Abstract
The majority of protein architectures exhibit elements of structural symmetry, and “gene duplication and fusion” is the evolutionary mechanism generally hypothesized to be responsible for their emergence from simple peptide motifs. Despite the central importance of the gene duplication and fusion hypothesis, experimental support for a plausible evolutionary pathway for a specific protein architecture has yet to be effectively demonstrated. To address this question, a unique “top-down symmetric deconstruction” strategy was utilized to successfully identify a simple peptide motif capable of recapitulating, via gene duplication and fusion processes, a symmetric protein architecture (the threefold symmetric β-trefoil fold). The folding properties of intermediary forms in this deconstruction agree precisely with a previously proposed “conserved architecture” model for symmetric protein evolution. Furthermore, a route through foldable sequence-space between the simple peptide motif and extant protein fold is demonstrated. These results provide compelling experimental support for a plausible evolutionary pathway of symmetric protein architecture via gene duplication and fusion processes.
Keywords: protein design, protein folding, protein symmetry
Symmetry is a central theme in protein structure, function, and evolution. Structural symmetry is observed in many different protein architectures, and gene duplication and fusion is the generally hypothesized mechanism for the emergence of symmetric architecture from simpler (i.e., archaic) peptide motifs (1–4). Such motifs, believed to represent the translational product of “genes of primordial life” (5), typically code for polypeptides of ∼40–60 residues in length (5, 6) and may define elemental folding units (i.e., “foldons”) (7). Two distinctly different evolutionary models for the emergence of symmetric protein architecture from a primordial peptide motif have been proposed (8–13). In the “emergent architecture” model, the primordial peptide autonomously folds to yield a simple architecture and the complex symmetric architecture emerges upon a final gene duplication and fusion event (Fig. 1A). In the conserved architecture model, the symmetric architecture (or integral units thereof) is present at each step of the evolutionary process (Fig. 1B).
Fig. 1.
Evolutionary models of symmetry protein architecture (e.g., the β-trefoil fold). (A) The emergent architecture model (8). The archaic peptide motif is autonomously folding, yielding simple single-domain architecture. Intermediate forms produced by gene duplication and fusion events have unique folds with increasing structural symmetry and complexity. The symmetric target architecture emerges upon the final gene duplication and fusion event. (B) The conserved architecture model (9). The archaic peptide is not an autonomously folding single-domain architecture; instead, it oligomerizes to yield the symmetric target architecture (e.g., β-trefoil fold). Intermediate forms produced by gene duplication and fusion events similarly oligomerize and reconstitute integral units of the symmetric target architecture. A final duplication and fusion event encodes the symmetric target architecture within a single polypeptide chain.
Experimental studies to probe the evolutionary pathway responsible for symmetric protein architecture have focused principally upon the behavior of subdomain fragments of extant symmetric proteins and have asked whether such fragments fold independently or oligomerize. A fragmentation study of the twofold symmetric aspartate racemase enzyme identified an independently folding monomeric subdomain, leading the authors to propose an emergent-architecture-type evolutionary model (10). Fragmentation studies of a fivefold symmetric “β-propeller” protein to identify an ancient folding motif showed evidence of multimeric assembly for ∼100 amino acid fragments comprising two copies of the repeating motif, but not for ∼50 amino acid single-motif fragments (11). Subsequent X-ray structure analysis showed that these duplicated fragments assembled to create two intact β-propeller pentamers, with each pentamer containing 2.5 dimeric domains (with one dimer spanning both pentamers) (14), supporting a conserved-architecture-type model. In the (βα)8-barrel architecture, studies of (βα)4 half-domain fragments showed that these subdomains assembled to form an intact (βα)8-barrel but could also exist as independently folded subdomains (15, 16). Computational and experimental studies designed to identify a corresponding (βα)2 “building block” identified a peptide capable of tetrameric oligomerization to create an intact (βα)8-barrel but could also exist as an independently folded subdomain (13), thus supporting both types of evolutionary models. However, such simple fragmentation studies of symmetric proteins have suspected limitations. Evolutionary divergence following a duplication/fusion event likely results in optimization of the unique interdomain interface, such that independent folding of fragments is unlikely; thus, solubility and folding properties of fragments of extant proteins are unlikely to recapitulate those of the archaic peptide motif (11).
In this report, we describe an experimental top-down symmetric deconstruction of symmetric protein architecture (the β-trefoil fold) with the goal of testing two competing hypotheses for its evolution from an archaic peptide motif. The starting point for this deconstruction is human fibroblast growth factor-1 (FGF-1), a 140 amino acid single-domain globular protein exhibiting the characteristic threefold symmetry of the β-trefoil architecture (where each ∼45 amino acid repeating subdomain is termed a trefoil-fold; ref. 17). As is commonly observed with symmetric proteins, the discernible tertiary structure symmetry of FGF-1 is substantially imperfect at both the primary and tertiary structure levels (Figs. 2B and 3A). The symmetric deconstruction involved sequential introduction of symmetric mutations (targeting core, reverse-turn, and β-strand secondary structure, respectively) until a purely threefold symmetric primary structure solution was achieved. This symmetric primary structure defines a repeating trefoil-fold polypeptide motif, whose folding and structural properties were studied along with dimer and monomer peptide fragments. The structure and folding properties for these polypeptides agree precisely with a previously hypothesized conserved architecture model of evolution; furthermore, each intermediate mutant in the symmetric deconstruction resides within foldable sequence space, providing thermodynamic support for the hypothesized evolutionary pathway.
Fig. 2.
Summary of the top-down symmetric deconstruction strategy and the result of its application to FGF-1. (A) The cumulative symmetric transforms applied to achieve a symmetric deconstruction of the β-trefoil architecture (beginning with FGF-1 and ending in the Monofoil and Difoil polypeptides). Details for the set of intermediary mutants comprising the deconstruction are provided in Table S1. (B) Primary structures of the FGF-1 starting protein and the resulting Symfoil-4P symmetric deconstruction. Positions of exact threefold primary structure symmetry are indicated in yellow. The Monofoil-4P and Difoil-4P polypeptides were generated by the introduction of stop codons at positions 53 and 94 of the Symfoil-4P protein, respectively.
Fig. 3.
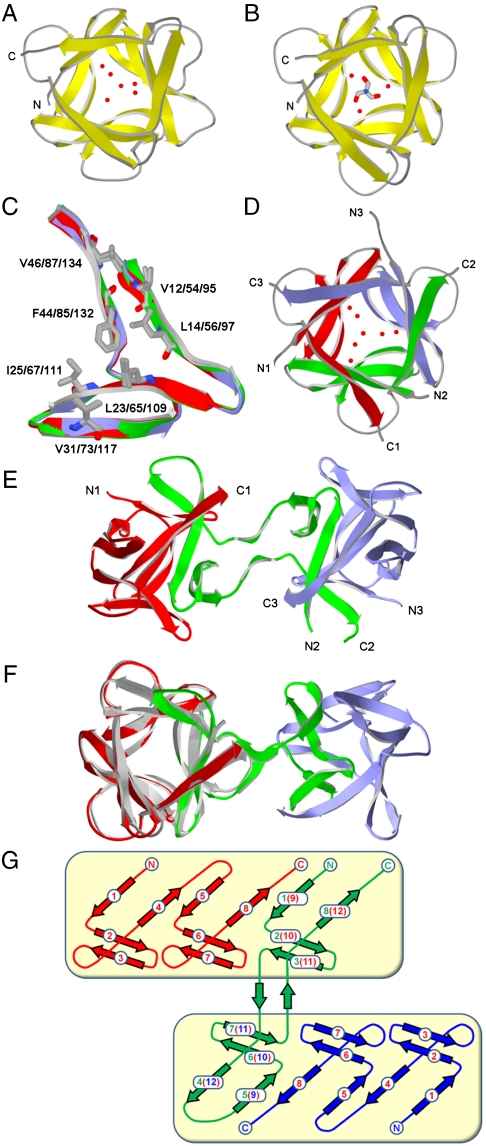
X-ray structures of FGF-1, Symfoil-1, Monofoil-4P, and Difoil-4P proteins. (A) Ribbon representation of FGF-1 oriented down the threefold axis of symmetry and including select solvent structure. (B) Similar representation of the Symfoil-1 mutant (also showing the location of a bound Tris molecule). (C) Overlay of the repeating trefoil-fold subdomains of Symfoil-1. The main-chain atoms (ribbon representation) are colored red, green, and blue for subdomains 1, 2, and 3, respectively. The set of 21 hydrophobic core residues (Corey, Pauling, Koltun coloring) are in shown in wireframe representation. (D) The Monofoil-4P structure with select solvent; the individual Monofoil-4P peptides are colored red, green, and blue and their respective N and C termini are indicated. (E) The Difoil-4P structure (individual polypeptides colored as in D) and with respective N and C termini indicated. The view is down a twofold axis of symmetry relating the two intact β-trefoil folds present in the homotrimer Difoil-4P structure. (F) Fig. 3E rotated to view down the threefold axis of symmetry within the first β-trefoil domain and overlaid with the Symfoil-4P structure (gray). (G) Secondary structure schematic diagram of Difoil-4P (colored as in E). The boxed regions indicate the two β-trefoil domains.
Results
Top-Down Symmetric Deconstruction.
A total of 18 mutant proteins were constructed and characterized in the process of successfully introducing a complete threefold symmetric primary structure constraint (involving 76 substitutions and 4 deleted positions total; Fig. 2 and Table S1). Detailed biophysical properties for each of these mutants, as well as details of unsuccessful mutations (principally resulting in poor folding cooperativity, solubility, or thermostability) will be reported elsewhere. The initial symmetric protein produced by the deconstruction is termed “Symfoil-1” (for symmetric β-trefoil protein 1). The Symfoil-1 protein has several notable properties in comparison to FGF-1. Symfoil-1 is more thermostable than FGF-1, and whereas FGF-1 irreversibly aggregates upon thermal denaturation, Symfoil-1 exhibits reversible two-state unfolding (Fig. S1). FGF-1 contains examples of all 20 common amino acids, whereas Symfoil-1 is devoid of Trp, Ala, Cys, and Met residues. Additionally, Symfoil-1 has no known FGF-1 functional properties (including FGF receptor-binding, heparin-binding, nuclear localization signal, mitogenic activity, etc.). Symfoil-1 is able to bind the C3 symmetric molecule Tris on a threefold axis of structural symmetry (Fig. S2). The Symfoil-1 synthetic protein was subsequently optimized by mutation to yield a hyperthermophilic variant (Symfoil-4P mutant, Table S1). Monomeric and dimeric versions of the repeating peptide motif within Symfoil-4P were subsequently created by the introduction of stop codons at the appropriate locations, producing the Monofoil-4P (for monomer trefoil fold) and Difoil-4P (for dimer trefoil fold) polypeptides, respectively (Fig. 2B).
Properties of the Monofoil-4P, Difoil-4P, and Symfoil-4P Polypeptides.
Both the Monofoil-4P and Difoil-4P polypeptides resolved as single peaks on calibrated size-exclusion chromatography, with apparent masses indicating a homotrimer (in both cases) and no detectable monomer (Fig. S3). Monofoil-4P and Difoil-4P proteins sedimented as homogeneous forms with s20,w values of 2.31 ± 0.036 s and 3.18 ± 0.021 s, respectively, and with corresponding molecular mass of 18.8 and 29.8 kDa, respectively. The molecular mass of the Monofoil-4P (6.6 kDa) and Difoil-4P (11.2 kDa) polypeptides therefore indicate homotrimer oligomerization for both polypeptides in solution. X-ray crystal structures were solved for Symfoil-1, 2, 4T, 4V, and 4P proteins as well as the Monofoil-4P and Difoil-4P polypeptides (Table S2). The peptide backbone of the Symfoil-1 protein exhibits a striking threefold rotational symmetry (Fig. 3B). The main-chain atoms of the three individual trefoil-fold subdomains of the Symfoil-1 protein overlay each other with an rms deviation of 0.2 Å, indicating essentially indistinguishable trefoil-fold subdomain structures. Furthermore, the side-chain rotamers for the set of core-packing residues (and essentially all surface residues with the exception of specific crystal contacts) are identical for a comparison of the three trefoil-fold subdomains (Fig. 3C); thus, the threefold symmetry of the Symfoil protein(s) appears exact. The Monofoil-4P structure exhibits a homotrimer assembly that recapitulates the intact β-trefoil architecture (Fig. 3D). An overlay of all main-chain atoms comprising the individual repeating trefoil-fold subdomains of the Monofoil-4P trimer with the Symfoil-4P structure yields an rms deviation of 0.5 Å, indicating essential structural identity. Similarly, analysis of side-chain rotamers indicates exact threefold symmetry in the Monofoil-4P structure. The Difoil-4P structure exhibits two intact β-trefoil folds related by a pseudo-twofold axis of symmetry and constructed from three polypeptide chains (Fig. 3E). Peptide chain B in this complex (colored green in Fig. 3E) adopts an entirely different conformation from the other two peptide chains. Despite this structural alteration, the main-chain atoms of the overall β-trefoil folds contained within the Difoil-4P homotrimer overlay those of the Symfoil-4P structure with an rmsd of ∼0.7 Å, indicating a highly conserved β-trefoil architecture (Fig. 3F). The two β-trefoil folds in Difoil-4P are constructed from three polypeptide chains via a “domain swapped” architecture (Fig. 3G). The first β-trefoil is comprised of chain “A” plus β-strands 1, 2, 3, and 8 of chain “B,” whereas the second β-trefoil is comprised of chain “C” plus β-strands 4, 5, 6, and 7 of chain B. In this second β-trefoil, contiguous β-strands 4 and 5 of chain B take the place of canonical β-strands 12 and 9, respectively (Fig. 3F).
Discussion
Support for the Conserved Architecture Model of Symmetric Protein Evolution.
Competing emergent architecture and conserved architecture models have been proposed for the evolutionary pathway of the threefold symmetric β-trefoil architecture. In the emergent architecture model of Mukhopadhyay (8), the ancient peptide motif was hypothesized to have been an autonomously folding single-domain, small globular protein, with a simple epidermal growth-factor-like fold. Dimerization driven by mutation(s) causing “domain-swapping” was postulated to produce a C2 symmetric molecule with an overall Ecotin-like fold. The β-trefoil architecture was hypothesized to have emerged after a final duplication and fusion event. A distinctly different conserved architecture model for the evolution of β-trefoil architecture was proposed at essentially the same time by Ponting and Russell (9). In this model, the ancient peptide motif was hypothesized to have existed as a homotrimer with overall β-trefoil architecture. Duplication and fusion resulted in a dimer repeat of this polypeptide which also folded as a homotrimer, producing two complete β-trefoil folds (although structural details of exactly how this would be accomplished were not proposed). A subsequent duplication and fusion event produced a triplicate repeat of the trefoil-fold peptide, yielding a single polypeptide with β-trefoil architecture.
Symmetric deconstruction of the β-trefoil architecture was undertaken with no preconceived ideas regarding the sequence or properties of the ultimate polypeptide; however, the results show that the oligomerization and structural details of the Monofoil, Difoil, and Symfoil polypeptides agree precisely with the conserved architecture model of β-trefoil evolution (Figs. 1B and 3). The symmetric deconstruction of the FGF-1 protein was accomplished in a punctuated stepwise manner with each of the 18 intermediate mutant forms being foldable, thermostable, and soluble (Fig. 4 and Table S3); thus, the results also present a thermodynamically tenable pathway, through foldable sequence space, extending from FGF-1 to the Monofoil-4P peptide. Sequence analysis suggests that all extant β-trefoil proteins evolved from a common ancestor (9), because no homotrimer examples of the β-trefoil architecture (either Monofoil-type or Difoil-type) have been reported, the experimentally supported evolutionarily model describes a likely primordial event. Whereas the emergent architecture model implies “simple gene–simple protein architecture,” the supported conserved architecture model enables complex protein architecture (e.g., a β-trefoil fold) via a simple gene (i.e., coding for a Monofoil-like 42-mer peptide). Thus, the results also support the hypothesis that complex protein architecture is possible in ancient life forms containing a substantially reduced genome complexity.
Fig. 4.
Isothermal equilibrium denaturation profiles for FGF-1 and mutants comprising the top-down symmetric deconstruction. (A) Denaturation profiles for FGF-1 and all single-polypeptide mutant proteins (culminating in the Symfoil-4P mutant). The starting FGF-1 protein is the least stable in comparison to all mutant proteins. (B) Denaturation profiles comparing the homotrimer assemblies of Monofoil-4P and Difoil-4P peptides (10 μM each) with FGF-1 and Symfoil-4P proteins.
Implications for de Novo Protein Design.
A long-standing goal in de novo protein design has been the exploitation of a “bottom-up” hierarchical design strategy utilizing peptide building blocks that can assemble (via oligomerization or concatenation) into target architecture (18–20) (in effect, paralleling aspects of the evolutionary pathway). The top-down symmetric deconstruction approach was remarkably successful in identifying a peptide building block solution (i.e., Monofoil-4P peptide) for the β-trefoil architecture. Engineered thermostable protein architecture can serve as a “function-competent” (21) scaffold for the incorporation of unique functionality (22, 23). Although the FGF-1 protein has low mesophilic stability (24, 25), inclusion of a stability screen in the deconstruction resulted in a hyperthermophilic β-trefoil (the Symfoil-4P protein), and may therefore be utilized as a departure point in bottom-up de novo protein design for the incorporation of unique functionality. The fundamental “symmetric deconstruction” concept is architecture independent and therefore applicable in identifying a peptide building block for other symmetric protein folds.
Materials and Methods
Top-Down Symmetric Deconstruction.
Top-down symmetric deconstruction is the introduction (via mutagenesis) of a symmetric constraint upon the primary and tertiary structure of a naturally evolved symmetric protein fold with the goal of achieving a purely symmetric primary structure. Details of the method will be published elsewhere, but briefly, the deconstruction is accomplished via a series of cumulative symmetric mutagenic transforms. In the case of FGF-1, this deconstruction involved a cumulative symmetric constraint upon core, reverse-turn, and β-strand secondary structure elements, respectively (Fig. 2A). Symmetric deconstruction of FGF-1 requires substantial change to both the primary and tertiary structure of the protein (Fig. 2B). Mutations were combined with a screen for protein folding, stability, and solubility, and were retained if they were neutral or improved upon such properties. Maximum use was made of previously published data (26–34) as well as sequence analysis to identify useful symmetric mutations in FGF-1. A summary of the application of specific transforms in pursuing a symmetric deconstruction of FGF-1 follows.
Transform 1: Symmetric deconstruction of the hydrophobic core.
Previously published work detailed the development of a series of symmetric core-packing mutations (SYM2-5, refs. 26 and 27; see Table S1 for mutant definitions) in FGF-1 that systematically introduced a symmetric primary structure constraint. Achieving a purely symmetric core while avoiding substantial destabilization required deletion of six amino acids within the third trefoil-fold subdomain, producing the SYM6ΔΔ mutant (28). Subsequent development of the SYM7ΔΔ mutant (29), involving symmetric mutations in additional buried hydrophobic positions of the protein represented essential completion of transform 1 and was the starting point for the application of transform 2.
Transform 2: Symmetric deconstruction of reverse turns.
The SYM7ΔΔ mutant was initially modified by the inclusion of two previously described stabilizing point mutations (Lys12Val and Pro134Val) (30) to produce the SYM9ΔΔ mutant. These mutations are located in the adjacent first and last β-strands of the β-barrel of the architecture, respectively, and represent a “discontinuous” β-turn within the threefold architecture. A His93Gly mutation, located within β-turn 8 and known to stabilize FGF-1 (31, 35), was introduced into the SYM9ΔΔ mutant to produce the SYM10ΔΔ mutant. Prior studies of the role of the statistically preferred β-turn motif Asx-Pro-Asx-Gly at individual β-turns 2, 6, and 10 in FGF-1 indicated a consistently favorable effect upon thermostability with Asn-Xxx-Asp-Gly (where Xxx represents the retained FGF-1 residue at this position) (34). The canonical Asn-Pro-Asp-Gly sequence was therefore substituted into the SYM10ΔΔ mutant at each of these symmetry-related β-turns to produce the SYM11ΔΔ mutant. The symmetric deconstruction of β-turn regions was not taken to completion at this stage, and the SYM11ΔΔ mutant was considered a successful “proof-of-concept” and was utilized as the starting point for transform 3.
Transform 3: Symmetric deconstruction of β-strands.
An Asn95Val mutation was introduced into SYM11ΔΔ resulting in a symmetric (Val) deconstruction of positions 12, 54, and 95 within β-strands 1, 5, and 9, respectively. Similarly, Leu46Val and Glu87Val mutations were introduced, resulting in a further symmetric constraint within symmetry-related β-strands 4, 8, and 12, respectively. The combination of these mutations resulted in the SYM12ΔΔ mutant. Ile56Leu and Tyr97Leu mutations were introduced into the SYM12ΔΔ mutant, resulting in a symmetric constraint upon symmetry-related β-strand positions 14, 56, and 97 and producing the SYM13ΔΔ mutant. The symmetric deconstruction of β-strand secondary structure was not taken to completion at this stage, and the SYM13ΔΔ mutant was considered a successful proof-of-concept. Completion of transforms 2 and 3 was achieved simultaneously by a chimera construct strategy chosen to eliminate reactive free thiols at positions Cys16 and Cys83. The resulting chimera sequence, yielding a complete symmetric deconstruction, is termed Symfoil-1.
Transform 4: Fragmentation of the repeating motif.
Optimization of Symfoil-1 stability was pursued prior to fragmentation of the repeating motif. Details of the optimization design will be published elsewhere, but involved development of the Symfoil-2, Symfoil-3, Symfoil-4T, 4V, and 4P mutants (Table S1). The Symfoil-4P mutant (the most stable Symfoil variant) was selected as the starting point for fragmentation studies of the repeating peptide motif. Monomer and dimer subdomains were constructed by mutagenesis to introduce stop codons at positions Glu53 or Glu94, respectively, of the Symfoil-4P construct (Fig. 2). The resulting monomeric and dimeric trefoil-fold peptides are referred to as Monofoil-4P and Difoil-4P, respectively (for monomeric trefoil fold and dimeric trefoil fold, respectively).
Mutagenesis, Protein Purification, and Characterization.
Mutagenesis and protein purification and details of structural and biophysical characterization of mutant proteins followed previously published methods and are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
This work was funded by Grant 0655133B from the American Heart Association and by additional research support provided by the Florida State University College of Medicine. Analytical ultracentrifugation calculations were performed on the UltraScan Laboratory Information Management System cluster at the Bioinformatics Core Facility at the University of Texas Health Science Center at San Antonio and the Lonestar cluster at the Texas Advanced Computing Center, supported by National Science Foundation Teragrid Grant MCB070038 (to B. Demeler). Use of the “mail-in crystallography” facility of Southeast Regional Collaborative Access Team for diffraction data collection is acknowledged. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract W-31-109-Eng-38. Use of the National Synchrotron Light Source, Brookhaven National Laboratory, is acknowledged, and financial support comes principally from the Office of Biological and Environmental Research and the Office of Basic Energy Sciences of the US Department of Energy, and the National Center for Research Resources of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3O49, 3O4A,3O4B, 3O4C, 3O4D, 3OL0, 3OGF).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015032108/-/DCSupplemental.
References
- 1.Sepulveda P, Marciniszyn JJ, Liu D, Tang J. Primary structure of porcine pepsin. III. Amino acid sequence of a cyanogen bromide fragment, CB2A, and the complete structure of porcine pepsin. J Biol Chem. 1975;250:5082–5088. [PubMed] [Google Scholar]
- 2.Tang J, James MN, Hsu IN, Jenkins JA, Blundell TL. Structural evidence for gene duplication in the evolution of the acid proteases. Nature. 1978;271:618–621. doi: 10.1038/271618a0. [DOI] [PubMed] [Google Scholar]
- 3.McLachlan AD. Three-fold structural pattern in the soybean trypsin inhibitor (Kunitz) J Mol Biol. 1979;133:557–563. doi: 10.1016/0022-2836(79)90408-x. [DOI] [PubMed] [Google Scholar]
- 4.Inana G, Piatigorsky J, Norman B, Slingsby C, Blundell T. Gene and protein structure of a β-crystallin polypeptide in murine lens: Relationship of exons and structural motifs. Nature. 1983;302:310–315. doi: 10.1038/302310a0. [DOI] [PubMed] [Google Scholar]
- 5.Tateno Y, et al. Evolutionary motif and its biological and structural significance. J Mol Evol. 1997;44:S38–S43. doi: 10.1007/pl00000056. [DOI] [PubMed] [Google Scholar]
- 6.Dorit RL, Schoenbach L, Gilbert W. How big is the universe of exons? Science. 1990;250:1377–1382. doi: 10.1126/science.2255907. [DOI] [PubMed] [Google Scholar]
- 7.Panchenko AR, Luthey-Schulten Z, Cole R, Wolynes PG. The foldon universe: A survey of structural similarity and self-recognition of independently folding units. J Biol Chem. 1997;272:95–105. doi: 10.1006/jmbi.1997.1205. [DOI] [PubMed] [Google Scholar]
- 8.Mukhopadhyay D. The molecular evolutionary history of a winged bean α-chymotrypsin inhibitor and modeling of its mutations through structural analysis. J Mol Evol. 2000;50:214–223. doi: 10.1007/s002399910024. [DOI] [PubMed] [Google Scholar]
- 9.Ponting CP, Russell RB. Identification of distant homologues of fibroblast growth factors suggests a common ancestor for all beta-trefoil proteins. J Mol Biol. 2000;302:1041–1047. doi: 10.1006/jmbi.2000.4087. [DOI] [PubMed] [Google Scholar]
- 10.Liu L, Iwata K, Yohda M, Miki K. Structural insight into gene duplication, gene fusion and domain swapping in the evolution of PLP-independent amino acid racemases. FEBS Lett. 2002;528:114–118. doi: 10.1016/s0014-5793(02)03264-7. [DOI] [PubMed] [Google Scholar]
- 11.Yadid I, Tawfik DS. Reconstruction of functional b-propeller lectins via homo-oligomeric assembly of shorter fragments. J Mol Biol. 2007;365:10–17. doi: 10.1016/j.jmb.2006.09.055. [DOI] [PubMed] [Google Scholar]
- 12.Akanuma S, Matsuba T, Ueno E, Umeda N, Yamagishi A. Mimicking the evolution of a thermally stable monomeric four-helix bundle by fusion of four identical single-helix peptides. J Biochem. 2010;147:371–379. doi: 10.1093/jb/mvp179. [DOI] [PubMed] [Google Scholar]
- 13.Richter M, et al. Computational and experimental evidence for the evolution of a (βα)8-barrel protein from an ancestral quarter-barrel stabilized by disulfide bonds. J Mol Biol. 2010;398:763–773. doi: 10.1016/j.jmb.2010.03.057. [DOI] [PubMed] [Google Scholar]
- 14.Yadid I, Kirshenbaum N, Sharon M, Dym O, Tawfik DS. Metamorphic proteins mediate evolutionary transitions of structure. Proc Natl Acad Sci USA. 2010;107:7287–7292. doi: 10.1073/pnas.0912616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hocker B, Beismann-Driemeyer S, Hettwer S, Lustig A, Sterner R. Dissection of a (βα)8-barrel enzyme into two folded halves. Nat Struct Biol. 2001;8:32–36. doi: 10.1038/83021. [DOI] [PubMed] [Google Scholar]
- 16.Akanuma S, Yamagishi A. Experimental evidence for the existence of a stable half-barrel subdomain in the (β/α)8-barrel fold. J Mol Biol. 2008;382:458–466. doi: 10.1016/j.jmb.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 17.Murzin AG, Lesk AM, Chothia C. β-Trefoil fold. Patterns of structure and sequence in the kunitz inhibitors interleukins-1β and 1α and fibroblast growth factors. J Mol Biol. 1992;223:531–543. doi: 10.1016/0022-2836(92)90668-a. [DOI] [PubMed] [Google Scholar]
- 18.DeGrado WF, Regan L, Ho SP. The design of a four-helix bundle protein. Cold Spring Harbor Symp Quant Biol. 1987;52:521–526. doi: 10.1101/sqb.1987.052.01.059. [DOI] [PubMed] [Google Scholar]
- 19.Regan L, DeGrado WF. Characterization of a helical protein designed from first principles. Science. 1988;241:976–978. doi: 10.1126/science.3043666. [DOI] [PubMed] [Google Scholar]
- 20.Richardson J, Richardson DC. The de novo design of protein structures. Trends Biochem Sci. 1989;14:304–309. doi: 10.1016/0968-0004(89)90070-4. [DOI] [PubMed] [Google Scholar]
- 21.Soskine M, Tawfik DS. Mutational effects and the evolution of new protein functions. Nat Rev Genet. 2010;11:572–582. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- 22.Gibney BR, Rabanal F, Dutton PL. Synthesis of novel proteins. Curr Opin Chem Biol. 1997;1:537–542. doi: 10.1016/s1367-5931(97)80050-6. [DOI] [PubMed] [Google Scholar]
- 23.Schafmeister CE, Stroud RM. Helical protein design. Curr Opin Biotechnol. 1998;9:350–353. doi: 10.1016/s0958-1669(98)80006-2. [DOI] [PubMed] [Google Scholar]
- 24.Blaber SI, Culajay JF, Khurana A, Blaber M. Reversible thermal denaturation of human FGF-1 induced by low concentrations of guanidine hydrochloride. Biophys J. 1999;77:470–477. doi: 10.1016/S0006-3495(99)76904-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Copeland RA, et al. The structure of human acidic fibroblast growth factor and its interaction with heparin. Arch Biochem Biophys. 1991;289:53–61. doi: 10.1016/0003-9861(91)90441-k. [DOI] [PubMed] [Google Scholar]
- 26.Brych SR, Blaber SI, Logan TM, Blaber M. Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core region of a β-trefoil. Protein Sci. 2001;10:2587–2599. doi: 10.1110/ps.ps.34701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brych SR, Kim J, Logan TM, Blaber M. Accommodation of a highly symmetric core within a symmetric protein superfold. Protein Sci. 2003;12:2704–2718. doi: 10.1110/ps.03374903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brych SR, et al. Symmetric primary and tertiary structure mutations within a symmetric superfold: A solution, not a constraint, to achieve a foldable polypeptide. J Mol Biol. 2004;344:769–780. doi: 10.1016/j.jmb.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 29.Dubey VK, Lee J, Blaber M. Redesigning symmetry-related “mini-core” regions of FGF-1 to increase primary structure symmetry: Thermodynamic and functional consequences of structural symmetry. Protein Sci. 2005;14:2315–2323. doi: 10.1110/ps.051494405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubey VK, Lee J, Somasundaram T, Blaber S, Blaber M. Spackling the crack: Stabilizing human fibroblast growth factor-1 by targeting the N and C terminus beta-strand interactions. J Mol Biol. 2007;371:256–268. doi: 10.1016/j.jmb.2007.05.065. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Brych SR, Lee J, Logan TM, Blaber M. Identification of a key structural element for protein folding within β-hairpin turns. J Mol Biol. 2003;328:951–961. doi: 10.1016/s0022-2836(03)00321-8. [DOI] [PubMed] [Google Scholar]
- 32.Kim J, Lee J, Brych SR, Logan TM, Blaber M. Sequence swapping does not result in conformation swapping for the b4/b5 and b8/b9 b-hairpin turns in human acidic fibroblast growth factor. Protein Sci. 2005;14:351–359. doi: 10.1110/ps.041094205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J, Blaber M. Structural basis for conserved cysteine in the fibroblast growth factor family: Evidence for a vestigial half-cystine. J Mol Biol. 2009;393:128–139. doi: 10.1016/j.jmb.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Lee J, Dubey VK, Longo LM, Blaber M. A logical OR redundancy with the Asx-Pro-Asx-Gly type I β-turn motif. J Mol Biol. 2008;377:1251–1264. doi: 10.1016/j.jmb.2008.01.055. [DOI] [PubMed] [Google Scholar]
- 35.Arakawa T, et al. Production and characterization of an analog of acidic fibroblast growth factor with enhanced stability and biological activity. Protein Eng. 1993;6:541–546. doi: 10.1093/protein/6.5.541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.