Abstract
During organogenesis, the final size of mature cell populations depends on their rates of differentiation and expansion. Because transient expression of Neurogenin3 (Neurog3) in progenitor cells in the developing pancreas initiates their differentiation to mature islet cells, we examined the role of Neurog3 in cell cycle control during this process. We found that mitotically active pancreatic progenitor cells in mouse embryos exited the cell cycle after the initiation of Neurog3 expression. Transcriptome analysis demonstrated that the Neurog3-expressing cells dramatically up-regulated the mRNA encoding cyclin-dependent kinase inhibitor 1a (Cdkn1a). In Neurog3 null mice, the islet progenitor cells failed to activate Cdkn1a expression and continued to proliferate, showing that their exit from the cell cycle requires Neurog3. Furthermore, induced transgenic expression of Neurog3 in mouse β-cells in vivo markedly decreased their proliferation, increased Cdkn1a levels, and eventually caused profound hyperglycemia. In contrast, in Cdkn1a null mice, proliferation was incompletely suppressed in the Neurog3-expressing cells. These studies reveal a crucial role for Neurog3 in regulating the cell cycle during the differentiation of islet cells and demonstrate that the subsequent down-regulation of Neurog3 allows the mature islet cell population to expand.
The mature structure of an organ depends on its constituent cell populations and how those populations expand and organize as the organ grows. Within the pancreas, the size of the islets of Langerhans and especially how many insulin-producing β-cells they contain is a critical determinant of pancreatic function and the risk of developing diabetes. The number of islet cells depends on the rate at which new endocrine cells [α-, β-, δ-, ε-, and pancreatic polypeptide (PP) cells] differentiate from progenitors, the size of the progenitor population, and the rates of proliferation of the progenitors and mature endocrine cells. Understanding the mechanisms that control these rates will help explain how distinct cell populations assemble into functional organs.
The coordinated activity of numerous transcription factors regulates the differentiation of the islet cells (1, 2). Among these factors, Neurogenin3 (Neurog3/Neurog3), a member of the basic helix–loop–helix (bHLH) transcription factor family, transiently marks the progenitor cells that will become islet cells and initiates endocrine differentiation during embryonic development, regeneration, and transdifferentiation (3–10).
Although we know that most descendants of Neurog3-expressing cells exit the cell cycle (6, 11), we do not know whether or how Neurog3 might drive cell cycle exit. To address these questions, we used several mouse models with loss- and gain-of-function mutations of Neurog3 and demonstrated that Neurog3 is both necessary and sufficient to promote cellular quiescence in pancreatic progenitors. Furthermore, transcriptome analysis with high time resolution using the Neurog3-Timer mouse model identified the cell cycle inhibitor Cdkn1a (p21/CIP1) as a downstream target of Neurog3 in the endocrine progenitors.
Results
Cell Cycle Exit Follows Neurog3 Induction.
To quantify cell division during endocrine differentiation, we performed triple-labeled immunohistochemistry with antibodies against Neurog3, eGFP, and BrdU at embryonic day (E)15.5 in pancreases from heterozygous mice in which one copy of the coding sequence for Neurog3 was replaced with eGFP (Neurog3eGFP/+ mice; ref. 12) (Fig. 1 A–D). Cells labeled by the anti-Neurog3 antibody but not anti-GFP antibody (Neurog3+/eGFP- cells) represent the newest endocrine progenitors that will shortly become Neurog3+/eGFP+ double-positive cells (13). Subsequently, the Neurog3+/eGFP+ cells lose Neurog3 immunoreactivity but temporarily retain eGFP immunoreactivity (Neurog3-/eGFP+ cells) because of the long half-life of green fluorescent protein (24 h or more; ref. 14) (Fig. 1E).
Fig. 1.
Proliferation in endocrine progenitor cells. After 3 h of BrdU labeling at E15.5, pancreases were dissected from Neurog3eGFP/+ embryos, and immunofluorescence costaining was performed for eGFP (green; A and D), Neurog3 (red; B and D), and BrdU (blue; C and D). Neurog3+/BrdU+/eGFP- and Neurog3+/BrdU+/eGFP+ cells are denoted by yellow arrows and white arrowheads, respectively, and quantified in a bar graph (E). Three embryos were analyzed, and data are presented as the mean ± SEM (**P < 0.01) between two cell populations, by two-tailed Student's t test. (F and G) Dissociated cells from E15.5 and E17.5 Neurog3-Timer embryos were stained with the DNA-specific dye Hoechst 33342 and analyzed by flow cytometry (F). The percentage of proliferating cells at S/G2/M phases at each population is shown in G. At least eight embryos were analyzed from three different litters for each stage, and data are presented as the mean ± SEM. **P < 0.01, E17.5 vs. E15.5, by two-tailed Student's t test.
As outlined at the bottom of Fig. 1E, when Neurog3 expression starts, the first cells are Neurog3+, but eGFP-, because maturation of the eGFP fluorophore takes time. These Neurog3+/eGFP- cells progress to Neurog3+/eGFP+ double-positive cells after eGFP maturation. Then a few hours later when these Neurog3+/eGFP+ cells lose Neurog3 expression, the long half-life eGFP leads to Neurog3-/eGFP+ cells. Three hours after BrdU labeling, 22.0% of Neurog3+/eGFP- cells, 7.2% of Neurog3+/eGFP+ cells, and 1.2% of Neurog3-/eGFP+ cells were labeled with BrdU (Fig. 1E). The differences in BrdU labeling rates in these three populations indicate that cells stop entering S-phase shortly after the initiation of Neurog3 expression.
To confirm the changes in cell cycle during endocrine maturation, embryonic pancreases were dissected from Neurog3-Timer embryos at E15.5 and E17.5, stained with the DNA dye Hoechst 33342, and analyzed by flow cytometry. The Timer (DsRed-E5) fluorescent protein shifts its fluorescence emission peak from green to red over time (15) and, thereby, overcomes the problems caused by the long half-life of GFP (16, 17) and allows the determination of the time because activation of Neurog3 in individual cells based on the ratio of green/red fluorescence (13). As shown in Fig. 1 F and G, at both E15.5 and E17.5, >98% of endocrine progenitors (gates A and B) were in G0/G1 phase and, thus, quiescent. In the mature endocrine cells in gate C, however, a significant increase in cells in S/G2/M demonstrates the reentry of cells into the cell cycle at E17.5, but not at E15.5 (Fig. 1G).
Neurog3 Is Required for Cell Cycle Exit.
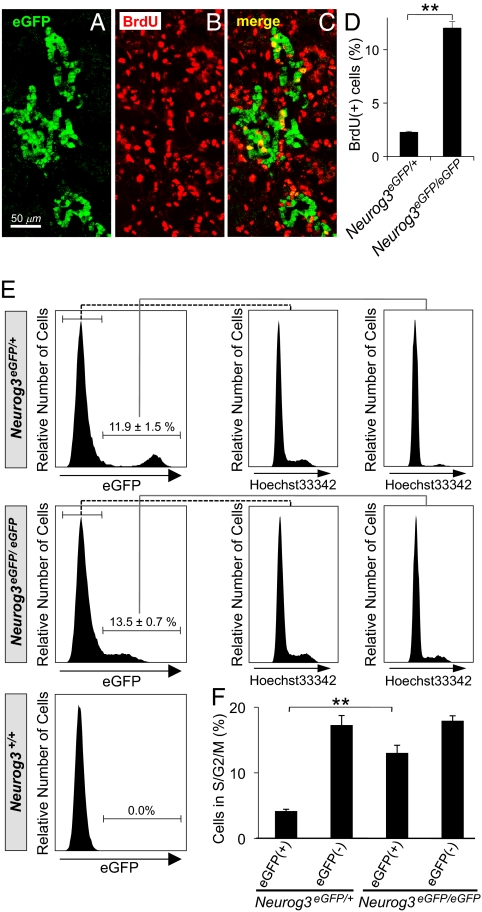
The studies in Fig. 1 correlate cell cycle exit with Neurog3 expression, but do not indicate whether Neurog3 drives the cells to exit the cell cycle, or whether, alternatively, cells already programmed to exit the cell cycle may preferentially activate Neurog3 expression. To address this question, we analyzed Neurog3-deficient mice by histology and flow cytometry. In Neurog3 null embryos (Neurog3eGFP/eGFP), eGFP expression identifies those pancreatic progenitor cells in which Neurog3 expression would have been activated. Immunostaining for eGFP and BrdU demonstrated higher rates of BrdU labeling in the eGFP-expressing cells of Neurog3eGFP/eGFP embryos at E15.5 than in those of Neurog3eGFP/+ heterozygous littermates (Fig. 2 A–D). In addition, the eGFP-expressing cells of Neurog3eGFP/eGFP embryos expand within the epithelium and do not delaminate from the epithelial layer as in the Neurog3eGFP/+ embryos at E15.5 (Fig. S1), although later in development some of these cells can adopt an alternate exocrine fate (18).
Fig. 2.
Proliferation of endocrine lineage cells in Neurog3-deficient embryos. (A–C) After 3 h of BrdU labeling at E15.5, the pancreas was dissected from Neurog3eGFP/eGFP embryos, and immunofluorescence costaining was performed for eGFP (green; A and C) and BrdU (red; B and C). Costaining cells, which appear yellow in the merged panel (C), were quantified, expressed as percentage of total eGFP+ cells, and compared with Neurog3eGFP/+ pancreases in the bar graph in D. In E, pancreatic cells were dissociated from Neurog3eGFP/+, Neurog3eGFP/eGFP, and Neurog3+/+ littermates at E14.5, stained with the DNA dye Hoechst 33342, and analyzed by flow cytometry. The percentage of eGFP+ cells proliferating in S/G2/M phases for each genotype is shown in F. Each data point represents the mean ± SEM of at least three independent experiments. **P < 0.01, Neurog3eGFP/eGFP vs. Neurog3eGFP/+, by two-tailed Student's t test.
In flow cytometric analysis, a significantly higher fraction of eGFP-expressing cells were proliferating (in S/G2/M phases) in Neurog3eGFP/eGFP embryos than in Neurog3eGFP/+ heterozygous littermates (Fig. 2 E and F; 13.0 vs. 4.2%; P < 0.01). Taken together, these data demonstrate that Neurog3 is necessary for the shift to cellular quiescence that occurs when pancreatic progenitors switch to endocrine progenitors and start the process of differentiation into islet cells.
Cell Cycle Regulators Controlled by Neurog3.
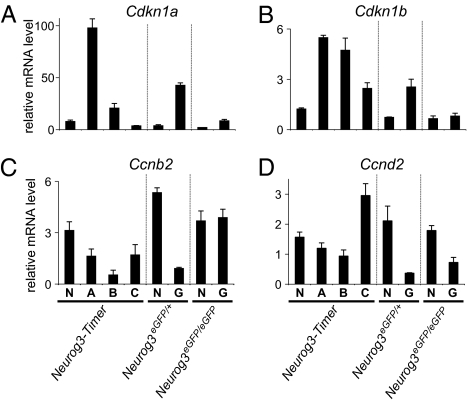
To gauge the expression of known cell cycle regulators in the differentiating endocrine cells, we performed TaqMan RT-PCR arrays with RNA from cells isolated by FACS from E17.5 Neurog3-Timer pancreases. Of the 78 cell cycle-related genes assayed, the mRNA most robustly induced in cells that recently activated Neurog3 (gate A) was Cdkn1a, which encodes the cyclin-dependent kinase inhibitor p21/CIP1 (Fig. 3 and Table S1). Cdkn1a increased 13.4-fold in gate A cells relative to the nonfluorescent cells (gate N) and then decreased again in the more mature endocrine cells in gate B and C (P < 0.001 by ANOVA). Moreover, the absolute expression level of Cdkn1a exceeded all other cell cycle-related mRNAs in gate A cells (Fig. S2). In parallel, other negative regulators of cell cycle progression, such as Cdkn1b (p27/Kip1), Trp53, Rb1, and Rbl1, also peaked in the endocrine progenitors in gate A (Fig. 3B, Fig. S2, and Table S1). In contrast, many positive regulators of cell cycle progression, such as Ccnb2, Ccnd2, Cdk6, and Cdc25c, and a cellular marker of proliferation, Mki67 (Ki67), decreased in the endocrine progenitors (Fig. S2 C–H and Table S1).
Fig. 3.
Expression of cell-cycle regulators in endocrine lineage cells. Pancreases were dissected from Neurog3-Timer embryos at E17.5, and fluorescent cells were sorted by FACS into the four gates shown in Fig. 1C. In parallel, NeurogeGFP/+ and Neurog3eGFP/eGFP embryos were dissected at E14.5, and GFP-positive cells (G) were separated from GFP-negative cells (N) by FACS. The sorted cell populations were analyzed by real-time RT-PCR for mRNA-encoding cell cycle-related genes, Cdkn1a (A), Cdkn1b (B), Ccnb2 (C), and Ccnd2 (D). All expression levels were normalized to glucuronidase, β (Gusb). Each data point represents the mean ± SEM of three independent experiments.
To determine whether these changes in gene expression require Neurog3, we used the same TaqMan arrays to assess gene expression in eGFP-positive and -negative cells sorted from Neurog3eGFP/+ and Neurog3eGFP/eGFP embryos at E14.5. Cdkn1a levels were significantly elevated in eGFP-expressing cells from the Neurog3eGFP/+ embryonic pancreases (P < 0.001; eGFP-expressing cells vs. nonfluorescent cells by Student's t test), but not from the Neurog3eGFP/eGFP pancreases (Fig. 3A and Table S2), indicating that Cdkn1a up-regulation in these cells requires Neurog3. In addition, Cdkn1b and Rb1 were robustly expressed only in endocrine populations of Neurog3eGFP/+, but not Neurog3eGFP/eGFP embryos (Fig. 3B, Fig. S2, and Table S2). The positive cell cycle regulators showed the opposite pattern (Fig. S2 and Table S2). Taken together, these data demonstrate that the changes in expression of the cell cycle regulators seen in the endocrine lineage in the embryonic pancreas require Neurog3.
Ectopic Expression of Neurog3.
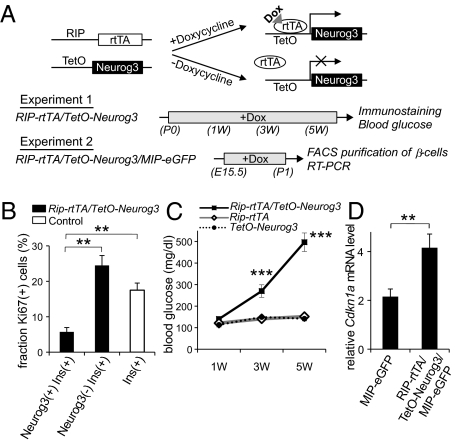
Normally, few Neurog3-expressing cells persist in the pancreas after birth (5, 19). Despite this waning of islet cell neogenesis, during the perinatal period, the β-cell population expands rapidly because of the rapid proliferation of the existing β-cells in both mice and humans (20, 21). During this period, starting from E16 and lasting through the first few weeks of life in mice, proliferation rates can exceed 25% per day (20). We hypothesized that the proliferation of mature endocrine cells requires down-regulation of Neurog3 and that continued expression of Neurog3 in terminally differentiated endocrine cells could inhibit cell cycle progression. To test this hypothesis, we generated a mouse line, Rip-rtTA/TetO-Neurog3 mice, in which the drug doxycycline can be used to induce the expression of Neurog3 in mature β-cells (Fig. 4A). After administration of doxycycline to Rip-rtTA/TetO-Neurog3 mice from birth (P0) to postnatal day 7 (P7), nuclear Neurog3 protein was detected in 71% of insulin-expressing cells, but was not detected in control littermates (Fig. S3). As assessed by Ki67 staining, the Neurog3-expressing β-cells proliferated at a significantly lower rate than the Neurog3-negative β-cells at P7 (Fig. 4B), indicating that exogenous expression of Neurog3 inhibited cell proliferation in β-cells. When doxycycline treatment of Rip-rtTA/TetO-Neurog3 neonatal mice was extended to 5 wk of age, blood glucose levels dramatically increased over time (Fig. 4C). At the age of 6 wk, the islets of the doxycycline-treated Rip-rtTA/TetO-Neurog3 mice contained markedly reduced numbers of insulin-expressing cells, and the serum insulin in the double mutant mice was below the level of detection by ELISA (Fig. S4).
Fig. 4.
Ectopic expression of Neurog3 in mature β-cells. As outlined in A, transgenic mice were generated with the tetracycline-regulated promoter driving Neurog3 expression (TetO-Neurog3 mice) and crossed with mice expressing the reverse-tetracycline receptor under the control of the rat insulin 2 promoter (RIP-rTtA mice; ref. 35) to generate Rip-rtTA/TetO-Neurog3 double transgenic mice, and these mice were crossed with mice expressing eGFP under the control of the mouse insulin 1 promoter (MIP-eGFP mice; ref. 35) to generate Rip-rtTA/TetO-Neurog3/MIP-GFP triple transgenic mice. In experiment 1, Rip-rtTA/TetO-Neurog3 double transgenic mice and controls (Rip-rtTA and TetO-Neurog3) received doxycycline from postnatal day 0 to 6 wk. The pancreases of some animals were harvested after 1 wk and stained for insulin, Neurog3, and the proliferation marker Ki67 (B). For the remaining animals, random blood glucose levels were checked every two weeks (C), and the pancreases were harvested at 6 wk for immunohistochemistry (Fig. S4). In experiment 2, Rip-rtTA/TetO-Neurog3/MIP-eGFP triple transgenic mice and MIP-eGFP controls (Rip-rtTA/MIP-eGFP and TetO-Neurog3/MIP-eGFP) received doxycycline from E15 for 5 d, and then on postnatal day 0, pancreases were harvested and sorted by FACS, and the levels of Cdkn1 AMRNA were analyzed by real-time RT-PCR (D). Each data point represents the mean ± SEM of at least three independent experiments. **P < 0.01, Neurog3+/Insulin+ vs. Neurog3-/Insulin+ cells (B), and MIP-eGFP controls vs. Rip-rtTA/TetO-Neurog3/MIP-eGFP mice (D), by two-tailed Student's t test. ***P < 0.001 Rip-rtTA/TetO-Neurog3 vs. control littermates by an analysis of variance (ANOVA).
To quantify the expression of cell-cycle regulators in the cells expressing transgenic Neurog3, we purified pancreatic β-cells by FACS from Rip-rtTA/TetO-Neurog3/MIP-eGFP triple transgenic mice after doxycycline treatment for 5 d between E15.5 and P1 (22) (Fig. 4A, Experiment 2). Measurement of mRNA levels by TaqMan RT-PCR demonstrated that the induction of transgenic Neurog3 increased the expression of Cdkn1a mRNA in the transgenic β-cells relative to control littermates (Fig. 4D). Taken together, this gain-of-function model revealed that persistent expression of Neurog3 up-regulates Cdkn1a expression, blocks β-cell proliferation, and results in severe hyperglycemia, thus demonstrating the critical importance of Neurog3 removal for the proper expansion of β-cell mass, especially during the critical perinatal period.
Role of Cdkn1a in Cell Cycle Exit.
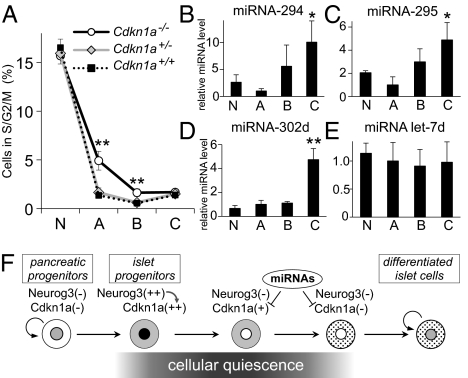
Having shown that Neurog3 induces the expression of the cell-cycle inhibitor Cdkn1, we next tested the importance of Cdkn1 in the cell-cycle exit driven by Neurog3. We generated Cdkn1a-deficient Neurog3-Timer reporter embryos (Neurog3-Timer/Cdkn1a−/−) (23) and quantified proliferating cells by flow cytometry with Hoechst 33342 staining. As shown in Fig. 5A, the number of proliferating cells in S/G2/M phases is significantly higher in the endocrine progenitors (gate A) of E15.5 Cdkn1a−/− embryos (5.00 ± 0.78% in gate A), compared with Cdkn1a+/− heterozygous and Cdkn1a+/+ wild-type embryos (1.65 ± 0.46 and 1.35 ± 0.26% in gate A, respectively). In the Cdkn1a−/− embryos, a higher fraction of the cells in gate B also were detected in S/G2/M phase, but the more differentiated cells in gate C showed no significant difference in proliferation rates between Cdkn1a−/− and wild-type embryos. These data demonstrate that Cdkn1a plays an essential role in promoting cell cycle exit in endocrine progenitors downstream of Neurog3.
Fig. 5.
Function and regulation of Cdkn1a in the pancreas. Pancreatic cells from Neurog3-Timer embryos with the genotypes shown were dissociated at E15.5, stained with the DNA dye Hoechst 33342, and analyzed by flow cytometry. The percentage of cells proliferating in S/G2/M phases is shown in A for each of the four gates from Fig. 1C. Data are presented as the mean ± SEM of at least five independent experiments. **P < 0.01, Cdkn1a−/− knockout vs. Cdkn1a+/− heterozygous or Cdkn1a+/+ wild-type embryos, by an analysis of variance (ANOVA). In B–E, dissociated cells from pancreases from Neurog3-Timer embryos at E17.5 were sorted by FACS into the four gates shown in Fig. 1C. Expression levels of miRNA-294 (B), miRNA-295 (C), and miRNA-302d (D), and control miRNA Let7d were determined by TaqMan miRNA assay, normalized to snoRNA-202, and shown relative to the level in gate A. Each data point represents the mean ± SEM of at least three independent experiments. *P < 0.05, **P < 0.01 vs. gate A, by two-tailed Student's t test. F shows a proposed model for cell cycle regulation in the pancreatic endocrine lineage.
Neurog3 Directly Binds to the Regulatory Region of the Cdkn1a Gene.
To investigate whether Neurog3 directly regulates transcription of the Cdkn1a gene in pancreatic cells, chromatin immunoprecipitation (ChIP) assay was performed in mouse pancreatic mPAC cells transfected with FLAG-tagged Neurog3 (Fig. S5). Within regulatory sequences upstream of the first exon of Cdkn1a, a 0.5-kb fragment between −2227 bp and −1691 bp containing an E-box was immunoprecipitated by the anti–FLAG-tag antibody. A highly conserved E-box was also identified at −39 Kb, and ChIP confirmed binding of Neurog3 to this site in mPAC cells. Taken together, these data show that Neurog3 directly controls the expression of the Cdkn1a gene by binding to upstream regulatory regions of the gene.
MicroRNAs in the Cell Cycle Control.
MicroRNAs miRNA-291a-3p, miRNA-291b-3p, miRNA-294, miRNA-295, and miRNA-302d promote the G1-S transition by directly targeting the 3′ UTR of the Cdkn1a mRNA and inhibiting translation (24). We asked whether these miRNAs might provide further regulation of Cdkn1a levels in differentiating endocrine cells in addition to the fall in Cdkn1a mRNA levels that occurs as new endocrine progenitor cells (gate A) differentiate (gates B and C) and reenter the cell cycle at E17.5 (Fig. 3A). Therefore, we measured the expression levels of these miRNAs in cells purified by FACS from pancreases of E17.5 Neurog3-Timer embryos and found that three of the miRNAs that target Cdkn1a, miRNA-294, miRNA-295, and miRNA-302d, were significantly enriched in differentiated endocrine cells (Fig. 5 B–E). Because the increase in proliferating rate (Fig. 1G) parallels the increase of these miRNAs in mature endocrine cells in area C (Fig. 5 B–D), these miRNAs may further inhibit the expression of Cdkn1a at the level of translation and, thereby, contribute to the reactivation of the cell cycle in mature endocrine cells (Fig. 5F).
Discussion
In these studies, several lines of evidence demonstrate that multipotent pancreatic progenitor cells exit the cell cycle when they become endocrine progenitor cells, and Neurog3 drives this exit. Although prior studies demonstrated that endocrine cells exit the cell cycle in the developing pancreas before E16 (6, 19), we used a combination of time-restricted BrdU labeling, Neurog3 staining, and transgene markers with different temporal expression patterns to demonstrate that Neurog3 expression initiates in rapidly cycling pancreatic progenitor cells, but these cells quickly exit the cell cycle while still expressing Neurog3. These cells initially remain quiescent after Neurog3 expression wanes, but late in fetal development, they renter the cell cycle.
The timing of cell cycle exit shortly after the initiation and during the peak of Neurog3 expression suggests that Neurog3 itself may force the endocrine progenitor cells out of the cell cycle, and the results with Neurog3-deficient embryos support this model. The eGFP-expressing cells in Neurog3eGFP/eGFP knockout embryos are programmed to transcribe the Neurog3 gene but never produce Neurog3 protein. In the Neurog3eGFP/+ heterozygous embryos, the GFP+ cells exit the cell cycle, but in the Neurog3eGFP/eGFP embryos that lack Neurog3 protein, these progenitor cells continue to proliferate within epithelium, demonstrating that cell cycle exit requires Neurog3 (Fig. 2 and Fig. S1).
In addition, data from the Rip-rtTA/TetO-Neurog3 mouse model demonstrate that Neurog3 expression alone is sufficient to drive endocrine cells to exit the cell cycle. Normally, endocrine progenitor cells only express Neurog3 briefly and rapidly reduce its expression before mature endocrine gene expression commences (4–7). Our data demonstrate the critical importance of this down-regulation because it permits the mature endocrine cells to reenter the cell cycle.
Transcriptome analysis for cell cycle-related genes in the Neurog3-expressing endocrine progenitor cells revealed sharp increases of several negative regulators of cell cycle progression (Fig. 3). Among these genes, Cdkn1a exhibited both the largest increase relative to the non-Neurog3–expressing cells, and the highest absolute expression level of any of the cell cycle regulators assessed in the endocrine progenitors. This up-regulation of Cdkn1a depended completely on Neurog3, because no increase was detected in the eGFP-expressing cells sorted from Neurog3eGFP/eGFP embryos. In addition, expression of Neurog3 in mature β-cell in the Rip-rtTA/TetO-Neurog3 transgenic mice induced the expression of Cdkn1a, but not the other negative regulators of cell cycle progression (Fig. S6).
These results strongly suggest that Cdkn1a functions downstream of Neurog3. Cdkn1a can function either as a downstream target of p53 (encoded by Trp53 in mice) or independently of p53 to inhibit cell cycle progression (23, 25). Because the up-regulation of Cdkn1a in the Rip-rtTA/TetO-Neurog3 transgenic mice did not correlate with the expression level of p53, we conclude that Neurog3 may induce Cdkn1a expression independently of p53.
Neurog3 activates the expression of both Cdkn1a and key factors required for endocrine differentiation, such as Pax4, Nkx2.2, NeuroD1, and Rfx6 (26–28), which suggests that cell cycle exit may play a role in differentiation. We saw small increases in some of the mRNA encoding islet hormones in the Cdkn1a−/− pancreases (Fig. S7), but these changes could simply result from the increase in proliferation of the progenitors. Expression levels of transcriptions factors Neurod1, Pax4, and Rfx6 did not change in the Cdkn1a−/− pancreases (Fig. S7), suggesting that the lack of Cdkn1a does not have a large impact on differentiation. However, the release of the block in proliferation was not complete in the Cdkn1a−/− progenitor cells, probably because other cell cycle inhibitors such as Cdkn1b remain, and this redundancy may prevent any deficiency in differentiation.
During normal development in both rodents and humans, β-cell proliferation increases dramatically during the perinatal period (21, 29), establishing the size of the β-cell population and the risk of diabetes. Our data demonstrate that without normal down-regulation of Neurog3, the β-cell population does not adequately expand and diabetes ensues. Interestingly, however, a very low level of Neurog3 expression persists in adult islets (30–33). Recent studies demonstrated that this sustained low-level expression of Neurog3 contributes to endocrine function (33). Considering these data together, we conclude that sustained, but tightly regulated, expression of Neurog3 at a very low level permits the proper expansion of endocrine cells as well as their normal function. Ultimately, this balance plays a critical role in determining the size of the β-cell population, the functional structure of the mature pancreas, and the risk of diabetes.
Experimental Procedures
Animal Experiments.
Mice were housed on a 12-h light–dark cycle in a controlled climate in the University of San Francisco vivarium. Timed matings were carried out with E0.5 being set as midday of the day of discovery of a vaginal plug. The UCSF Institutional Animal Care and Use Committee approved all studies involving mice.
The generation of the Neurog3-Timer mice was described (13). Neurog3eGFP+ knock-in mice (12) were obtained through the Mutant Mouse Regional Resource Centers (University of California, Davis, CA). The TetO-Neurog3 transgenic mouse line was generated by standard methods using DNA generated by inserting the mouse Neurog3 cDNA into pTRE2 (Clontech). Rip-rtTA (29) and Cdkn1a−/− (23) mice were purchased from the Jackson Laboratory. Rip-rtTA/TetO-Neurog3 double transgenic mice and their littermates were administered chow with added doxycycline or standard chow. MIP-eGFP mice were obtained from M. Hara (University of Chicago) (22).
Immunostaining.
Tissues were fixed in 4% paraformaldehyde in PBS at 4 °C, washed in PBS, immersed in sucrose solution, and embedded and frozen in Tissue-Tek (OCT Compound; Sakura). Sections were stained with antibodies against GFP (rabbit, 1:500; MBL International), BrdU (rat, 1:200, Serotec), Ki67 (mouse, 1:50; BD Pharmingen), Neurog3 (guinea pig, 1:1,000; made in our laboratory), insulin (guinea pig, 1:2,000; Millipore), glucagon (rabbit, 1:2,000; Millipore), E-cadherin (rat, 1:200; Abcam), laminin (rabbit, 1:1,000; Sigma), and cleaved caspase-3 (rabbit, 1:200; Cell Signaling). For BrdU experiments, BrdU (Sigma; 100 mg/kg of body weight) was injected into pregnant mice 3 h before killing, and tissues were pretreated with 1 M HCl for 30 min at room temperature before immunofluorescence. Slides were imaged on Leica SP2 AOBS confocal laser scanning microscope.
Pancreatic Cell Dispersion and Flow Cytometry.
After manual dissection, pancreases were treated with 0.05% trypsin/0.53 mM EDTA (Invitrogen) at 37 °C for 5 min, and the digestion was inactivated by addition of FBS. For cell cycle analysis, the dissociated cells were incubated with medium containing 5 μg/mL Hoechst 33342 dye at 37 °C for 60 min, washed with cold PBS, then analyzed by using an LSRII flow cytometer (PerkinElmer). For sorting the fluorescent cells, a MoFlo cell sorter was used (Dako Cytomation). Dead cells were excluded with DNA dye TO-PRO-3 (Molecular Probes).
Real-Time PCR.
Total RNA was extracted from FACS-sorted cells, linearly amplified, and converted into cDNA with the NuGEN WT-Ovation RNA Amplification system (NuGen) as described (13). Individual cDNAs were quantified by real-time PCR using a TaqMan Low Density array system (Applied Biosystems), designed for cell cycle-related genes (Table S1 and Table S2). Gene expression levels of the assayed genes were normalized to the expression levels of glucuronidase, β (Gusb). The mean values and SE are shown in Table S1 and Table S2. For quantitative miRNA assays, extracted RNA was converted into cDNA, amplified, and quantified with TaqMan miRNA assays according to the manufacturer's protocol (Applied Biosystems). The expression levels of miRNAs were normalized to snoRNA202, and the relative level of miRNAs from each gate was determined with respect to the value in nonfluorescent cells (gate A).
Chromatin Immunoprecipitation (ChIP).
ChIP assays were performed as described (34) Mouse pancreatic mPAC cells were transiently transfected with plasmid vectors expressing eGFP or FLAG-tagged human NEUROG3 by using Lipofectamine PLUS reagent (Invitrogen). Cells were harvested 48 h after transfection. Proteins were cross-linked to DNA with 1% formaldehyde for 10 min at 37 °C. The cells were washed and lysed with SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris at pH 8.1). Chromatin complexes were immunoprecipitated overnight with 5 μg of preimmune serum or antibody specific for FLAG at 4 °C, and pulled down by using protein A magnetic beads (Invitrogen). Putative binding regions were amplified from precipitated DNA fragments by PCR. The Neurog3 binding site in Pax4 gene was used as a positive control (27), and the Tph1 gene promoter was used as a negative control. PCR primer sequences are available upon request.
Supplementary Material
Acknowledgments
We thank Gerold Grodsky, Francis Lynn, Chester Chamberlain and members of the German laboratory for helpful discussions; Yi Zhang and Shuhong Zhao for assistance with mouse husbandry and genotyping; and M. Hara (University of Chicago, Chicago) for generously providing MIP-eGFP mice. This work was supported by grants from the Larry L. Hillblom Foundation (to M.S.G.), the Juvenile Diabetes Research Foundation (to T.M. and M.S.G.), the Nora Eccles Treadwell Foundation (to M.S.G.), the American Diabetes Association (M.S.G.), and the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (to M.S.G.).
Footnotes
Conflict of interest statement: M.S.G. is an inventor on patents held by the University of California covering Neurog3 and its use.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004842108/-/DCSupplemental.
References
- 1.Zaret KS. Genetic programming of liver and pancreas progenitors: Lessons for stem-cell differentiation. Nat Rev Genet. 2008;9:329–340. doi: 10.1038/nrg2318. [DOI] [PubMed] [Google Scholar]
- 2.Oliver-Krasinski JM, Stoffers DA. On the origin of the beta cell. Genes Dev. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apelqvist A, et al. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 4.Schwitzgebel VM, et al. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- 5.Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci USA. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen J, et al. Independent development of pancreatic alpha- and beta-cells from neurogenin3-expressing precursors: A role for the notch pathway in repression of premature differentiation. Diabetes. 2000;49:163–176. doi: 10.2337/diabetes.49.2.163. [DOI] [PubMed] [Google Scholar]
- 7.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 8.Gasa R, et al. Proendocrine genes coordinate the pancreatic islet differentiation program in vitro. Proc Natl Acad Sci USA. 2004;101:13245–13250. doi: 10.1073/pnas.0405301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X, et al. Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell. 2008;132:197–207. doi: 10.1016/j.cell.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desgraz R, Herrera PL. Pancreatic neurogenin 3-expressing cells are unipotent islet precursors. Development. 2009;136:3567–3574. doi: 10.1242/dev.039214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee CS, Perreault N, Brestelli JE, Kaestner KH. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 2002;16:1488–1497. doi: 10.1101/gad.985002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyatsuka T, Li Z, German MS. Chronology of islet differentiation revealed by temporal cell labeling. Diabetes. 2009;58:1863–1868. doi: 10.2337/db09-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12:1035–1040. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- 15.Terskikh A, et al. “Fluorescent timer”: Protein that changes color with time. Science. 2000;290:1585–1588. doi: 10.1126/science.290.5496.1585. [DOI] [PubMed] [Google Scholar]
- 16.Davis I, Girdham CH, O'Farrell PH. A nuclear GFP that marks nuclei in living Drosophila embryos; maternal supply overcomes a delay in the appearance of zygotic fluorescence. Dev Biol. 1995;170:726–729. doi: 10.1006/dbio.1995.1251. [DOI] [PubMed] [Google Scholar]
- 17.Wu YL, et al. Development of a heat shock inducible gfp transgenic zebrafish line by using the zebrafish hsp27 promoter. Gene. 2008;408:85–94. doi: 10.1016/j.gene.2007.10.027. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, et al. Neurog3 gene dosage regulates allocation of endocrine and exocrine cell fates in the developing mouse pancreas. Dev Biol. 2010;339:26–37. doi: 10.1016/j.ydbio.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sander M, et al. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- 20.Teta M, Long SY, Wartschow LM, Rankin MM, Kushner JA. Very slow turnover of beta-cells in aged adult mice. Diabetes. 2005;54:2557–2567. doi: 10.2337/diabetes.54.9.2557. [DOI] [PubMed] [Google Scholar]
- 21.Meier JJ, et al. Beta-cell replication is the primary mechanism subserving the postnatal expansion of beta-cell mass in humans. Diabetes. 2008;57:1584–1594. doi: 10.2337/db07-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara M, et al. Transgenic mice with green fluorescent protein-labeled pancreatic beta -cells. Am J Physiol Endocrinol Metab. 2003;284:E177–E183. doi: 10.1152/ajpendo.00321.2002. [DOI] [PubMed] [Google Scholar]
- 23.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbas T, Dutta A. p21 in cancer: Intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith SB, et al. Rfx6 directs islet formation and insulin production in mice and humans. Nature. 2010;463:775–780. doi: 10.1038/nature08748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith SB, et al. Neurogenin3 and hepatic nuclear factor 1 cooperate in activating pancreatic expression of Pax4. J Biol Chem. 2003;278:38254–38259. doi: 10.1074/jbc.M302229200. [DOI] [PubMed] [Google Scholar]
- 28.Watada H, Scheel DW, Leung J, German MS. Distinct gene expression programs function in progenitor and mature islet cells. J Biol Chem. 2003;278:17130–17140. doi: 10.1074/jbc.M213196200. [DOI] [PubMed] [Google Scholar]
- 29.Finegood DT, Scaglia L, Bonner-Weir S. Dynamics of beta-cell mass in the growing rat pancreas. Estimation with a simple mathematical model. Diabetes. 1995;44:249–256. doi: 10.2337/diab.44.3.249. [DOI] [PubMed] [Google Scholar]
- 30.Huang HP, et al. Regulation of the pancreatic islet-specific gene BETA2 (neuroD) by neurogenin 3. Mol Cell Biol. 2000;20:3292–3307. doi: 10.1128/mcb.20.9.3292-3307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dror V, et al. Notch signalling suppresses apoptosis in adult human and mouse pancreatic islet cells. Diabetologia. 2007;50:2504–2515. doi: 10.1007/s00125-007-0835-5. [DOI] [PubMed] [Google Scholar]
- 32.Wilson ME, et al. The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes. 2005;54:3402–3409. doi: 10.2337/diabetes.54.12.3402. [DOI] [PubMed] [Google Scholar]
- 33.Wang S, et al. Sustained Neurog3 expression in hormone-expressing islet cells is required for endocrine maturation and function. Proc Natl Acad Sci USA. 2009;106:9715–9720. doi: 10.1073/pnas.0904247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynn FC, et al. Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc Natl Acad Sci USA. 2007;104:10500–10505. doi: 10.1073/pnas.0704054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milo-Landesman D, et al. Correction of hyperglycemia in diabetic mice transplanted with reversibly immortalized pancreatic beta cells controlled by the tet-on regulatory system. Cell Transplant. 2001;10:645–650. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.