Abstract
The activated B-cell–like (ABC) subtype of diffuse large B-cell lymphoma (DLBCL) represents a very aggressive human lymphoma entity. Constitutive NF-κB activation caused by chronic active B-cell receptor (BCR) signaling is common feature of many ABC DLBCL cells; however, the pathways linking BCR signaling to the NF-κB prosurvival network are largely unknown. Here we report that constitutive activity of PI3K and the downstream kinase PDK1 are essential for the viability of two ABC DLBCL cell lines that carry mutations in the BCR proximal signaling adaptor CD79B. In these cells, PI3K inhibition reduces NF-κB activity and decreases the expression of NF-κB target genes. Furthermore, PI3K and PDK1 are required for maintaining MALT1 protease activity, which promotes survival of the affected ABC DLBCL cells. These results demonstrate a critical function of PI3K-PDK1 signaling upstream of MALT1 protease and NF-κB in distinct ABC DLBCL cells and provide a rationale for the pharmacologic use of PI3K inhibitors in DLBCL therapy.
A complex consisting of CARMA1 (also known as CARD11), B-cell lymphoma 10 (BCL10), and mucosa-associated lymphoid tissue lymphoma translocation gene 1 (MALT1) bridges antigenic stimulation initiated by B-cell receptors (BCRs) or T-cell receptors (TCRs) to the canonical NF-κB signaling pathway (1). Diffuse large B-cell lymphoma (DLBCL) represents the largest group of non-Hodgkin's lymphomas, and distinct subtypes have been classified based on gene expression profiling. Constitutive anti-apoptotic and pro-proliferative NF-κB activity via the CARMA1-BCL10-MALT1 (CBM) complex is a characteristic of the activated B-cell–like (ABC) subtype of DLBCL that constitutes an aggressive lymphoma entity (2–4). MALT1 encodes for a cystein protease whose activity is required for optimal T-cell activation (5–7) as well as survival of ABC DLBCL cells (8, 9). Distinct molecular aberrations have been suggested to contribute to pathological activation of the CBM complex in ABC DLBCL cells. Whereas oncogenic CARMA1 mutations are found in ≈10% of all ABC DLBCL patients (10), most ABC DLBCL cells display chronic active BCR signaling, and mutations have been identified in the BCR proximal regulators CD79A and B (11).
The PI3K pathway is active in all DLBCL cell lines tested, as well as in many primary DLBCL tumor samples independent of classification (11–13). Class I PI3Ks convert phosphatidylinositol-4,5-diphosphates to phosphatidylinositol-3,4,5-triphosphates, leading to activation of the effector kinases PDK1 (putative 3-phosphoinositide-dependent kinase 1) and protein kinase B (AKT). In B lymphocytes, the PI3K pathway is activated after antigenic engagement of BCRs. Deficiency of the PI3K regulatory subunit p85α impairs BCR-triggered NF-κB activation (14, 15). In line with this, chronic active BCR signaling promotes constitutive PI3K/AKT signaling in ABC DLBCL cells (11), but whether PI3K signaling contributes to NF-κB–dependent prosurvival signaling in these cells remains unclear. Here we provide evidence that PI3K-PDK1 signaling is essential for viability, MALT1 protease activity, and NF-κB activation in ABC DLBCL cells that carry mutations in the BCR proximal signaling adaptor CD79B.
Results
PI3K-PDK1 Signaling Controls Viability of a Subset of ABC DLBCL Cell Lines.
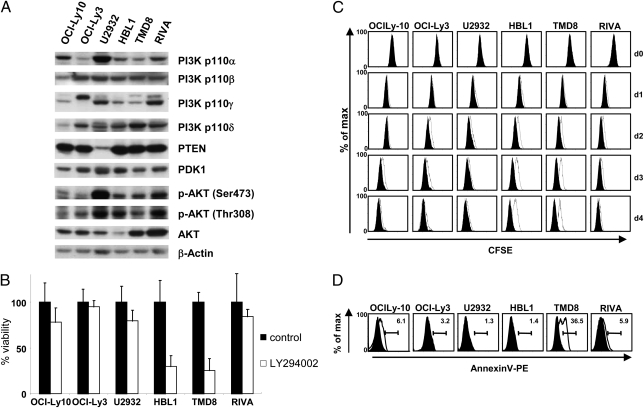
To monitor whether PI3K signaling is activated in ABC DLBCL cells, we first assessed the phosphorylation status of AKT in the well-characterized ABC DLBCL cell lines OCI-Ly10, OCI-Ly3, U2932, HBL1, TMD8, and RIVA (Fig. 1A). Constitutive AKT phosphorylation, a common feature of all ABC DLBCL cells, is monitored using anti–phospho-S473 and anti–phospho-T308 AKT antibodies. AKT phosphorylation is detected despite the high expression of the PI3K antagonist PTEN in most ABC DLBCL cells, except U2932 cells. The catalytic isoforms PI3K p110α/β/γ/δ, as well as the effector kinase PDK1, are expressed in ABC DLBCL cells.
Fig. 1.
PI3K-PDK1 inhibitors are toxic to a subset of ABC DLBCL cell lines. (A) Expression of PI3K pathway components in the indicated ABC DLBCL cells were measured by Western blot analysis. Proteins were detected with the indicated antibodies. Data are representative of at least two independent experiments. (B) ABC DLBCL cells were treated with 10 μM PI3K inhibitor LY294002 or solvent for 4 d. Counts of viable cells were done by trypan blue exclusion. Data are the mean from three independent experiments. Error bars indicate SD. (C) After incorporation of CFSE, ABC DLBCL cells were treated with 10 μM LY294002 or solvent, and the population of viable cells was analyzed for CFSE dilution every 24 h by FACS analysis. FACS data are representative of three independent experiments. Quantification for the three experiments is shown in Fig. S2A. (D) ABC DLBCL cells were treated with 10 μM LY294002 as in C. Apoptotic cells were identified by annexin V–PE staining of 7AAD− (necrotic) cells and analyzed by FACS analysis. The experiment shown is representative of three independent experiments. Quantification for the three experiments is shown in Fig. S2C. Error bars indicate SD.
To test whether PI3K activity delivers a survival signal in ABC DLBCL cells, we determined cell viability after 4 d of incubation with the pan-PI3K inhibitor LY294002 (Fig. 1B). We first determined 10 μM LY294002 as an optimal concentration that effectively blocked AKT phosphorylation in all ABC DLBCL cells (Fig. S1 A and B). This treatment strongly reduced the viability of the two ABC DLBCL cell lines HBL1 and TMD8, but had a minimal affect on OCI-Ly10, OCI-Ly3, U2932, and RIVA cells (Fig. 1B). To examine the physiological consequences of PI3K inhibition, we measured cell proliferation and apoptosis in ABC DLBCL cells (Fig. 1 C and D). For proliferation assays, carboxyfluorescein succinimidyl ester (CFSE) was incorporated into the cells, and cell divisions were tracked by measuring the dilution of the cellular CFSE label from the viable cells by FACS (Fig. 1C). CFSE dilution from three independent experiments was also quantified after 4 d of PI3K inhibition (Fig. S2 A and B). Congruent with cell viability tests, LY294002 incubation selectively impaired proliferation of HBL1 and TMD8 cells, but had minimal effects on the growth of all other ABC DLBCL cells. In addition, we determined the effect of PI3K inhibition on apoptosis by measuring annexin V+/7AAD− cells after 4 d of PI3K inhibitor treatment (Fig. 1D). We also quantified the rates of apoptosis from three independent experiments (Fig. S2C). PI3K inhibition selectively induced apoptosis in TMD8 cells, but had no significant effect on HBL1 cells or any other ABC DLBCL cells. These results indicate that PI3K inhibitors are toxic to some ABC DLBCL cells, and that toxicity results from decreasing proliferation and/or increasing apoptosis of these cells.
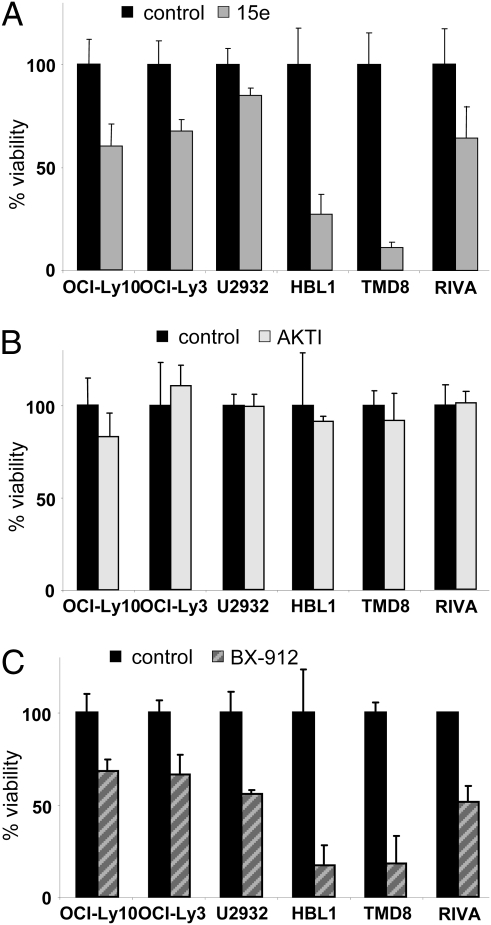
To provide further evidence for a critical role of PI3K signaling in the viability of HBL1 and TMD8 cells, we used the PI3K inhibitor 15e, which most potently inhibits p110α activity (IC50 = 2 nm) but also strongly impairs other isoforms, especially p110β (IC50 = 16 nm) (16). We found that 0.4 μM p110α inhibitor 15e blocked AKT phosphorylation in ABC DLBCL cells (Fig. S1C) and reduced the viability of HBL1 and TMD8 cells, but had little effect on the numbers of living OCI-Ly3, OCI-Ly10, U2932, and RIVA cells (Fig. 2A). Again, we examined proliferation and apoptosis in the four different ABC DLBCL cell lines after inhibition with 15e. Similar to LY294002, 15e inhibition impaired cell division most strongly in HBL1 and TMD8 cells, and had little effect on the growth of OCI-Ly3 and U2932 cells (Fig. S3 A and B). Apoptosis was significantly increased after 15e treatment only in TMD8 cells, not in any of the other ABC DLBCL cell lines (Fig. S3 C and D).
Fig. 2.
PI3K and PDK1, but not AKT, are required for viability of HBL1 and TMD8 ABC DLBCL cells. ABC DLBCL cells were treated with 0.4 μM PI3K inhibitor 15e (A), 2.5 μM AKTI (B), 0.25 μM PDK1 inhibitor BX-912 (C), or solvent for 4 d. Counts of viable cells were done by trypan blue exclusion. Data are the mean from three independent experiments. Error bars indicate SD.
We used pharmacologic AKT and PDK1 inhibitors to test which downstream effector is responsible for mediating PI3K-dependent viability of ABC DLBCL cells HBL1 and TMD8. We found that 2.5 μM AKT inhibitor (AKTI) VIII blocked AKT phosphorylation in ABC DLBCL cells (Fig. S1 D and E). In addition, the selective PDK1 inhibitor BX-912 (17) inhibited phosphorylation on Thr308 and Ser473 of AKT (Fig. S1 F and G), in agreement with previous findings that PDK1 also acts upstream of AKT (18). Although AKTI was not toxic to the ABC DLBCL cells after 4 d of treatment, the PDK1 inhibitor BX-912 strongly affected the viability of HBL1 and TMD8 cells compared with other ABC DLBCL cell lines (Fig. 2 B and C). These data suggest a critical role of PI3K-PDK1 signaling in maintaining the viability of distinct ABC DLBCL cell lines.
PI3K Activity Maintains Constitutive NF-κB Signaling in HBL1 and TMD8 Cells.
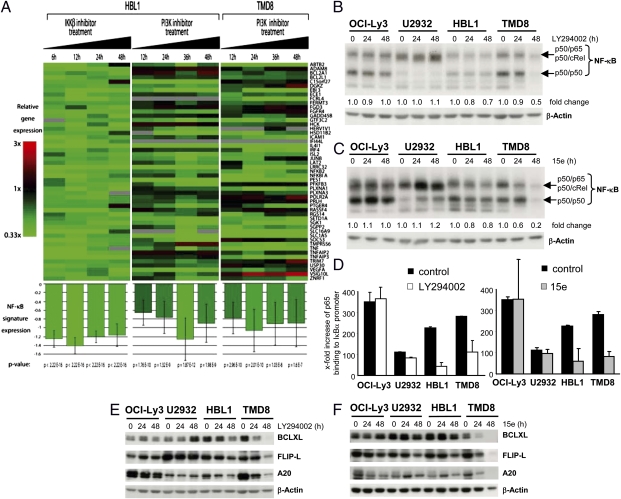
The growth and survival of ABC DLBCL cells depend on the constitutive activation of canonical NF-κB signaling. In most ABC DLBCL cells, including HBL1 and TMD8, high nuclear NF-κB levels are caused by chronic BCR upstream signaling, which also promotes activation of the PI3K pathway (11). Given our results suggesting that HBL1 and TMD8 cells are sensitive to inactivation of PI3K-PDK1 signaling, we wanted to assess whether PI3K contributes to NF-κB prosurvival signaling in these cells. We first asked whether NF-κB–driven gene expression might be influenced by PI3K inhibition. We determined relative changes in the gene expression after increasing times of treatment with the PI3K inhibitor 15e in HBL1 and TMD8 cells by genome-wide expression arrays, and applied these data to two independent NF-κB gene signatures. The first signature comprised all genes that were down-regulated in HBL1 by at least 50% after treatment with the selective inhibitor of nuclear factor kappa-B kinase subunit beta (IKK-β) inhibitor MLN120b at three of four time points (Fig. 3A). The second NF-κB gene signature consisted of genes that were both down-regulated by at least 1.4-fold after expression of an inhibitor of NF-κB (IκB) super-repressor in OCI-Ly3 and OCI-Ly10 and that were significantly down-regulated in HBL1 cells after MLN120b treatment (19) (Fig. S4). After PI3K inhibition, the overall expression as well as the proportion of NF-κB target genes from both signatures was significantly down-regulated (Tables S1 and S2), clearly implicating that PI3K inhibition suppresses activation of the NF-κB pathway in HBL1 and TMD8 cells.
Fig. 3.
PI3K signaling is required for NF-κB activation and target gene expression in HBL1 and TMD8 cells. (A) Gene expression profiling of the ABC DLBCL cell lines HBL1 and TMD8 after treatment with the IKKβ inhibitor MLN120b (for 6 h, 12 h, 24 h, and 48 h) and after incubation with the PI3K inhibitor 15e (for 12 h, 24 h, 36 h, and 48 h). Gene expression changes were assessed using DNA microarrays and depicted according to the color scale shown. A gene was selected as an NF-κB target gene in HBL1 cells if MLN120b decreased the expression of the gene by at least 50% at three time points. This signature of 51 genes was subsequently applied to the gene expression data after treatment with the PI3K inhibitor 15e. Gene expression measurements for the component genes in the NF-κB signature were averaged for each sample and are graphed at the bottom. Error bars depict SEM. The significance of the decrease in the NF-κB signature average in each treated sample was verified by the paired t test. (B and C) NF-κB DNA binding was analyzed by EMSA analysis from extracts of OCI-Ly3, U2932, HBL1, and TMD8 treated with 10 μM LY294002 (B) or 0.4 μM 15e (C) for the indicated times. Migration of the NF-κB heterodimers p50/p65 is indicated. The fold change of NF-κB activity was quantified by densitometric analysis and normalized to β-Actin amounts. (D) Recruitment of p65 to the IκBα promoter was determined by quantitative real-time PCR after anti-p65 ChIP. Values represent the x-fold increase of p65 bound to the IκBα promoter over an IgG control ChIP. Normalization was calculated using IκBα promoter DNA amplified from input. Error bars depict SDs from three experiments. (E and F) Expression of NF-κB target genes BCL-XL, FLIP-L, and A20 in ABC DLBCL cells after treatment with 10 μM LY294002 (E) or 0.4 μM 15e (F) for the indicated times was examined by Western blot analysis. EMSA and Western blot data are representative of two independent experiments.
To determine whether PI3K inhibition directly affects NF-κB activation, we measured NF-κB DNA binding by EMSA. NF-κB binding was confirmed by supershift analysis (Fig. S5A). Intriguingly, cell treatment with either PI3K inhibitors LY294002 or 15e or PDK1 inhibitor BX-912 for 24 h or 48 h specifically decreased the amount of NF-κB DNA binding in HBL1 and TMD8, but not in the other ABC DLBCL cell lines (Fig. 3 B and C and Fig. S5 B and D). We performed anti-p65 ChIP assays to verify that PI3K inhibition impedes the recruitment of NF-κB to endogenous target gene promoters. For this, we used quantitative real-time PCR to determine the amount of precipitated IκBα promoter as a prototype NF-κB target gene highly expressed in all ABC DLBCL cells (Fig. 3D). Ly294002 and 15e selectively impaired the association of NF-κB to the IκBα promoter in HBL1 and TMD8 cells but not in OCI-Ly3 and U2932 cells, indicating that the high level of nuclear NF-κB in HBL1 and TMD8 cells is controlled by PI3K activity.
NF-κB promotes cellular transformation through up-regulation of anti-apoptotic and pro-proliferative genes (20). Gene expression profiling revealed that PI3K signaling is required to maintain the NF-κB gene signature in HBL1 and TMD8 cells (Fig. 3A). To confirm these results, we determined the expression of BCL-XL, FLIP-L, and A20, three well-defined NF-κB target genes, by Western blot analysis in ABC DLBCL cells after PI3K or PDK1 inhibition (Fig. 3 E and F and Fig. S5 C and E). After 24 h or 48 h of inhibitor treatment, expression of all three proteins was reduced in HBL1 cells and even more severely so in TMD8 cells. In contrast, no reductions were detectable in other ABC DLBCL cell lines using LY294002 or BX-912 (Fig. 3E and Fig. S5E). PI3K inhibition by 15e led to a decrease in A20 in OCI-Ly3 and a slight reduction of FLIP-L in OCI-Ly3 and U2932 (Fig. 3F). We noted decreased expression of the NF-κB target genes JUNB and IL-10 after 15e treatment in all ABC DLBCL cells (Fig. S6 A and B), indicating that PI3K activity is also required to maintain the expression of some NF-κB target genes without directly affecting nuclear NF-κB DNA binding. Taken together, our findings show that expression of many NF-κB target genes in HBL1 and TMD8 is more sensitive to PI3K or PDK1 inhibition compared with other ABC DLBCL cells, in agreement with a differential effect of PI3K inhibitors on ABC DLBCL cell viability. Clearly, the strongest effect of PI3K-PDK1 inhibition on expression of the anti-apoptotic NF-κB target genes is seen in TMD8 cells, which correlates well with the strong induction of apoptosis in these cells compared with HBL1 cells.
PI3K and PDK1 Are Required for Constitutive MALT1 Protease Activity in HBL1 and TMD8 Cells.
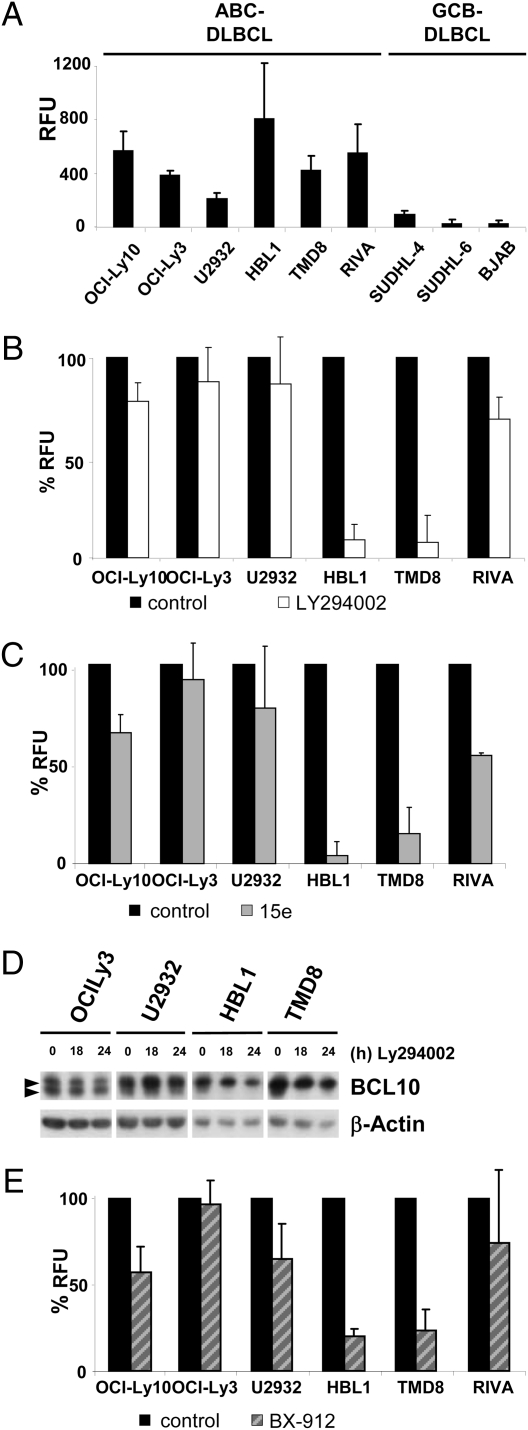
The MALT1 protein encodes for a mammalian cystein protease that can cleave BCL10 and A20 and is activated in T cells or ABC DLBCL cells (5–9). MALT1 inhibition is toxic to ABC DLBCL cells (8, 9). To examine whether PI3K activity could control constitutive MALT1 activity, we first conducted an assay to measure cellular MALT1 protease activity (Fig. S7 A–D). For this, we used the fluorogenic substrate AC-LRSR-AMC derived from the C-terminal BCL10 cleavage site, which is a substrate of recombinant purified GST-MALT1, but not of GST-MALT C453A, which carries a mutation in the catalytic center (Fig. S7 A and B). Cleavage activity of GST-MALT1 is blocked in a dose-dependent manner by the previously characterized antagonistic tetrapetide Z-VRPR-FMK (Fig. S7C). We next determined the activity of the cellular MALT1 protease after MALT1 immunoprecipitation from extracts of ABC and germinal center B-cell (GCB) DLBCL cells (Fig. 4A). Congruent with the previously observed constitutive cleavage of the MALT1 substrates BCL10 and A20 in ABC DLBCL cells (8, 9), we found increased constitutive MALT1 activity in all ABC DLBCL cell lines compared with three GCB DLBCL cell lines, despite comparable amounts of MALT1 in the different DLBCL cells (8). Similar to the recombinant GST-MALT1 protease, cellular MALT1 activity was completely blocked by the addition of 50 nM Z-VRPR-FMK to the cleavage reaction (Fig. S7D), providing evidence that substrate cleavage in ABC DLBCL cells indeed results from enhanced activity of the MALT1 protease.
Fig. 4.
PI3K-inhibition interferes with MALT1 protease activity in ABC DLBCL cells HBL1 and TMD8. (A) Equal protein amounts of ABC or GCB DLBCL cell extracts were used for IP with anti-MALT1 antibody. MALT1 protease activity was measured by a fluorescence-based cleavage assay using AC-LRSR-AMC as a substrate. (B and C) ABC DLBCL cells were either control-treated or incubated with PI3K inhibitors LY294002 (10 μM) or 15e (0.4 μM) for 7 h, and MALT1 cleavage activity from cellular lysates was measured as in A. For comparison, MALT1 activity in the absence of the inhibitor was set to 100%. (D) ABC DLBCL cells were incubated with the PI3K inhibitor LY294002, and BCL10 full-length as well as cleaved BCL10 (upper and lower arrows, respectively) were detected by Western blot analysis. (E) ABC DLBCL cells were either solvent- treated or incubated with 0.25 μM PDK1 inhibitor BX-912 for 7 h, and determination of MALT1 cleavage activity from cellular lysates was performed as in A. All MALT1 activity data in A, B, C, and E are the mean from three independent experiments. Error bars indicate SD.
To investigate whether PI3K signaling is involved in regulation of the MALT1 protease in ABC DLBCL cells, we determined cellular MALT1 activity after incubation with the PI3K inhibitors LY294002 and 15e (Fig. 4 B and C). Both inhibitors strongly impaired constitutive MALT1 activity in HBL1 and TMD8 cells, but had only a minimal effect on MALT1 activity in all other ABC DLBCL cells, suggesting that PI3K signaling is selectively involved in triggering the activation of the MALT1 protease in these distinct ABC DLBCL cells. We confirmed these data by demonstrating that PI3K inhibition also strongly impairs cleavage of the known MALT1 substrates BCL10 in HBL1 and TMD8 cells, but not in OCI-Ly3 and U2932 cells (Fig. 4D). Furthermore, PDK1 inhibition by BX-912 significantly impaired MALT1 protease activity selectively in HBL1 and TMD8 cells (Fig. 4E), whereas AKT inhibition by AKTI VIII had no effect (Fig. S7E). MALT1 expression was not reduced by PI3K or PDK1 inhibition, indicating that PI3K signaling is directly controlling MALT1 activity in these cells (Fig. S7 F–H). Thus, our data demonstrate that PI3K and PDK1 are essential for maintaining high MALT1 protease activity in ABC DLBCL cells that depend on PI3K-PDK1–mediated prosurvival signaling.
Discussion
We have shown that constitutive activation of the PI3K pathway is a common feature of ABC DLBCL cells. PI3K or PDK1 inhibition affects viability, MALT1 protease activity, and NF-κB activation in two ABC DLBCL cells. Because PI3K signaling depends on chronic active BCR signaling in these cells (11), PI3K and PDK1 link proximal BCR signaling to NF-κB–dependent prosurvival signaling in a subgroup of ABC DLBCL cell lines. Thus, our data provide evidence that the ABC DLBCL subtype encompasses a heterogeneous group of lymphoma entities that can be further subdivided based on distinct molecular aberrations.
Mutations in the immunoreceptor tyrosine-based activation motif of the BCR proximal adaptor CD79B were identified in ∼18% of patients with ABC DLBCL. The PI3K-PDK1–sensitive HBL1 and TMD8 cells carry heterozygous missense mutations that affect the first Tyr in the immunoreceptor tyrosine-based activation motif of CD79B (Y196F and Y196H, respectively). Mutation of Y196 in CD79B impairs association of the negative regulatory Lyn kinase, suggesting that this mutation is causing a gain of function (11, 21). All other ABC DLBCL cells that are less sensitive to PI3K inhibition are WT for CD79B. Even though we cannot exclude the possibility of involvement of other molecular aberrations in HBL1 and TMD8 cells, our data indicate that the CD79B mutations might be responsible for preventing the action of a negative regulator that specifically interferes with BCR-PI3K-PDK1-MALT1-NF-κB–dependent prosurvival signaling. Despite these similarities between HBL1 and TMD8 cell, there are clear differences, especially with respect to induction of apoptosis after PI3K inhibition. The stronger repression of anti-apoptotic genes like BCL-XL and FLIP-L might explain the increased sensitivity of TMD8 cells toward PI3K-PDK1 inhibition.
Tumor-specific somatic mutations have been detected in the p110α gene PIK3CA (22, 23). Even though PI3K inhibitor 15e is more selective for PI3K p110α (16), other isoforms are efficiently inhibited as well. Which PI3K isoforms are responsible for NF-κB activity and survival of HBL1 and TMD8 cells, and whether oncogenic mutations in PI3K isoforms are also found in patients with ABC DLBCL, remains to be determined. AKT and PDK1 are direct downstream effector kinases of PI3K. Intriguingly, we found that HBL1 and TMD8 cells are insensitive to AKT inhibition, but that viability and MALT1 activity is affected by a selective PDK1 inhibitor. In other human cancer cell lines, oncogenic p110α signaling has been shown to promote transformation independent of AKT, but to require PDK1 (24). Furthermore, PDK1 has been shown to directly recruit PKCθ to CARMA1 in T cells to allow CARMA1 phosphorylation, a crucial step in CBM activation in response to TCR/CD28 costimulation (25). Our data indicate that the PI3K-PDK1 pathway, which is required for costimulation in T cells, also provides a pathological signal in some ABC DLBCL entities.
PI3K inhibition in HBL1 and TMD8 cells affects the NF-κB gene signature and exerts toxic effects resembling the changes seen after inhibition of MALT1 protease activity (8, 9), suggesting that PI3K-PDK1–mediated MALT1 activity is responsible for the observed effects. It has been shown that in T cells, MALT1 protease activity is required for optimal NF-κB activation and induction of target genes, but is dispensable for IKK/NF-κB upstream signaling (6, 7). Thus, the stage of NF-κB activation controlled by PI3K-PDK1-MALT1 remains to be determined. In addition, PI3K signaling acts as a pleiotropic regulator of many cellular processes, and its inactivation is not expected to be restricted to MALT1-triggered NF-κB activation. In line with this, expression of some NF-κB target genes (e.g., JUNB, IL-10) is also reduced in ABC DLBCL cell lines that are less sensitive to PI3K inhibition. In this scenario, PI3K may contribute to gene expression by modulating other pathways, such as mTOR or MAPK signaling (22). Interestingly, constitutively active PI3K p110α*, but not IKK-βCA, rescues survival of BCR-deficient mature B cells, indicating that NF-κB activation alone is not sufficient to substitute for the lack of a BCR signal (26). Even though ABC DLBCLs originate from activated B lymphocytes and display chronic BCR signaling, we found a critical role of PI3K activity in only two CD79B mutated cell lines. In these cells, NF-κB activation is apparently required, but not necessarily sufficient, to mediate PI3K-dependent tumor cell viability.
Clinical trials for cancer therapy have been started with many compounds that target the PI3K signaling pathway (22). We have identified a critical role of PI3K and PDK1 in cell growth and survival of a subset of ABC DLBCL cell lines characterized by CD79B mutations. Our data suggest that PI3K inhibition may be a promising strategy for treating these aggressive lymphomas, and that mutation in CD79B could potentially serve as a molecular marker to predict PI3K dependency. Further studies are needed to confirm this prediction and to develop precise markers to monitor PI3K dependency in ABC DLBCL tumor samples.
Methods
Cell Culture and Reagents.
The ABC DLBCL cell lines used were OCI-Ly10, OCI-Ly3, U2932, HBL1, TMD8, and RIVA. The GCB DLBCL cell lines used were SUDHL-4, SUDHL-6, and BJAB. All DLBCL cell lines except OCI-Ly10 were cultured in RPMI medium 1640 supplemented with 20% FCS, L-glutamine, penicillin, and streptomycin. OCI-Ly10 was cultured in Isocove's modified essential medium with 20% heparinized human plasma, penicillin, streptomycin, and β-mercaptoethanol. LY294002, AKTI VIII (Merck Biosciences), 15e (Alexis Biochemicals), and BX-912 (Axon Medchem) were solved in DMSO. The following antibodies were used: AKT, p-AKT (Ser473), p-AKT (Thr308), PDK1, PI3K p110α, PI3K p110γ, PTEN, BCL-XL (Cell Signaling); PI3K p110β, PI3K p110δ (Millipore); MALT1 (H300, B12), BCL10 (331.3), β-Actin (I-19), anti-p65 (sc-372) (Santa Cruz Biotechnology); FLIPL/S (Alexis Biochemicals); and A20 (eBioscience).
Viability, Apoptosis, and Proliferation Assays.
ABC DLBCL cells were incubated with the different inhibitors in the indicated final concentrations and for the indicated times. Cell viability was quantified by counting cells after trypan blue staining. Cell proliferation rates were determined after CFSE staining (Sigma-Aldrich) at the indicated times by FACS. Apoptosis rates were determined after PE–annexin V and 7AAD staining (BD Pharmingen) at the indicated times by FACS. FACS was performed on an LSRII flow cytometer (BD), and data were analyzed using FlowJo software (Treestar).
MALT1 Protease Activity Assay.
Solvent-treated or inhibitor-treated (for 7 h) DLBCL cells were lysed in immunoprecipitation lysis buffer [50 mM Hepes (pH 7.5), 10% glycerol, 0.1% (vol/vol) Triton X-100, 1 mM DTT, 150 mM NaCl, 2 mM MgCl2, and protease inhibitors]. For the immunoprecipitation, 4 μL of anti-Malt1 antibody (H-300) was added to 400 μL of the precleared lysate. After overnight immunoprecipitation and washing, the beads were resuspended in cleavage assay buffer [50 mM MES (pH 6.8), 150 mM NaCl, 10% (wt/vol) sucrose, 0.1% (wt/vol) CHAPS, 1 M ammonium citrate, 10 mM DTT], and the substrate AC-LRSR-AMC (20 μM) was added. After an initial preincubation, the accumulation of AMC fluorescence was measured for 1 h at 30 °C. Fluorescence of the cleaved substrates was measured using a Synergy 2 Microplate Reader (Biotek). Protease activity is expressed in relative fluorescence units (RFU).
Western Blot Analysis and EMSA.
For Western blot analysis and EMSA, cells were lysed in whole cell lysis buffer [20 mM Hepes (pH 7.9), 350 mM NaCl, 20% glycerin, 1 mM MgCl2, 0.5 mM EDTA, 0.1 EGTA, 1% Nonidet P-40, 0.5 M NaF, 1 M DTT, 1 M β-glycerophosphate, 200 mM Na vanadate, and 25× Protease Inhibitor Mixture (Roche)]. For EMSA, 2 μg of protein extract (determined by the Bradford method) were incubated with a 32P-dATP–labeled, double-stranded NF-κB oligonucleotide probe (5′-CAGGGCTGGGGATTCCCCATCTCCACAGG-3′) and separated on native polyacrylamide gel electrophoresis before autoradiography.
Gene Expression Profiling.
Gene expression profiling for the ABC DLBCL cell lines HBL1 and TMD8 was performed after treatment with DMSO; the PI3K p110α-specific inhibitor 15e for 12 h, 24 h, 36 h, and 48 h; or the IKKβ inhibitor MLN120b (for HBL1 only) for 6 h, 12 h, 24 h, and 48 h. Gene expression was measured using whole-genome 4 × 44K gene expression arrays (Agilent Technologies) following the manufacturer's protocol. Signals from DMSO-treated HBL1 cells (labeled with Cy3) were compared with signals from the respective MLN120b- and 15e-treated cells (labeled with Cy5). A gene was selected as an NF-κB target gene in HBL1 cells if MLN120b treatment decreased the expression of the gene by at least 50% at three time points. In cases of multiple probes per gene, we chose the one that was the most significantly down-regulated under MLN120b treatment (paired t test). We subsequently applied this gene signature to the gene expression data after treatment with PI3K-specific inhibitor 15e. In addition, we applied a previously developed NF-κB target gene signature (NF-κB_all_OCILy3_Ly10 signature: http://lymphochip.nih.gov/cgi-bin/signaturedb/signatureDB_DisplayGenes.cgi?signatureID=83) to the gene expression data, comparing those genes that were significantly inhibited by MLN120b (P < 0.05).
ChIP.
Cells were treated with solvent, 10 μM Ly294002, or 0.4 μM 15e for 24 h. Cross-linking and ChIP were performed according to the recommendations in the Active Motif ChIP-IT Kit. Chromatin shearing was done by sonification, and immunoprecipitation was carried out with control IgG or anti-p65 antibody overnight. After reversal of the crosslink and protease K treatment, chromatin was purified using the QIAquick DNA Purification Kit (Qiagen). Quantification of precipitated IκBα promoter was done by real-time PCR in triplicate using the standard LightCycler protocol (Roche) and LC-480 SybrGreen PCR Mix (Roche). The IκBα promoter primers were as described previously (27). X-fold enrichment of IκBα promoter DNA precipitated with anti-p65 antibody over DNA precipitated by IgG antibody was calculated as the ratio after normalization to input control IκBα promoter DNA.
Supplementary Material
Acknowledgments
We thank Katrin Demski, Kerstin Dietze, and Benjamin Storek for their excellent technical assistance. This work was supported by grants from the Deutsche Krebshilfe (to D.K.) and the German Research Foundation (to G.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008969108/-/DCSupplemental.
References
- 1.Schulze-Luehrmann J, Ghosh S. Antigen-receptor signaling to nuclear factor kappa B. Immunity. 2006;25:701–715. doi: 10.1016/j.immuni.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell–like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 4.Rosenwald A, et al. Lymphoma/Leukemia Molecular Profiling Project The use of molecular profiling to predict survival after chemotherapy for diffuse large B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 5.Coornaert B, et al. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat Immunol. 2008;9:263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- 6.Düwel M, et al. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J Immunol. 2009;182:7718–7728. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- 7.Rebeaud F, et al. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat Immunol. 2008;9:272–281. doi: 10.1038/ni1568. [DOI] [PubMed] [Google Scholar]
- 8.Ferch U, et al. Inhibition of MALT1 protease activity is selectively toxic for activated B cell–like diffuse large B cell lymphoma cells. J Exp Med. 2009;206:2313–2320. doi: 10.1084/jem.20091167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hailfinger S, et al. Essential role of MALT1 protease activity in activated B-cell–like diffuse large B-cell lymphoma. Proc Natl Acad Sci USA. 2009;106:19946–19951. doi: 10.1073/pnas.0907511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 11.Davis RE, et al. Chronic active B-cell receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasselblom S, et al. High immunohistochemical expression of p-AKT predicts inferior survival in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Br J Haematol. 2010;149:560–568. doi: 10.1111/j.1365-2141.2010.08123.x. [DOI] [PubMed] [Google Scholar]
- 13.Uddin S, et al. Role of phosphatidylinositol 3′-kinase/AKT pathway in diffuse large B-cell lymphoma survival. Blood. 2006;108:4178–4186. doi: 10.1182/blood-2006-04-016907. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki H, et al. PI3K and Btk differentially regulate B cell antigen receptor–mediated signal transduction. Nat Immunol. 2003;4:280–286. doi: 10.1038/ni890. [DOI] [PubMed] [Google Scholar]
- 15.Deane JA, Fruman DA. Phosphoinositide 3-kinase: Diverse roles in immune cell activation. Annu Rev Immunol. 2004;22:563–598. doi: 10.1146/annurev.immunol.22.012703.104721. [DOI] [PubMed] [Google Scholar]
- 16.Hayakawa M, et al. Synthesis and biological evaluation of 4-morpholino-2-phenylquinazolines and related derivatives as novel PI3 kinase p110alpha inhibitors. Bioorg Med Chem. 2006;14:6847–6858. doi: 10.1016/j.bmc.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 17.Feldman RI, et al. Novel small molecule inhibitors of 3-phosphoinositide–dependent kinase-1. J Biol Chem. 2005;280:19867–19874. doi: 10.1074/jbc.M501367200. [DOI] [PubMed] [Google Scholar]
- 18.Alessi DR, Cohen P. Mechanism of activation and function of protein kinase B. Curr Opin Genet Dev. 1998;8:55–62. doi: 10.1016/s0959-437x(98)80062-2. [DOI] [PubMed] [Google Scholar]
- 19.Lam LT, et al. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-kappaB pathways in subtypes of diffuse large B-cell lymphoma. Blood. 2008;111:3701–3713. doi: 10.1182/blood-2007-09-111948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greten FR, Karin M. The IKK/NF-kappaB activation pathway: A target for prevention and treatment of cancer. Cancer Lett. 2004;206:193–199. doi: 10.1016/j.canlet.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 21.Gauld SB, Cambier JC. Src-family kinases in B-cell development and signaling. Oncogene. 2004;23:8001–8006. doi: 10.1038/sj.onc.1208075. [DOI] [PubMed] [Google Scholar]
- 22.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 24.Vasudevan KM, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park SG, et al. The kinase PDK1 integrates T cell antigen receptor and CD28 coreceptor signaling to induce NF-kappaB and activate T cells. Nat Immunol. 2009;10:158–166. doi: 10.1038/ni.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srinivasan L, et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.