346. [PubMed] [Google Scholar]
- 28.Armstrong JA, Froelich EJ. Inactivation of viruses by benzalkonium chloride. Appl Microbiol. 1964;12:132–137. doi: 10.1128/am.12.2.132-137.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oxford JS, Potter CW, McLaren C, Hardy W. Inactivation of influenza and other viruses by a mixture of virucidal compounds. Appl Microbiol. 1971;21:606–610. doi: 10.1128/am.21.4.606-610.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabanov VA, Yaroslavov AA. What happens to negatively charged lipid vesicles upon interacting with polycation species? J Controlled Release. 2002;78:267–271. doi: 10.1016/s0168-3659(01)00496-5. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda T, Lee B, Yamaguchi H, Tazuke S. Time-resolved fluorescence anisotropy studies on the interaction of biologically active polycations with phospholipid membranes. Biochim Biophys Acta. 1990;1021:56–62. [Google Scholar]
- 32.Polozov IV, Bezrukov L, Gawrisch K, Zimmerberg J. Progressive ordering with decreasing temperature of the phospholipids of influenza virus. Nat Chem Biol. 2008;4:248–255. doi: 10.1038/nchembio.77. [DOI] [PubMed] [Google Scholar]
- 33.Scheiffele P, Rietveld A, Wilk T, Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J Biol Chem. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- 34.Rothman JE, Tsai DK, Dawidowicz EA, Lenard J. Transbilayer phospholipid asymmetry and its maintenance in membrane of influenza-virus. Biochemistry. 1976;15:2361–2370. doi: 10.1021/bi00656a018. [DOI] [PubMed] [Google Scholar]
- 35.Shaw ML, Stone KL, Colangelo CM, Gulcicek EE, Palese P. Cellular proteins in influenza virus particles. PLoS Pathog. 2008;4:e1000085. doi: 10.1371/journal.ppat.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.