Abstract
In mammals, the canonical histone H3 and the variant H3.3 are assembled into chromatin through replication-coupled and replication-independent (RI) histone deposition pathways, respectively, to play distinct roles in chromatin function. H3.3 is largely associated with transcriptionally active regions via the activity of RI histone chaperone, HIRA. However, the precise role of the RI pathway and HIRA in active transcription and the mechanisms by which H3.3 affects gene activity are not known. In this study, we show that HIRA is an essential factor for muscle development by establishing MyoD activation in myotubes. HIRA and Asf1a, but not CHD1 or Asf1b, mediate H3.3 incorporation in the promoter and the critical upstream regulatory regions of the MyoD gene. HIRA and H3.3 are required for epigenetic transition into the more permissive chromatin structure for polymerase II recruitment to the promoter, regardless of transcription-associated covalent modification of histones. Our results suggest distinct epigenetic management of the master regulator with RI pathway components for cellular differentiation.
Keywords: myogenesis, histone exchange
The eukaryotic genome is packed in chromatin by formation of nucleosomes that consist of two copies of histones H2A, H2B, H3, and H4. Although canonical H3 (H3.1/H3.2) is predominantly assembled into a nucleosome during S phase via replication-coupled (RC) histone H3/H4 chaperone CAF1, H3.3 is expressed and deposited throughout the cell cycle via replication-independent (RI) histone chaperone HIRA (1–3). The cell-cycle-independent expression pattern of H3.3 and its specific amino acid substitution around residues 87–90 are important in order for this evolutionarily conserved H3 variant to incorporate into specific chromatin domains according to biological function (4, 5).
Recent analysis of the genome-wide distribution pattern of H3.3 in embryonic stem cells showed that H3.3 is enriched in both transcriptionally active and inactive regions (6). As for nontranscriptional function, H3.3 or HIRA contributes to mammalian sex chromosome inactivation (7), germline cell development (8), and fertilization (9) through a massive genome-wide nucleosome assembly. Chromatin remodeling factor CHD1, which belongs to the chromodomain family of SNF2-like adenosine triphosphatases, is also responsible for H3.3 incorporation into the male pronucleus, presumably in couple with HIRA (10). Besides, HIRA is involved in the formation of senescence-associated heterochromatic foci (SAHF) (11) and the maintenance of gene silencing (12–14). Formation of SAHF is driven by HIRA and another H3/H4 chaperone Asf1a and involves repression of proliferation-promoting genes.
Simultaneously, many aspects of H3.3 indicate its involvement in active transcription; posttranslational modification patterns enriched on H3.3 such as H3 lysine (K) 4, K36, K79 methylation all linked to active transcription (15–17), strong resemblance of the genome-wide distribution profile of H3.3 to that of polymerase II or methylated H3 K4 (18), and a role as a structural component of H2A.Z-containing nucleosomes that mark nucleosome-free regions of active promoters (19). Indeed, transcription induction coincides with H3.3 accumulation at several individual genes (20–22). However, recent reports based on detailed analysis of transcriptional and developmental phenotypes of flies that have been genetically manipulated to deplete H3.3 have challenged its functional significance in active transcription (23, 24), because H3.3 was dispensable for most transcriptional events throughout Drosophila development. One of the proposed mechanisms for H3.3 replacement is that canonical H3 may be able to serve as a functionally equivalent substrate for the transcription process (5). It is thus uncertain whether H3.3 is simply deposited via the RI pathway as a consequence of restoration of chromatin structure that was perturbed by transcription or whether it has a specific regulatory role in transcription.
Recently, with regard to its role in euchromatin, H3.3 was reported to be associated with epigenetic memory of transcription state during muscle development (25). During skeletal myogenesis, upon receiving differentiation cues, proliferating myoblasts irreversibly exit the cell cycle and begin to differentiate into multinucleated myotubes, which is accompanied by a coordinated transition of gene activities (26). In this study, we examined H3.3 deposition and the role of HIRA, Asf1, and CHD1 during myogenesis and showed that RI pathway-H3.3-HIRA/Asf1a plays an important role in myogenic differentiation through cell type-specific activation of MyoD at the level of transcription. Our data show that the RI pathway operates during myogenic transcription to deposit H3.3, which cannot be substituted by canonical histones.
Results
Differential Expression of RC and RI Components During Myoblast Differentiation.
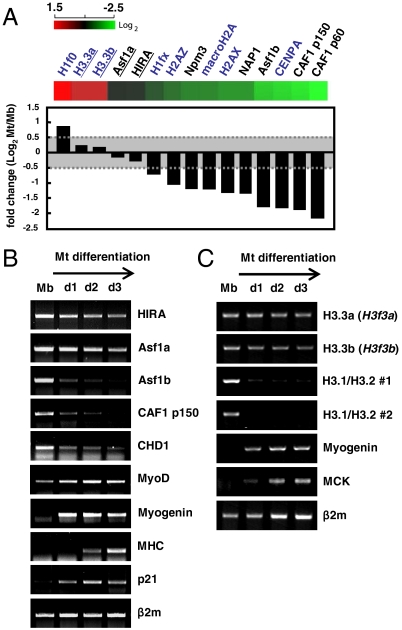
To investigate the role of variant histones and various histone chaperones, we first analyzed the shift of global gene expression profiles in skeletal muscle C2C12 cells during myoblast differentiation using the mouse 24K gene expression microarray chip (Illumina). C2C12 myoblasts differentiate into multinucleated myotubes upon serum withdrawal (Fig. S1A). Microarray analysis revealed that expression of HIRA and Asf1a was largely maintained during myogenesis, whereas the RC histone chaperone CAF1 (p150 and p60) or other basal nucleosome assembly factors such as Nap1 and nucleoplasmin were all reduced (Fig. 1A). The expression patterns of the above genes were examined individually at the mRNA and the protein levels (Fig. 1B and Fig. S1B). HIRA was maintained, but CAF1 decreased as cells differentiated into myotubes, during which other muscle markers were appropriately induced [MyoD, MHC (myosin heavy chain), myogenin, or p21] or decreased (Ezh2, SIRT1) as reported (26). Interestingly, the expression fate of two isotypes of Asf1 diverged during differentiation such that Asf1a persisted, similar to HIRA, while Asf1b decreased, similar to CAF1, consistent with their pairwise interaction (Fig. 1B) (1, 27). Consistent with the expression pattern of HIRA, two mouse H3.3 genes, H3.3a and H3.3b, were continuously expressed as previously reported in chicken myogenesis (28), whereas canonical H3s (H3.1s and H3.2s) and other variant histones, macroH2A, H2A.X, H2A.Z, and CENP-A, were down-regulated after withdrawal from the cell cycle (Fig. 1 A and C and Fig. S1C). Unexpectedly, the expression of CHD1, another chromatin remodeling factor implicated in H3.3 deposition (10), was decreased at the onset of differentiation (Fig. 1B and Fig. S2). These results suggest that H3.3 and the RI pathway involving HIRA and Asf1a might play a central role in the remodeling of chromatin structure associated with muscle differentiation.
Fig. 1.
The expression of RI pathway chaperones and H3.3 is maintained throughout skeletal myogenesis. (A) The mRNA levels of histone chaperone (black) and various histone (blue) genes during skeletal myogenesis were analyzed by microarray. Genes whose fold difference (Log2) is less than 0.5 are underlined. (B and C) RNA extracted from C2C12 cells grown to each differentiation day (d1 ∼ d3) was reverse transcribed with oligo dT (B) or random hexamers (C) to synthesize cDNA. The mRNA level of each gene was analyzed by RT-PCR. Two independent primer pairs used to detect canonical H3 (H3.1/H3.2 #1 and #2) recognize most of the H3.1 and H3.2 genes in the mouse genome, whereas each H3.3 primer pair is specific to H3.3a (H3f3a) and H3.3b (H3f3b), respectively. β2m (beta2-microglobulin) was used as a control. MHC (myosin heavy chain), MyoD, MCK (muscle creatine kinase), myogenin, and p21 are myogenesis markers. Mb, myoblasts; Mt, myotubes.
HIRA Is Necessary for Transcriptional Activation of MyoD in a Cell Type-Specific Manner.
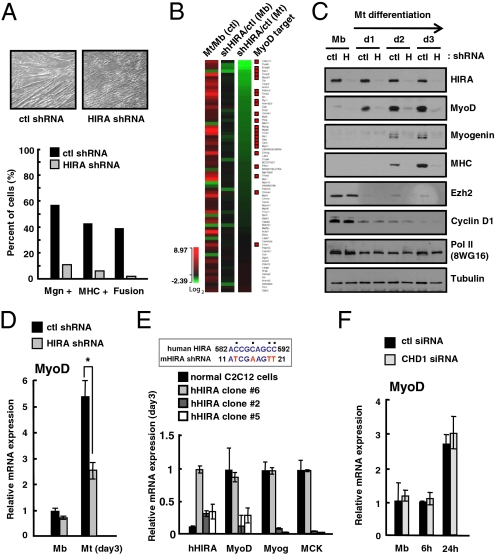
To directly ascertain the role of HIRA in skeletal myogenesis, we depleted HIRA in C2C12 cells using the shRNA-mediated knockdown system (Fig. S3 A and B). The differentiation efficiency, as measured by the percentage of myogenin+, MHC+ cells, or cell fusion, showed that C2C12 myoblasts stably expressing HIRA shRNA were not able to differentiate efficiently into myotubes (Fig. 2A and Fig. S3C). Those cells remained only partially differentiated as a cell-cycle marker (Cyclin D1) or a negative differentiation-regulatory factor (Ezh2) was appropriately down-regulated (Fig. S4A) but induction of myotube-specific muscle markers (myogenin and MHC) was markedly reduced. To obtain a genome-wide description of the effect of HIRA on cellular differentiation, we performed global gene expression profiling with control (ctl) or HIRA knockdown cells (shHIRA). A total of 412 genes were either up- or down-regulated by more than 1.5-fold in a differentiation-induced condition when HIRA was depleted (Fig. S3D). Notably, among 71 genes involved in muscle-specific structural, functional, and transcriptionally regulatory functions, 15 were dramatically affected by HIRA depletion (Fig. 2B). Of note, many HIRA-dependent genes such as Cdkn1c were also reported to be under the control of MyoD (29, 30), a key regulator of myogenesis (26), indicating that HIRA shares its target genes with MyoD or functions upstream of MyoD. To assess these two possibilities, we evaluated the expression of the MyoD gene and other marker genes in control shRNA and shHIRA C2C12 cells. Interestingly, the expression of MyoD and other myogenesis markers was greatly reduced in the shHIRA C2C12 cells at both the protein and mRNA levels (Fig. 2C and Fig. S4A). To avoid the possibility of clonal variation, C2C12 cells were treated transiently with two independent siRNAs that target different regions of HIRA mRNA (siRNA-1 and siRNA-2). These experiments also showed that the expression of MyoD and myogenesis markers was affected by HIRA depletion (Fig. S4 B and C). In particular, the expression of MyoD was dramatically reduced in differentiating conditions but was barely affected in proliferating myoblasts (Fig. 2D), suggesting that HIRA specifically affects MyoD expression during terminal differentiation of muscle cells. To confirm the role of HIRA in MyoD expression, we generated three independent shHIRA clones that express different levels of human HIRA. Human HIRA is able to escape from the mouse-specific RNA interference. Here, the impaired expression of MyoD [myogenin, MCK (muscle creatine kinase), and MHC as well] was partially rescued by the ectopic expression of human HIRA in correlation with their expression levels (Fig. 2E and Fig. S5A). In addition, ectopic expression of FLAG-tagged MyoD in shHIRA cells suppressed differentiation defects without affecting the level of endogenous MyoD gene (Fig. S5 B and C). In contrast, reducing CHD1 with specific siRNA affected neither MyoD nor myogenin expression in proliferating or early differentiating cells (Fig. 2F and Fig. S2D). These results indicate that HIRA is an essential factor for myogenesis and likely an upstream regulator of cell type-specific activation of MyoD transcription.
Fig. 2.
HIRA is an upstream regulator of MyoD and essential for skeletal myogenesis. (A) Myotube formation of control shRNA (ctl) or shHIRA C2C12 cells evaluated by phase contrast microscopy. Stable polyclonal shHIRA cells that express reduced HIRA were analyzed for myogenic differentiation. Myogenin+, MHC+ cells, and the extent of cell fusion were analyzed by immunofluorescence microscopy. (B) Genes known to be involved in muscle differentiation were selected and their expression changes were compared in the ctl and shHIRA cells. The heat map shows genes sorted in the order of the effect of HIRA depletion in myotubes; shHIRA/ctl (Mt). Genes reported as MyoD targets are marked by red squares on the right column. (C) The protein level of individual genes was monitored by immunoblotting analysis of whole cell lysate from ctl or shHIRA (H) cells on each differentiation day. Ezh2, Cyclin D1; myogenesis markers, pol II, Tubulin; loading controls. (D) Relative mRNA level of MyoD in control (ctl) and shHIRA cells analyzed by quantitative RT-PCR. Error bars represent SD, n = 3. The significance of difference was evaluated (* P value < 0.05). (E) shHIRA C2C12 cells were rescued by human HIRA. Three independent clones that express different levels of supplemented human HIRA were selected for analysis. Control C2C12 and three clones were subjected to quantitative RT-PCR for the indicated genes. The expression of human HIRA was monitored by human-specific primers. The approximate expression level of human HIRA of clone #6 was estimated by immunoblotting assay as ∼60% of that of mouse HIRA of control cells (Fig. S5A). (Upper) Part of the nucleotide sequence of human HIRA that deviates from the shRNA used for targeting mouse HIRA. (F) Quantitative RT-PCR analysis of MyoD mRNA levels in control (ctl) or siCHD1-treated C2C12 cells at each differentiation time point.
HIRA Is Responsible for H3.3 Deposition onto the Transcriptionally Regulatory Elements of MyoD During Differentiation.
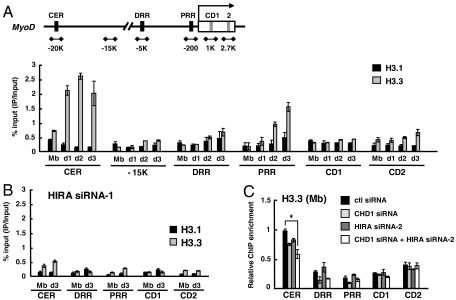
We next asked whether HIRA-dependent and differentiation-associated MyoD activation was relevant to its H3.3-specific histone chaperone activity. To this end, C2C12 cells stably expressing either epitope-tagged H3.1 or H3.3 (eH3.1 or eH3.3) were established and confirmed to differentiate normally (Fig. S6 A and B). H3.1 and H3.3 were expressed in similar amounts and seemed to support in vivo function as the differential histone modification pattern was recapitulated such that a repressive mark (H3K27me3 and H3K9me2) was associated with H3.1, whereas an active mark (H3K4me2) was associated with H3.3 (Fig. S6C). In addition, H3.3 interacted preferentially with HIRA, whereas H3.1 interacted with the H3K27 methyltransferase, Ezh2 (Fig. S6D). We then analyzed the relative incorporation of H3.1 or H3.3, spanning upstream regulatory elements (CER: the core enhancer region; DRR: the distal regulatory region; PRR: the proximal regulatory region) or two coding regions (CD1 and CD2) of MyoD with an irrelevant region (∼-15 K) as a control, by chromatin immunoprecipitation (ChIP) followed by quantitative real time PCR analysis. Dramatic accumulation of H3.3, but not H3.1, especially around the CER, DRR, and PRR, which are known to be critical for MyoD expression, was apparent as differentiation proceeded (Fig. 3A). In particular, the H3.3 level in the CER was relatively high compared to that in other regions already in myoblasts and this level rapidly increased further upon differentiation while the H3.3 level in the PRR increased more gradually. The level of H3.1 was largely unchanged throughout differentiation. In contrast, cells treated with siRNA targeting HIRA were not only impaired in MyoD activation, but also failed to accumulate H3.3 specifically in the differentiated condition (Fig. 3B). The level of H3.3 in the CER of proliferating cells was barely affected by HIRA knockdown (Fig. 3C). Instead, combined depletion of CHD1 and HIRA reduced it to ∼50%, indicating that H3.3 is deposited by redundant contribution of HIRA, CHD1, and other unknown factors (Fig. 3C). Thus our data indicate that prominent differentiation-associated H3.3 deposition around the upstream regulatory and promoter regions is mediated by HIRA and might be important for MyoD activation.
Fig. 3.
H3.3 is incorporated into MyoD regulatory regions in a HIRA-dependent manner. (A and B) ChIP was performed to monitor incorporation of eH3.1 or eH3.3 at MyoD regulatory and coding regions. Chromatin solution prepared at each indicated time point was immunoprecipitated with anti-FLAG and analyzed in duplicate by quantitative real-time PCR to measure the relative enrichment of eH3.1 or eH3.3 at different loci. The data represent the percentage of ChIP (IP/Input). eH3 ChIP signal was normalized to H3 ChIP. (C) eH3.3 levels in the growing myoblasts treated with various siRNAs were analyzed by ChIP and represented as a relative enrichment (the ChIP value obtained from the CER of control siRNA sample was arbitrarily set to 1). For HIRA k.d. (knock down), data with HIRA siRNA-1 and siRNA-2 were shown for B and C, respectively. The sequences of each siRNA are described in the SI Materials and Methods. The k.d. efficiency of two siRNAs is similar as shown in Fig. S4. Error bars represent standard deviation of three independent experiments. *, P < 0.05. eH3, epitope-tagged H3; CER, core enhancer region; DRR, distal regulatory region; PRR, proximal regulatory region; CD, coding region.
Myotube-Specific and Differentiation-Associated H3.3 Deposition Is Mediated by HIRA and Asf1a.
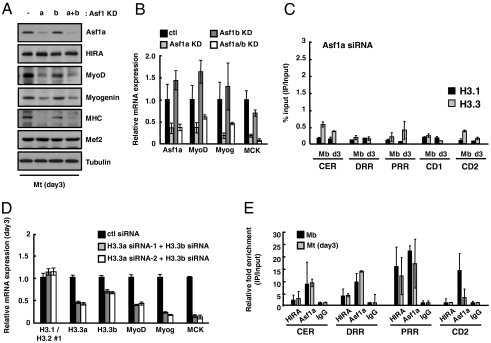
Histone chaperone Asf1 is expressed as two isotypes, Asf1a and Asf1b, in the mouse genome. Asf1a is the form that preferentially interacts with HIRA (27), is detected in the H3.3 complex (1), and is continuously expressed during myogenesis (Fig. 1). As Asf1a serves as an essential partner for HIRA in the senescence-associated heterochromatin foci formation pathway (11), we asked whether Asf1a also plays a role in HIRA/H3.3 mediated MyoD activation and muscle differentiation. Indeed, the expression of MyoD and marker genes such as myogenin and MHC was reduced at both the mRNA and the protein levels in C2C12 cells treated with siRNA targeting Asf1a but not Asf1b (Fig. 4 A and B). Knockdown of Asf1a did not affect the level of HIRA, excluding the possibility that Asf1a reduced the MyoD expression indirectly by affecting the stability of HIRA (Fig. 4A and Fig. S7). Importantly, specific enrichment of H3.3 in differentiated myotubes could not be detected upon RNA interference of Asf1a (Fig. 4C). However, Asf1a siRNA had no discernible effect on the level of H3.3 in the myoblast CER, indicating that Asf1a participates in differentiation-associated H3.3 accumulation. Our data suggest that Asf1a, together with HIRA, has a role in MyoD activation and this ability might be related to their H3.3 deposition activity. To directly ask whether H3.3 incorporation is critical for MyoD activation, we treated cells with a mixture of siRNAs that target H3.3a and H3.3b without perturbing the expression of histone H3.1 or H3.2 (Fig. 4D). Importantly, myogenic differentiation of these cells was partially impaired as observed in HIRA- or Asf1a-depleted cells. The mRNA levels of MyoD, myogenin, and MCK were down-regulated in a differentiation condition, reminiscent of HIRA or Asf1a depletion (Fig. 4D). Next, we examined the recruitment of histone chaperones using specific antibodies against HIRA (WC15) and Asf1a. Consistent with the H3.3 incorporation around the MyoD gene, occupancy of both HIRA and Asf1a was detected in the regulatory regions where H3.3 incorporation takes place actively (Fig. 4E). Asf1a was additionally detected in the coding region consistent with its role in transcriptional elongation (31, 32). However, unexpectedly, their occupancy was either not changed or marginally decreased, depending on the loci, as myoblasts differentiated into myotubes. Although ChIP was useful to detect their occupancy, the efficiency of cross-linking of histone chaperones to DNA did not correlate with their activity, potentially suggesting that the way they interact with chromatin may be altered while histones are actively exchanged and/or their activity rather than their recruitment may be targeted by differentiation signals.
Fig. 4.
H3.3 incorporation is mediated by HIRA/Asf1a for MyoD activation. (A and B) The expression level of each gene was analyzed by immunoblotting (A) or quantitative RT-PCR analysis (B) after cells were shifted to the differentiation condition for 3 d (day3). To knock down Asf1a and Asf1b separately, either nontargeting control siRNA or Asf1a-specific siRNA was transfected into C2C12 cells expressing control (lanes 1,2) or Asf1b-specific shRNA (lanes 3,4) (Fig. S7A). (C) ChIP was performed to monitor incorporation of eH3 at MyoD regulatory and coding regions in Asf1a siRNA-treated C2C12 cells. ChIP was analyzed as described in Fig. 3. (D) The assessment of the relative mRNA levels of myogenic markers in C2C12 cells treated with a mixture of siRNAs targeting H3.3a and H3.3b transcripts. Control and siH3.3-treated cells were induced to differentiate for 3 d and subjected to quantitative RT-PCR for the analysis of the indicated genes. (E) Relative occupancy of HIRA and Asf1a was analyzed by ChIP. Chromatin prepared from myoblasts or myotubes was immunoprecipitated with anti-HIRA(WC15), anti-Asf1a or normal IgG as a control. Acquired signals were normalized to input and IgG. Error bars represent SD, n = 2 or more.
Chromatin Changes Mediated by RI Histone Deposition.
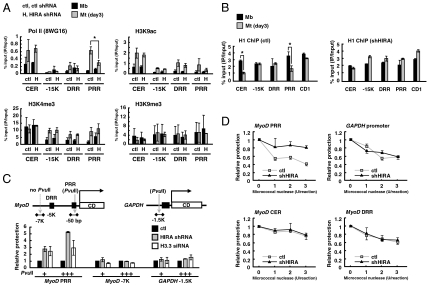
To gain insight into the mechanism by which H3.3 regulates MyoD transcription, we first analyzed the covalent modification patterns of H3 in the MyoD elements by ChIP. The H3 K4 methylation (H3K4me3) and the H3 K9 acetylation (H3K9ac) were increased at multiple regions (except for H3K4me3 at the CER) when transcription was activated as determined by increased occupancy of RNA polymerase II, whereas the H3 K9 methylation (H3K9me3) was relatively unchanged (Fig. 5A). As expected by reduced transcription, C2C12 cells expressing HIRA shRNA showed decreased occupancy of polymerase II at the PRR compared to the control cells, but unexpectedly, the levels of histone markers (H3K4me3, K9ac, and K9me3) were barely affected by HIRA depletion. Only H3 K9ac was slightly decreased at the PRR, probably reflecting the ongoing transcription state. This implies that a large part of transcription-associated histone modification is acquired in a post-histone-deposition step, independently of histone specificity, which means that the defect in MyoD activation by lack of H3.3 deposition was not attributed to the failure of induction of appropriate histone modification. To explore any chromatin change in the CER and PRR, we next investigated the binding of linker histone H1. Histone H1 is necessary for folding chromatin into a more compact structure that becomes refractory to transcription (33). In particular, binding of H1b (one of the H1 subtypes) to the CER via interaction with Msx1 homeodomain protein is necessary for repression of MyoD until differentiation is initiated (34). We used a subtype-insensitive H1 antibody for the ChIP assay. Notably, the H1 occupancy was significantly and specifically diminished at both CER and PRR in myotubes compared to that in proliferating myoblasts (Fig. 5B). Importantly, this H1 loss accompanied by cellular differentiation was not observed in shHIRA cells. Given the opposite distribution behavior of H3.3 and H1 (35), our data suggest that HIRA and H3.3 deposition might facilitate rapid removal of repressive H1. Additional structural differences were examined by DNA accessibility analysis using PvuII or micrococcal nuclease (converted into the relative protection value). Compared to the control, HIRA and H3.3 depletion led to increased PvuII protection (decreased accessibility) around the PRR but not around the GAPDH gene, which also harbors a single PvuII recognition site (Fig. 5C). In addition, Fig. 5D shows that the PRR gained increased protection from micrococcal nuclease in HIRA-depleted cells compared to control cells. Unlike PRR, DNase sensitivity of CER was not affected by HIRA, partly because of a nuclease sensitive feature of CER already established in myoblasts during cell specification (36). Taken together, H3.3 incorporation around the CER and the PRR is very likely the key step in epigenetic transition of chromatin structure necessary for the polymerase II to gain access to the MyoD promoter. Therefore, chromatin transition with H3.3 itself is indispensible, even though posttranslational histone modification is achieved.
Fig. 5.
HIRA and H3.3 promote polymerase II recruitment via chromatin change around the MyoD CER and PRR. (A) ChIP was performed to monitor the recruitment of polymerase II and various H3 modifications at MyoD regulatory regions. Chromatin solution prepared from myoblasts (Mb) or myotubes (Mt) was immunoprecipitated with the indicated antibodies. (B) ChIP with histone H1 antibody. (C and D) Chromatin accessibility was monitored by ChART-PCR (chromatin accessibility by real-time PCR) using PvuII (C) or micrococcal nuclease (D). The PvuII or micrococcal nuclease was added to nuclei prepared from control (ctl), shHIRA, or siH3.3 (H3.3 a siRNA-2 + H3.3b siRNA) treated C2C12 cells. The digestion efficiency was analyzed by quantitative PCR (qPCR) with appropriate primers. Amplified DNA intensity obtained from digested sample was converted into reciprocal numbers to represent relative protection. Error bars represent SD, n = 3. *, P < 0.05.
Discussion
MyoD is expressed in steps during acquisition of myogenic identity and activation of terminal differentiation under distinct regulatory networks to meet the cell state-specific protein amounts and activities. Here, our findings provide insights into the molecular mechanisms that mediate MyoD activation upon terminal differentiation. In response to differentiation cues, rapid accumulation of H3.3 is achieved in the CER, which has been premarked by H3.3 for the proliferation stage; this is followed by de novo incorporation of H3.3 in the PRR, which eventually leads to removal of H1, opening of chromatin, and acceleration of transcription initiation (Fig. S8). Dramatic changes in H3.3 and the essential role of RI histone chaperones during myogenesis seem to be relevant to the neuromuscular defective phenotype of H3.3 knockdown mice (37).
How could the components of RI histone deposition pathway contribute to MyoD transcription? In the PRR region, which contains a conserved TATA box and serves as a promoter of the MyoD gene (38), HIRA/Asf1a seem to mediate the replacement of promoter nucleosomes with those containing H3.3, affecting chromatin structure in favor of transcriptional activation. As evident in yeast PHO activation driven by Asf1-dependent nucleosome loss (39), H3.3 substitution could cause intrinsic nucleosome instability (19), which might be a prerequisite for polymerase recruitment and MyoD activation. As for the CER, which is known to be critical for both cell specification and differentiation (36), H3.3 induction in the CER does not seem to be aimed at polymerase recruitment toward the CER. A polymerase was able to enter this element in the absence of HIRA or additional induction of H3.3 (Fig. 5A). Instead, in accordance with CER’s role in temporal control of MyoD during terminal differentiation by being targeted by H1, HIRA/Asf1a-dependent rapid accumulation of H3.3 in response to a differentiation signal appears to correlate with H1 loss from the CER. The relative content of H3 variants could potentially further influence transcriptional environment by affecting relocation of MyoD gene within the nucleus (40, 41). Interestingly, recent genome-wide analysis of H3.3 has revealed many upstream regulatory enhancers enriched with H3.3 (6), suggesting that site-specific and regulated deposition of H3.3 via the RI pathway might play a more general role in gene expression by providing an instructive chromatin environment. Nonetheless, because many enhancer elements also produce substantial amounts of noncoding RNAs (42), it is important to delineate whether H3.3 is the cause or the consequence of enhancer transcription rather than (or in addition to) target gene regulation.
With present study and reports by others (41, 43), we propose functionally distinct roles of RC and RI pathways. The RC pathway operates for faithful maintenance of current chromatin states and cellular identity during cell proliferation. With the decision to differentiate, the RC pathway and chromatin factors involved in the massive nucleosome assembly seem to decline to a level necessary for maintenance of genome integrity, which might be one of the mechanisms for keeping differentiated cells in cell-cycle arrest, whereas the RI pathway, the function of which has been minimally required for marking CER with H3.3 and securing the specific gene state (25, 43), becomes active, driving epigenetic reprogramming of a cell state by activating MyoD expression as one of its targets. H3.3 in the myoblast CER persisted during cell proliferation, which could be partially explained by preferential association of HIRA/Asf1a in this region (43). Although their binding monitored by ChIP was not further increased upon differentiation, there are possibilities that the activities of RI histone chaperone could be regulated under various signaling network by phosphorylation (44, 45). This model provides that the epigenetic reprogramming mechanism underlying the progression of multipotent progenitors into differentiated cells might be driven by the timely and regulated contribution of RC vs. RI pathways. Considering many other developmental steps that involve precise regulation of proliferation and differentiation such as generation of neurons, adipocytes, and hematopoietic cells (46), coordinated regulation of histone deposition pathways might be important for both maintaining a particular cellular stage and reprogramming into another one. Future studies will determine how various histone chaperones are targeted to specific regions and how their activities are regulated in the context of cellular differentiation networks.
Materials and Methods
C2C12 cells were induced to differentiate in DMEM containing 2% (vol/vol) horse serum (differentiation medium) for 3 d. To monitor differentiation, cells were paraformaldehyde-fixed and stained with anti-myogenin and anti-MHC to calculate myogenin+, MHC+ cells, and the fusion index. For production of stable cell lines, retroviral or lentiviral systems were used for transduction and selected with appropriate drugs for a week. Transfection of siRNAs was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. To analyze the relative expression levels of mRNA, cDNAs prepared using TRIzol reagent (Invitrogen) and cDNA synthesis kit (Promega) were amplified by real time qPCR. Primers used in this study are listed in Table S1. Further detailed methods are contained in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Dr. Peter D. Adams (Beatson Institute, Glasgow, UK) for HA-HIRA constructs and HIRA antibodies, Dr. Yoshihiro Nakatani (Dana-Farber Cancer Institute, Boston, MA) for pOZ H3.1 and H3.3, and Dr. Kwang Youl Lee (Chonnam National University, Gwangju, South Korea) for MyoD construct. This work was supported by Mid-career Researcher Program (ROA-2008-0060084 and 2009-0080410 to E.-J.C.) and Basic Science Research Program (2010-0028646 to E.-J.C. and J.-W.H.) through the National Research Foundation of Korea grant funded by the Ministry of Education, Science and Technology and the Korea Healthcare Technology R&D Project (A080181 to E.-J.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009830108/-/DCSupplemental.
References
- 1.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 2.Ray-Gallet D, et al. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- 3.Koning LD, Corpet A, Haber JE, Almouzni G. Histone chaperoens: An escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 4.Sarma K, Reinberg D. Histone variants meet their match. Nat Rev Mol Cell Bio. 2005;6:139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- 5.Elsaesser S, Goldberg AD, Allis CD. New functions for an old variant: No substitute for histone H3.3. Curr Opin Genet Dev. 2010;20:1–8. doi: 10.1016/j.gde.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldberg AD, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Heijden GW, et al. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat Genet. 2007;39:251–258. doi: 10.1038/ng1949. [DOI] [PubMed] [Google Scholar]
- 8.Hajkova P, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loppin B, et al. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- 10.Konev AY, et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317:1087–1090. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang R, et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 12.Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley MA. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol Cell. 1999;4:75–83. doi: 10.1016/s1097-2765(00)80189-6. [DOI] [PubMed] [Google Scholar]
- 13.Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–373. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 14.Anderson HE, et al. The fission yeast HIRA histone chaperone is required for promoter silencing and the suppression of cryptic antisense transcripts. Mol Cell Biol. 2009;29:5158–5167. doi: 10.1128/MCB.00698-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci USA. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hake SB, et al. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- 17.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 18.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 19.Jin C, et al. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Placek BJ, et al. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J Virol. 2009;83:1416–1421. doi: 10.1128/JVI.01276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutcliffe EL, et al. Dynamic histone variant exchange accompanies gene induction in T cells. Mol Cell Biol. 2009;29:1972–1986. doi: 10.1128/MCB.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura T, et al. Inducible deposition of the histone variant H3.3 in interferon-stimulated genes. J Biol Chem. 2009;284:12217–12225. doi: 10.1074/jbc.M805651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodl M, Basler K. Transcription in the absence of histone H3.3. Curr Biol. 2009;19:1221–1226. doi: 10.1016/j.cub.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 24.Sakai A, Schwartz BE, Goldstein S, Ahmad K. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr Biol. 2009;19:1816–1820. doi: 10.1016/j.cub.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 26.Sartorelli V, Caretti G. Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr Opin Genet Dev. 2005;15:528–535. doi: 10.1016/j.gde.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang Y, et al. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat Struct Mol Biol. 2006;13:921–929. doi: 10.1038/nsmb1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wunsch AM, Lough J. Modulation of histone H3 variant synthesis during the myoblast-myotube transition of chicken myogenesis. Dev Biol. 1987;119:94–99. doi: 10.1016/0012-1606(87)90210-7. [DOI] [PubMed] [Google Scholar]
- 29.Bergstrom DA, et al. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- 30.Bean C, et al. The Ankrd2, Cdkn1c and calcyclin genes are under the control of MyoD during myogenic differentiation. J Mol Biol. 2005;349:349–366. doi: 10.1016/j.jmb.2005.03.063. [DOI] [PubMed] [Google Scholar]
- 31.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 32.Kim HJ, Seol JH, Han JW, Youn HD, Cho EJ. Histone chaperones regulate histone exchange during transcription. EMBO J. 2007;26:4467–4474. doi: 10.1038/sj.emboj.7601870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laybourn PJ, Kadonaga JT. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- 34.Lee H, Habas R, Abate-Shen C. Msx1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- 35.Braunschweig U, Hogan GJ, Pagie L, van Steensel B. Histone H1 binding is inhibited by histone variant H3.3. EMBO J. 2009;28:3635–3645. doi: 10.1038/emboj.2009.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldhamer DJ, et al. Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development. 1995;121:637–649. doi: 10.1242/dev.121.3.637. [DOI] [PubMed] [Google Scholar]
- 37.Couldrey C, Carlton MB, Nolan PM, Colledge WH, Evans MJ. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum Mol Genet. 1999;8:2489–2495. doi: 10.1093/hmg/8.13.2489. [DOI] [PubMed] [Google Scholar]
- 38.Tapscott SJ, Lassar AB, Weintraub H. A novel myoblast enhancer elements mediates MyoD transcription. Mol Cell Biol. 1992;12:4994–5003. doi: 10.1128/mcb.12.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adkins W, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 40.Gasser SM. Positions of potential: Nuclear organization and gene expression. Cell. 2001;104:639–642. doi: 10.1016/s0092-8674(01)00259-8. [DOI] [PubMed] [Google Scholar]
- 41.Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: The “H3 barcode hypothesis”. Proc Natl Acad Sci USA. 2006;103:6428–6435. doi: 10.1073/pnas.0600803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng RK, Gurdon JB. Maintenance of epigenetic memory in cloned embryos. Cell Cycle. 2005;4:760–763. doi: 10.4161/cc.4.6.1743. [DOI] [PubMed] [Google Scholar]
- 44.Sillje HH, Nigg EA. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr Biol. 2001;11:1068–1073. doi: 10.1016/s0960-9822(01)00298-6. [DOI] [PubMed] [Google Scholar]
- 45.Ye X, et al. Downregulation of Wnt signaling is a trigger for formation of facultative heterochromatin and onset of cell senescence in primary human cells. Mol Cell. 2007;27:183–196. doi: 10.1016/j.molcel.2007.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu L, Skoulchi AI. Coordinating cell proliferation and differentiation. Curr Opin Genet Dev. 2001;11:91–97. doi: 10.1016/s0959-437x(00)00162-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.