Abstract
Posttranscriptional regulation is emerging as a key factor in glucocorticoid (GC)-mediated gene regulation. We investigated the role of the human glucocorticoid receptor (GR) as an RNA-binding protein and its effect on mRNA turnover in human airway epithelial cells. Cell treatment with the potent GC budesonide accelerated the decay of CCL2 mRNA (t1/2=8±1 min vs. 62±17 min in DMSO-treated cells) and CCL7 mRNA (t1/2=15±4 min vs. 114±37 min), but not that of CCL5 mRNA (t1/2=231±8 min vs. 266±5 min) in the BEAS-2B cell line. This effect was inhibited by pre-incubation with an anti-GR antibody, indicating that GR itself plays a role in the turnover of these transcripts. Co-immunoprecipitation and biotin pulldown experiments showed that GR associates with CCL2 and CCL7 mRNAs, but not CCL5 mRNA. These methods confirmed CCL2 mRNA targeting by GR in human primary airway epithelial cells. Association of the GR was localized to the 5’UTR of CCL2 mRNA, and further mapped to nucleotides 44–60. The collection of transcripts associated with GR, identified by immunoprecipitation of GR-mRNA complexes followed by microarray analysis, revealed 479 transcripts that associated with GR. Computational analysis of the primary sequence and secondary structures of these transcripts yielded a GC-rich motif, which was shown to bind to GR in vitro. This motif was used to predict binding of GR to an additional 7889 transcripts. These results indicate that cytoplasmic GR interacts with a subset of mRNA through specific sequences and can regulate turnover rates, suggesting a novel posttranscriptional role for GR as an RNA-binding protein.
Keywords: Glucocorticoid receptor, messenger RNA, RNA-binding protein, posttranscriptional regulation, inflammation, airway epithelium
Introduction
Synthetic glucocorticoids (GC)3 are the mainstay in the treatment of allergic and inflammatory diseases due to their potent anti-inflammatory effect. The anti-inflammatory action arises from a complex mechanism of action, in which innate and adaptive immune functions are modulated through changes in gene expression for critical immune and pro-inflammatory genes such as TNF-α, granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukins and chemokines (1). When used in the treatment of allergic airway diseases such as asthma and rhinitis, the effect of GC is deemed critical due to their efficacy in inhibiting the mucosal inflammation. This effect is chiefly driven by the inhibition of epithelial-derived cytokines and chemokines that support the recruitment and activation of eosinophils and other inflammatory cells (2).
The mechanism of GC–mediated gene regulation entails an integrated set of modifications spanning from early signaling events to posttranscriptional processes occurring in the cytoplasm, well after transcriptional changes have been implemented. In this context, modulation of gene transcription is considered to be the central feature of GC-mediated gene regulation (3). Binding of GC to the glucocorticoid receptor (GR) induces its dissociation from a cytoplasmic multimeric complex of chaperone proteins and its translocation to the nucleus, where it dimerizes and acts as a transcription factor, via binding to a glucocorticoid response element (GRE) within the 5′ promoter region of target genes (4, 5). While this mechanism predominantly drives gene transactivation (6), protein-protein interaction between GR and a host of transcription factors - such as AP-1, NF-κB subunits and members of STAT and Forkhead box families - chiefly results in transcriptional inhibition of proinflammatory genes and modulation of the adaptive immune responses (3, 5).
Post-transcriptional gene regulation (PTR) has a pivotal physiological role in the immune response and in key phases of the establishment and resolution of inflammation (7). PTR regulates the rate of mRNA transport, turnover and translation, thus ultimately determining the duration and amplitude of the elicited cellular response (8). Among these mechanisms, the control of mRNA turnover is critical in regulating the levels of inflammatory and immune-mediated gene expression (9–11). As a result of seemingly small changes, such as doubling in mRNA half-life, 100-1000-fold differences in mRNA abundance can occur (12, 13). The effect of GCs has been increasingly recognized to occur not only through transcriptional control, but also through regulation of mRNA turnover (14). Genes regulated by GCs via PTR include TNFα, GM-CSF, COX-2, IL-4Rα, IL-6, iNOS, vascular endothelial growth factor (VEGF), CCL11, CCL2, CCL7 and many others (2, 14–16).
Since their generation, mRNA molecules are carried throughout the different cellular compartments within ribonucleoprotein (RNP) complexes, in dynamic association with a host of RNA-binding proteins (RBP). These regulatory factors bind to conserved regulatory cis-elements shared by subsets of transcripts, and direct the bound targets either towards the ribosomal assembly for translation, or towards cytoplasmic foci dedicated to translational silencing and mRNA decay, such as stress granules and P bodies (17–19). Multiple regulatory mRNA regions, defined as USER (Untranslated Sequence Elements for Regulations), are preferentially located in the mRNA untranslated regions (UTR), particularly – but not exclusively – in the 3’UTR (10, 18, 20). The adenylate-uridylate-rich elements (ARE) are the most conserved USER for immune genes (21), but research in the last decade has identified a growing number of additional non-ARE elements within the 5’ and 3’UTR of many immune genes, such as IL-2, IL-4 and more (20, 22, 23). While some RBPs, such as the ubiquitous HuR, mainly act as a positive regulator of mRNA stability, others, such as the zinc finger RBP tristetraprolin (TTP) limits the inflammatory response by accelerating the mRNA decay of its targets (24). Importantly, GC induce TTP expression and loss of TTP significantly affects GC-mediated gene regulation in vitro (15, 25), underscoring the importance of PTR in the mechanism of anti-inflammatory action of GC.
Proteins functionally characterized as transcription factors, such as the GR, and RBPs have several features in common, such as stimulus- or ligand-induced nucleocytoplasmic shuttling function and the ability to coordinately regulate multiple genes through binding to conserved nucleic acid sequences. Some regulatory factors can bind to both DNA and RNA, thus influencing both transcription and mRNA fate. For instance, the zinc finger protein NF-90 can regulate transcription of IL-2, but can also bind to and stabilize the IL-2 mRNA (26). Recently, it was shown in rat smooth muscle cells that the GR can interact with CCL2 mRNA, and that its presence in the RNP complex is necessary for its degradation to occur in a cell-free mRNA stability assay (27). This work identified a novel role of GR in mRNA turnover, but raised questions about the scope and mechanisms underlying this process.
As we have previously shown that RBPs, such as TTP, are critical for GC-mediated gene regulation, we sought to investigate whether the GR would act as an RBP in human airway epithelial cells treated with GC. According to an established paradigm, RNA-binding proteins can coordinately regulate subsets of functionally related transcripts that share a common recognition motif (18). In this report, we first identified an interaction between GR and the CCL2 and CCL7 mRNAs by immunoprecipitation of ribonucleoprotein complex (RNP-IP) and biotin pull-down in the human airway epithelial cell line BEAS-2B. With these methods, we confirmed CCL2 mRNA association with GR in human, non-transformed airway epithelial cells as well. In the case of CCL2, the binding site was mapped to the 5’UTR of the transcript. Furthermore, we aimed at identifying the full complement of transcripts that could bind to GR using a ribonomic approach, and computationally searched for a common GR recognition motif within the identified GR target transcripts. We demonstrate that the GR in human lung epithelial cells is capable of associating with almost 500 transcripts, and we validated such association for a subset of them. Finally, we identified by computational analysis a novel, GC-rich motif, for which we confirmed GR association, present in the 5’ UTRs of 7889 predicted mRNA targets, or in the entire sequences of 25,672 predicted mRNA targets (21% of the UniGene transcript pool). These data indicate that the GR can mediate GC action beyond its nuclear functions in transcriptional gene control, and that it may directly participate, via association with mRNA, in GC-mediated control of cytoplasmic posttranscriptional mechanisms of gene expression.
Methods
Cell culture
The BEAS-2B cell line is derived from human tracheal epithelium transformed by an adenovirus 12-SV40 hybrid virus (28) (ATCC, Manassas, VA). Cell cultures were maintained in F12/DMEM (Gibco/Invitrogen, Frederick, MD) containing 5% heat-inactivated fetal calf serum, 2 mM L-glutamine, penicillin (100 U/ml) and streptomycin (100 mg/ml) (29). Cells were cultured at 37°C in humidified air containing 5% CO2. BEAS-2B cells were used from passages 36 to 45. Human Primary bronchial epithelial cells (PBEC) were isolated by pronase digestion from bronchi of cadaveric lungs, as described (29). PBECs were cultured on collagen-coated flasks and maintained in serum-free LHC-9 medium (Biofluids, Rockville, MD). Cultures of PBEC were used during the first passage only. All cells were cultured at 37°C in humidified air containing 5% CO2.
Generation of biotinylated transcripts
Full-length CCL2, CCL7, and CCL5 transcripts were generated by amplifying cDNA from BEAS-2B cells using forward primers that contained a T7 RNA polymerase promoter sequence [CCA AGC TTC TAA TAC GAC TCA CTA TAG GGA GA] as described (30). A complete list of primers is shown in supplemental table S1. Briefly, biotinylated transcripts were synthesized from cDNA in the presence of a 5:1 ratio of CTP to biotin-CTP, using the MAXIscript in vitro transcription kit (Ambion) per the manufacturer's instructions. Fragments corresponding to nucleotides 10–26, 27–43, and 44–60 were chemically synthesized with a biotin group at the 5' end (Integrated DNA Technologies).
Cell-free mRNA decay assay
BEAS-2B cells were grown to 85% confluence, treated with 10−7 M budesonide (bud) or DMSO for 3 h, and lysed with polysomal lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM Hepes pH 7.0, 0.5% NP-40) for 10 min at 4°C. Assessment of decay of biotinylated RNA following incubation with protein lysate has been described previously (27, 31). Briefly, two µg of total protein from cytoplasmic cell lysate was mixed with biotinylated RNA (1 µg) and incubated for increasing time points up to 30 min. For experiments utilizing GR-specific antibody to inhibit transcript degradation, cytoplasmic cell lysate (2 µg) was incubated with three µg of a mouse monoclonal antibody against the GR (H-300 clone, Santa Cruz) or rabbit IgG (Santa Cruz) for 30 min prior to the addition of biotinylated RNA. Samples were separated on a 2% agarose gel and transferred to a nylon membrane by capillary transfer. RNA was fixed to the membrane by exposure to UV light, blocked with 3% bovine serum albumin (Sigma) in TBS-T (20 mM Tris pH 7.4, 150 mM NaCl, 0.1% Tween-20) for 60 min, and incubated with a 1:5000 dilution of streptavidin-horse radish peroxidase (SA-HRP, GE healthcare) for 60 min at room temperature. Membranes were developed with chemiluminescence substrate and exposed to film.
Immunoprecipitation of ribonucleoproteins (RNP-IP) with an anti-GR antibody
BEAS-2B cells were grown to 85% confluence and lysed by the addition of PLB supplemented with RNAse inhibitor (10 U/µl final concentration, New England Biolabs) and protease inhibitors (Complete Mini, EDTA-free, Roche) as described (25). For isolation of GR-associated transcripts, cytoplasmic lysates obtained from 1.6×107 cells/condition were generated as described (25) and verified to be free of nuclear contamination by Western blot analysis of cytoplasmic (β-tubulin, Santa Cruz Biotechnology) and nuclear proteins (PCNA, HNRNP C1/C2, Santa Cruz Biotechnology). Cytoplasmic lysates were mixed with protein A sepharose beads (Sigma) coated with 30 µg of a specific GR antibody (H-300 clone) or rabbit IgG. Beads were washed three times with NT2 buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl2, 1 mM MgCl2, and 0.05% Nonidet P-40), incubated with 20 U of RNase-free DNase I (15 min at 30°C), and further incubated in 100 µl NT2 buffer containing 0.1% SDS and 0.5 mg/ml proteinase K (30 min at 55°C). RNA was extracted by the addition of acid phenol-chloroform (Ambion), followed by the addition of 3M sodium acetate, pH 5.2 (10% v/v) and precipitation with ethanol. Isolated total RNA was treated with DNase and was reverse transcribed using MuLV reverse transcriptase and oligo dT primers (Applied Biosystems). Expression of gene targets was obtained using real-time qPCR employing SYBR green PCR master mix (Applied Biosystems) and specific primers shown in supplemental Table S1. For global analysis of GR-mRNA interactions, 5×107 cells/condition were lysed and RNP complexes were immunoprecipitated as described above. Isolated RNA was reverse-transcribed and hybridized to an Illumina ref-8 v3 microarray as described (32, 33). Data were extracted with the BeadStudio software v3.4.0 (Illumina) and normalized by Z score transformation to calculate differences in signal intensities. The data were calculated from four independent experiments, and significant values were tested using a two-tailed Z test and P < 0.01. Array data was uploaded to the GEO database (http://www.ncbi.nlm.nih.gov/geo/; Accession no. GSE24748. For single-gene RNP-IP, the specific association of mRNA with GR was calculated as the difference of PCR cycles (CT) between the anti-GR antibody IP and the isotype-matched control antibody IP (ΔCT), and quantified as fold enrichment of mRNA in the GR IP over the control IP (2−(ΔCT)) as described (25).
RNA biotin pulldown
Biotinylated transcripts (1 µg) were mixed with BEAS-2B cell lysates (40 µg) for 30 min at room temperature in TENT buffer (10 mM Tris (pH 8.0), 250 mM NaCL, 0.5% v/v Triton X-100) and incubated with streptavidin-dynabeads (Invitrogen) for 30 min as previously described (30). The beads were washed three times with cold PBS and protein was eluted by boiling in 1% SDS for 4 min. Samples were loaded onto an 8% acrylamide gel, separated by SDS-PAGE, and analyzed by Western blot using the H-300 GR antibody.
Electrophoretic mobility gel shift assays (EMSA) and RNA cross-linking
Gel shift assays were performed by incubating purified GR (25 ng) (34) in reaction buffer (20 mM HEPES pH7.4, 200 mM NaCl, 1 mM DTT, 5 mM MgCl2, 2.5% glycerol, 0.05% NP-40) with poly (dI-dC) at a final concentration of 0.05 mg/ml for 5 min at room temperature. Biotinylated RNA (0.75 ng) was added, incubated at room temperature for 30 min, and the products were subjected to a native 6% polyacrylamide gel in 0.5X TBE (45 mM Tris, 45 mM Boric acid, 0.1 mM EDTA) for 1 h at 100 V at 4° C. The DNA was transferred to a nylon membrane using a Trans-blot semi-dry transfer apparatus (Bio-Rad) at 19 V for 60 min. RNA was fixed to the membrane by exposure to UV light. The membrane was blocked and developed using the Pierce chemiluminsecent nucleic acid detection module (Thermo Scientific) per manufacturer's instructions.
For detection of GR-mRNA interaction by crosslinking, purified GR (1.8 ng) (kindly provided by Dr. K. Pearce, GlaxoSmithKline, Inc.) was mixed with each RNA oligonucleotide (2 µg) for 30 min at room temperature and then subjected to UV light (120 mJ/cm2). Samples were separated by SDS-PAGE on an 8% gel under denaturing and reducing conditions, and a Western blot was performed to detect GR (25).
Computational analysis
The top ~500 human transcripts enriched in the IP were utilized as the experimental dataset (Supplementary Table 1) for the computational identification of the GR motif, as described for the identification of other RBP motifs (32, 35, 36). The transcript sequences of the experimental dataset were obtained from the recent UniGene database and repetitive sequences were removed using RepeatMasker (www.repeatmasker.org). The complete, high-quality 5′UTR sequences were selected from the dataset for further analysis. The selected sequences were first divided into 100-base-long subsequences with a 50-base overlap between consecutive sequences and were organized into 50 sub-datasets. Common RNA motifs in each of the 50 sub-datasets were then identified, respectively. The top 10 candidate motifs from each sub-dataset were selected and used to build the stochastic context-free grammar (SCFG) model, which summarizes the folding, pairing and additional secondary structure. Next, the SCFG model of each candidate motif was used to search against the experimental 5′UTR dataset as well as the entire UniGene 5′UTR dataset to obtain the number of hits for each motif. The motif with the highest frequency of hits per kb in the experimental dataset when compared to the entire UniGene mRNA sequence database was considered to be the top GR candidate motif. The identification of the RNA motif in unaligned sequences of each sub-dataset was conducted using FOLDALIGN software (37), and the constucture of the SCFG model and the search against the transcript dataset were conducted using the COVE and COVELS software packages (38). The motif logo was constructed using WebLogo (http://weblogo.berkeley.edu/). RNAplot was used to depict the secondary structure of the representative RNA motifs. The computation was performed using the NIH Biowulf computer farm.
Statistical analysis
Results from cell-free RNA decay and Western blot analysis were quantified by densitometry. All data were subjected to t-test analysis and when appropriate, to ANOVA with multiple post-hoc testing; p<0.05 was used for statistical difference.
Results
GCs accelerate the decay of CCL2 and CCL7 mRNAs
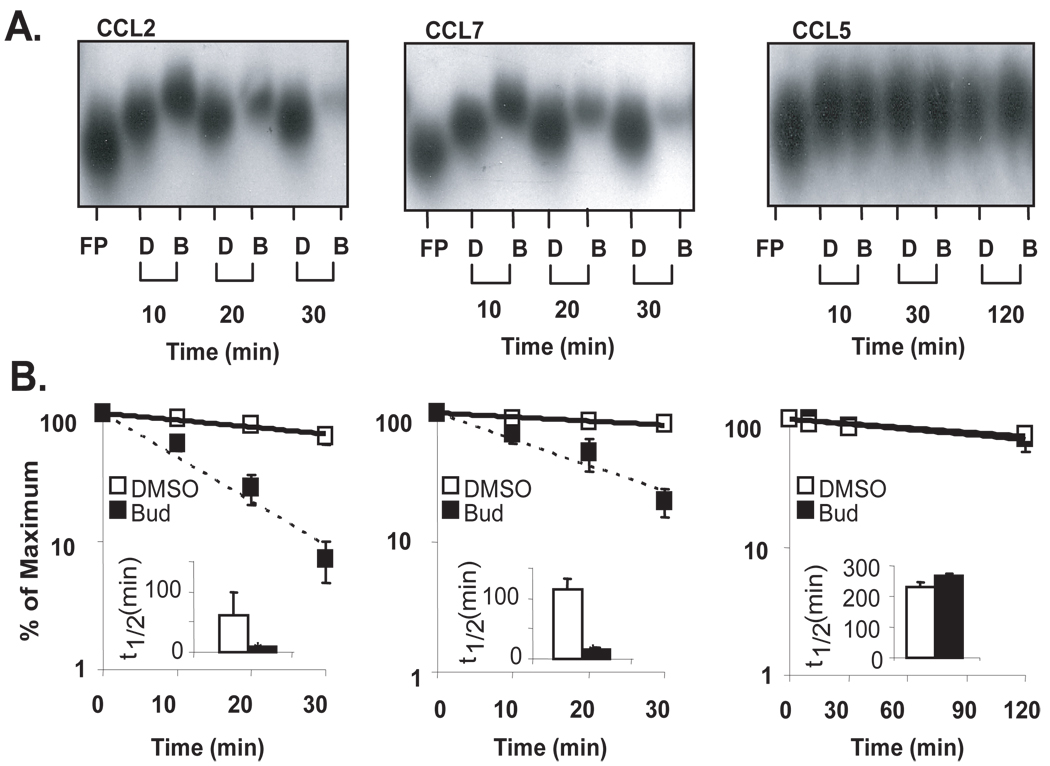
We have previously shown that GCs selectively accelerate the decay of numerous transcripts that are targeted by TTP, an RBP that is induced also by GC (25). Using the transcriptional inhibitor actinomycin D, we identified CCL2 and CCL7 mRNAs as transcripts whose decay was accelerated by GC treatment, while the expression of the CCL5 mRNA, which does not contain AREs, was inhibited by GC without changes in half-life (25). Use of transcriptional inhibitors has disadvantages, as the global blockade of gene transcription can prevent the synthesis of components necessary in mRNA decay, diminishing the observed effects on mRNA turnover. To integrate these findings, we probed the role of GR in the effect of GCs on mRNA stability using a cell-free assay (27, 31), in which we followed the degradation of labeled, full-length synthetic transcripts spanning the human CCL2, CCL7, and CCL5 mRNA after incubation with airway epithelial cell lysates (Fig. 1A). The biotinylated mRNAs were mixed with cytoplasmic lysates (2 µg) obtained from BEAS-2B cells treated with budesonide (10−7 M) or DMSO (diluent for budesonide) for 3 hours. Samples were incubated at RT for increasing times, separated by PAGE, and mRNA was detected by streptavidin linked to horseradish peroxidase. Decay curves were generated based on the densitometric reading of the bands (Fig. 1B), allowing the determination of half-lives (Fig. 1B, bargraph insets). Compared to the DMSO-treated lysates, the GC-treated lysates significantly accelerated the decay of CCL2 mRNA (t1/2= 8 ± 1 min compared to 62 ± 17 min in DMSO-treated cells, p<0.05, n=3) and CCL7 mRNA (t1/2= 15 ± 4 min compared to 114 ± 37min, p<0.05, n=3), but not of CCL5 mRNA (t1/2= 231 ± 8 min compared to 266 ± 5 min, n=3), in agreement with data obtained using actinomycin D assay (25).
Figure 1. Glucocorticoids promote selective degradation of CCL2 and CCL7 mRNAs in a cell-free assay of mRNA decay.
A. Non-denaturing PAGE (representative of n=3) of in vitro transcribed, biotinylated human CCL2, CCL7, or CCL5 full-length mRNAs alone [lane 1, Free Probe (FP)] or mixed with cytoplasmic lysates of BEAS-2B cells (pre-treated with budesonide (B, 10−7 M) or DMSO (D) for 3h) for the indicated times. B. Densitometric measurement of the amount of mRNA at each time point; mean ± SEM, n=3. Inset bargraphs: Half-lives of chemokine mRNA in each condition (calculated from plots in B, as Ln(0.5)/slope, indicating the time at which 50% of the initial mRNA is left); * p<0.05.
GR plays a role in CCL2 and CCL7 mRNA turnover
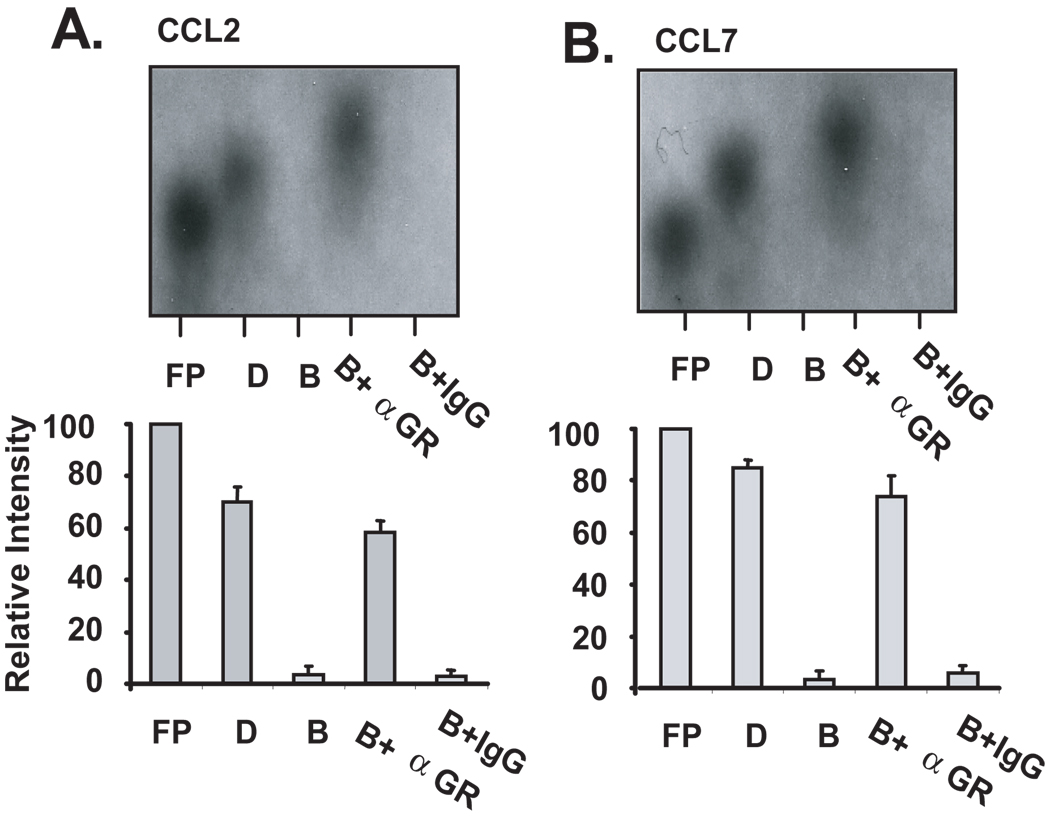
The results obtained with the cell-free mRNA decay assay suggested the association of regulatory proteins with mRNA. In fact, a mobility shift was observed for both CCL2 and CCL7 mRNA probes after incubation with Bud-treated cell lysates (Fig. 1A), indicating the possible association of proteins, among which we hypothesize to be the GR. To investigate the presence of GR in this complex and its potential role in mediating the observed changes in mRNA decay, the cell-free mRNA decay assay was performed following a preincubation with a mouse monoclonal antibody against the GR (Fig. 2) or an isotype control antibody. Consistent with the results shown in Figure 1, cytoplasmic lysates from budesonide-treated BEAS-2B cells (n=3) induced the degradation of CCL2 (Fig. 2A, lane ‘B’) and CCL7 mRNA (Fig 2B, lane ‘B’). However, a 30-min pre-incubation of the cytoplasmic lysates with 3 µg of the anti-GR Ab prevented the decay of both transcripts (Fig. 2A and 2B, Lane ‘B+α GR’), while the control antibody had no effect on GC-induced degradation (Lane ‘B+IgG’). These data indicate that GR is present in the protein complex associated with the chemokine RNA and participates in the GC-induced increase in chemokine mRNA decay, as anti-GR reversed this action, possibly by blocking interactions of GR with other proteins of the mRNA degradation machinery.
Figure 2. Treatment of BEAS-2B cell lysates with a specific antibody against hGR inhibits GC-mediated decay of CCL2 and CCL7 mRNAs.
Non-denaturing PAGE (representative of n=3) of biotinylated human CCL2 (A) or CCL7 (B) mRNA incubated with cytoplasmic lysates (2 µg) of DMSO-(D) or budesonide-treated (B) BEAS-2B cells. Budesonide-treated lysates were pre-incubated with 3 µg of an anti-GR antibody (B+αGR) or isotype control antibody (B+IgG) for 30 minutes before addition of labeled RNA. Samples were further incubated for 30 minutes and then subjected to PAGE. The relative intensity of each band compared to the free probe was determined by densitometry (shown in bar graphs below is densitometric mean ± SEM of n=3 for each chemokine).
Association of GR with endogenous CCL2 and CCL7 mRNAs
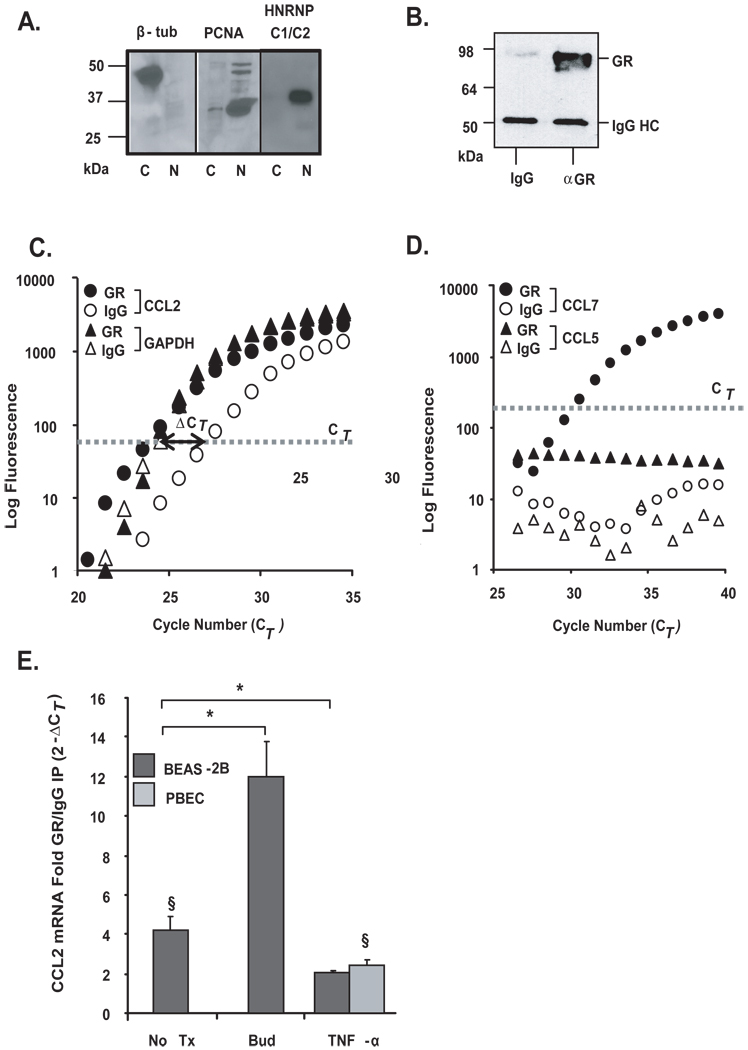
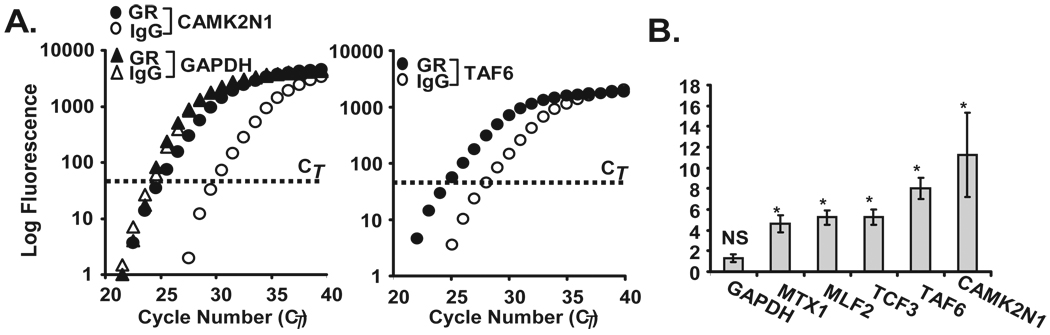
In light of the observed GR-mediated effect on chemokine mRNA decay, we investigated whether the GR would associate with the endogenous CCL2 and CCL7 mRNAs by RNP-IP analysis (39), similarly to what we reported for TTP (25). BEAS-2B cytoplasmic lysates were verified to be free of nuclear proteins by Western blot analysis (Fig. 3A), and were incubated with the anti-GR antibody to immunoprecipitate endogenous GR-mRNA complexes, or with an IgG isotype control to measure background binding (Fig. 3B). Higher levels of CCL2 mRNA were consistently detected in the GR-IP over the control-IP (Fig. 3C), with an overall statistically significant 4.1 fold enrichment of CCL2 mRNA in the GR-IP relative to the control-IP, indicating a specific association of GR with the cellular CCL2 mRNA (Fig. 3C). No enrichment of GAPDH mRNA was noted, which was expected as this gene is known to not be a target of several other RNA binding proteins (Fig. 3C). No CCL7 mRNA was detected in the isotype IgG control-IP likely due to low baseline expression in BEAS-2B, but CCL7 mRNA was detected in the GR-IP, indicating that this transcript also associates with GR (Fig. 3D). Importantly, no CCL5 mRNA was detected in either the transcript pool of either the GR- or control-IP (Fig. 3D) despite detectable baseline levels, in line with the previous results suggesting the absence of a direct posttranscriptional regulation of CCL5 by GC (16). We then explored whether association of GR with CCL2 would be susceptible to modulation. In BEAS-2B cells, incubation with budesonide (10−7 M for 30 min) substantially increased the enrichment of CCL2 mRNA in the GR-IP (12.0-fold vs. IgG IP), while treatment with TNFα (10 ng/ml for 3 h) showed a level of CCL2 mRNA still significantly lower (2.0-fold enrichment) than in the unstimulated group.
Figure 3. Association of chemokine transcripts with GR in airway epithelial cells.
A. The purity of cytoplasmic lysates was verified by Western blot analysis of a cytoplasmic protein [β-tubulin (β-tub)] and nuclear proteins (PCNA, HNRNP C1/C2), detected exclusively in cytoplasmic (c) and nuclear (n) extracts, respectively. B. Western Blot analysis of GR after immunoprecipitation (IP) of protein-mRNA complexes obtained using the monoclonal anti-GR antibody (αGR) or the IgG-control antibody control (IgG), showing selectivity of the IP. Bands corresponding to GR and IgG heavy chain (IgG HC) are indicated. C. Real-time PCR amplification plot of CCL2 and GAPDH mRNA (representative of n=10) and D. of CCL7 and CCL5 mRNA (representative of n=5), both after GR (filled circles) or IgG control (open circles) IP. The bold line with arrow points indicates the difference in Ct (ΔCT) between the CCL2 mRNA detected in the GR-IP versus the IgG-IP. E. Mean ± SEM fold GR/IgG IP enrichment for CCL2 mRNA (expressed as 2−ΔCT) in BEAS-2B and PBEC cells unstimulated (n=10) or treated with budesonide (bud, n=4) or TNFα (n=3). * p < 0.03 for treated samples vs. untreated, § p < 0.03 vs. GR-IP control.
In non-transformed PBEC, CCL2 mRNA expression at baseline was at the limit of detection by real-time PCR (> 35 CT) and therefore was not detectable in any of the IP RNA pools isolated from unstimulated and budesonide-treated cells. However, as treatment with TNFα induced its expression, CCL2 mRNA became significantly enriched (2.5 fold) in the GR-IP vs. the IgG-IP in these cells.
Interaction of GR with in vitro transcribed CCL2 and CCL7 RNAs
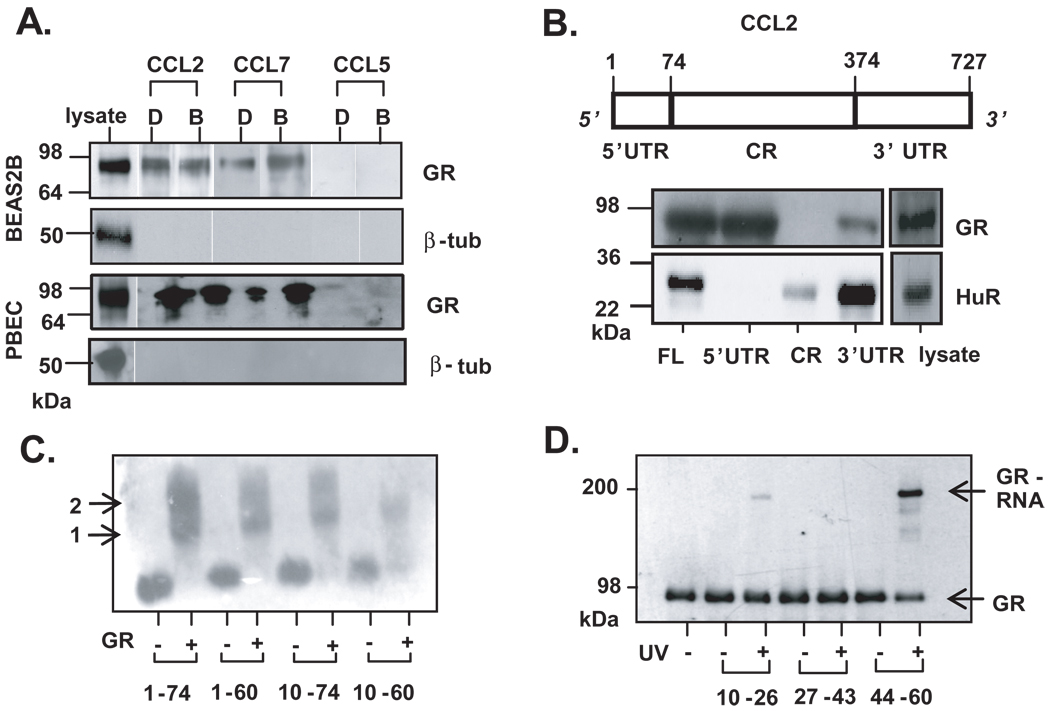
We confirmed the GR-mRNA association in both BEAS-2B (n=3) and PBEC (n=2) by an in vitro approach using a biotin pull-down assay (30). Figure 4A shows the detection of GR associated with the CCL2 and CCL7 transcripts, in either the absence or presence of budesonide treatment. No association of GR with CCL5 was observed, consistent with the results of the RNP-IP experiment.
Figure 4. Localization of GR binding on the 5’UTR of CCL2 mRNA.
A. Western blot analysis of biotin pull-down assay showing association of GR from cytoplasmic extracts of BEAS-2B (n=3) and PBEC (n=2) treated with budesonide (B, 10−7 M) or DMSO (D), incubated with biotinylated transcripts spanning human CCL2 or CCL7 full-length mRNA. In both cell types, lack of β-tubulin detection was used to rule out non-specific binding. B. Above: schematic of CCL2 mRNA. Indicated are the nucleotide numbers corresponding to untranslated regions (UTR) and coding region (CR). Below, detection of GR by Western blot after biotin pulldown assay from unstimulated BEAS-2B cell lysate, using biotinylated transcripts encompassing the CCL2 full-length RNA (FL), the 5’ and 3’ UTRs, or the CR. The RBP HuR is detected as positive control for association with CCL2 3’UTR. C. EMSA of purified, recombinant GR incubated with mRNA probes encompassing discrete regions of CCL2 5’UTR (indicated by nucleotide number). Arrows represent putative GR monomer (1) and dimer (2) binding. C. UV cross-linking of purified GR protein to 15-nt fragments of the CCL2 5’UTR, followed by detection by Western blot of the covalently-linked GR via mobility shift of the RNA-bound GR protein (GR-RNA).
Mapping of the GR-mRNA interaction site
Having established the association of GR with mRNAs in multiple independent assays, we sought to define the RNA regions involved in this interaction. Inflammatory genes are regulated posttranscriptionally mainly, but not exclusively, through 3’UTR, ARE-mediated processes (21). In the case of rat CCL2, a GC-responsive sequence was mapped on the 5’UTR, suggesting that this region could harbor a regulatory element involved in GR binding (31). We performed biotin pulldown experiments (n=3) to locate the binding site of GR using the full-length CCL2 mRNA, as well as truncated transcripts containing the 5'UTR, coding region (CR) and 3’UTR sections (Fig. 4B). Besides the expected association with full-length CCL2 mRNA, association of GR was observed predominantly with the 5'UTR construct, minimally with the 3’UTR, and it was absent with the probe encompassing the CR. As a control, we analyzed binding of each construct with HuR, an RNA binding protein known to associate with the 3'UTR of CCL2. As expected, HuR selectively associated with the 3’UTR and full-length mRNA (Fig. 4B).
To further map the GR binding site within the 5'UTR of CCL2, besides the full length 5’UTR [nucleotides (nt) 1–74] we synthesized additional 5' UTR truncations (nt 1–60, 10–74, and 10–60). As assessed by EMSA, purified (> 95%) (34), recombinant GR bound to the full length 5'UTR as well as to each of the specific regions (Fig. 4C), n=3. Two bands were visible using the probe spanning the full-length 5'UTR and those encompassing nucleotides 1–60 and 10–74, indicating that GR may bind as a dimer. The double band is not observed in association with the 10–60 nt probe. The region corresponding to nucleotides 10–60 was further fragmented, and 15 nt-long sequences spanning nt 10–26, 27–43, and 44–60 were synthesized. Each oligonucleotide was mixed with GR and subjected to UV light, which induces a protein-RNA covalent crosslink if an interaction is present (n=2). The protein-RNA linkage, identified as an electrophoretic mobility shift by Western blot using an anti-GR antibody, was observed for fragment 44–60 (Fig 4D, ‘GR-RNA’).
Profiling of transcripts associated with GR by gene array analysis
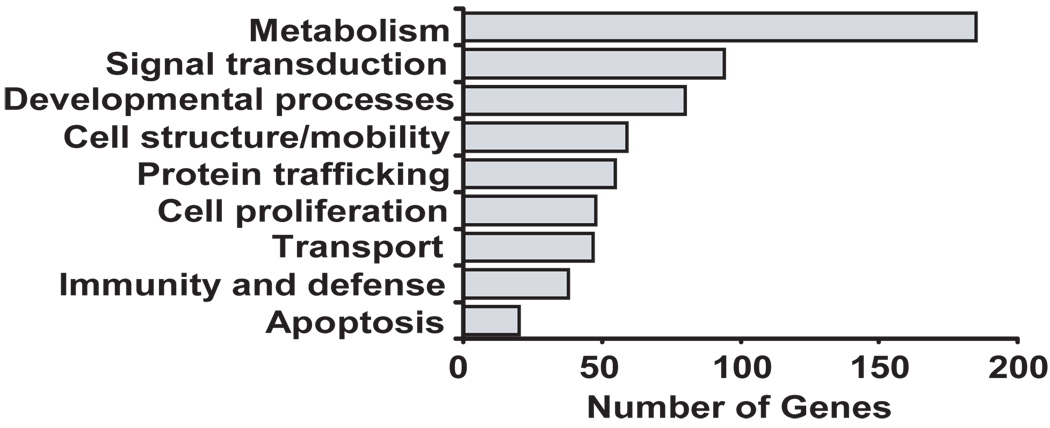
Based on the data described, the GR appears to function as an RNA-binding protein in the cytoplasm, associating with select mRNAs and participating in the GC-induced acceleration of mRNA decay. One characteristic of RBPs is their ability to associate with discrete subsets of mRNAs through shared regulatory elements in their UTRs, as demonstrated for HuR, TIA, TIAR, TTP, NF-90 and others (32, 33, 35, 36, 40). Therefore, we aimed at determining the full complement of transcripts associated with GR and to identify the sequence motif shared by the transcripts associated with the GR. To do so, RNA was isolated from a set of RNP-IP experiments using the GR and control IgG antibodies (n=4) in unstimulated BEAS-2B cells, and the global profile of GR targets was identified by hybridization of the corresponding cDNAs to an Illumina human cDNA array containing more than 25,000 genes. A comparison of signal in GR-IP arrays versus IgG control-IP arrays yielded 477 transcripts with Z ratios of >1.5, indicating their specific association with GR (full array data is available at the GEO website as described in the methods section). A list of the transcripts most enriched by GR co-IP is shown in Table 1 (Full list provided as Supplemental Table 2). In agreement with the enrichment of CCL2 mRNA we observed by real-time PCR, this transcript was also found to be enriched in the GR-IP array. The GR association was validated for five (out of five tested) of the most enriched targets (MTX1, MLF2, TCF3, TAF6, and CAMK2N1) and found to be statistically significant by single-gene RNP-IP analysis (Fig. 5) (n=3, p<0.05 compared to IgG control IP). Besides CAMK2N1, which displayed the highest enrichment, the other four targets were chosen without a z-ratio bias to better validate the range of significant enrichment. Gene ontology analysis demonstrated a wide variety of functional classes of interacting transcripts, covering areas such as development, signal transduction and metabolism (Fig. 6).
Table 1.
Top GR-associated transcripts by RNP-IP (full list in Supplemental Table 2).
| Name | Symbol | Accession No. | Z-ratio |
|---|---|---|---|
| Calcium/calmodulin-dependent protein kinase II inhibitor 1 | CAMK2N1 | NM_018584.5 | 4.46 |
| SMT3 suppressor of mif two 3 homolog 2 | SUMO2 | NM_001005849 | 3.95 |
| Uncoupling protein 2 | UCP2 | NM_003355.2 | 3.94 |
| Zinc finger protein 219 | ZNF219 | NM_016423.1 | 3.80 |
| Myeloid leukemia factor 2 | MLF2 | NM_005439.1 | 3.39 |
| Eukaryotic translation initiation factor 4H | EIF4H | NM_022170.1 | 3.36 |
| TAF6 RNA pol II, TATA box binding protein-associated factor | TAF6 | NM_005641.2 | 3.04 |
| Hepatoma-derived growth factor (high-mobility group protein 1-like | HDGF | NM_004494.1 | 3.04 |
| Transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | TCF3 | NM_003200.1 | 3.00 |
| Zinc finger protein 395 | ZNF395 | NM_018660.2 | 2.97 |
| Cyclin-dependent kinase inhibitor 2A | CDKN2A | NM_058195.2 | 2.95 |
| Anterior pharynx defective 1 homolog A (C. elegans) | APH1A | NM_001077628 | 2.94 |
| Programmed cell death 5 | PDCD5 | NM_004708.2 | 2.93 |
| Biglycan | BGN | NM_001711.3 | 2.91 |
| Cysteine-rich, angiogenic inducer, 61 | CYR61 | NM_001554.3 | 2.90 |
| WAS protein family, member 2 | WASF2 | NM_006990.2 | 2.88 |
| RAB43, member RAS oncogene family | RAB43 | NM_198490.1 | 2.87 |
| 1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) | AGPAT1 | NM_006411.2 | 2.85 |
| N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 | NDST1 | NM_001543.3 | 2.84 |
| Metallothionein 1X | MT1X | NM_005952.2 | 2.83 |
| Acylphosphatase 1, erythrocyte (common) type | ACYP1 | NM_001107.3 | 2.83 |
| Ribosomal protein L21 | RPL21 | NM_000982.3 | 2.83 |
| Neuroblastoma, suppression of tumorigenicity 1 | NBL1 | NM_005380.4 | 2.82 |
| Dpy-30 homolog (C. elegans) | DPY30 | NM_032574.2 | 2.81 |
| General transcription factor IIA, 2 | GTF2A2 | NM_004492.1 | 2.80 |
| Chemokine (C-C motif) ligand 2 | CCL2 | NM_002982.3 | 2.77 |
| Mindbomb homolog 2 (Drosophila) | MIB2 | NM_080875.1 | 2.75 |
| Striatin, calmodulin binding protein 4 | STRN4 | NM_001039877 | 2.70 |
| Calcium binding protein P22 | CHP | NM_007236.3 | 2.69 |
| Calpain, small subunit 1 | CAPNS1 | NM_001749.2 | 2.68 |
| Hypothetical protein HSPC152 | HSPC152 | NM_016404.1 | 2.64 |
| Mitochondrial ribosomal protein L34 | MRPL34 | NM_023937.2 | 2.64 |
| ADP-ribosylation factor 3 | ARF3 | NM_001659.1 | 2.63 |
| Splicing factor 3b, subunit 4 | SF3B4 | NM_005850.3 | 2.62 |
| Barrier to autointegration factor 1 | BANF1 | NM_003860.2 | 2.57 |
Figure 5. Validation of GR-associated targets identified by gene array.
A. Real-time PCR amplification plot (representative of n=3) after GR (filled circles) or control (open circles) IP for CAMK2N1and TAF6, identified as GR-associated transcripts by the GR-IP array study, and GAPDH as control. B. Bargraph represent the mean ± SEM fold GR/IgG IP enrichment for MTX1, MLF2, TCF3, TAF6, and CAMK2N1 (n=3; *, p<0.05 compared to IgG IP, NS=not significant for GAPDH).
Figure 6. Genome Ontology Analysis of GR-associated genes.
The analysis was performed on the full list of 477 putative GR targets (listed in full in the Supplemental Table2) identified by hybridization of unstimulated BEAS-2B cDNAs to an Illumina human cDNA array using GR-IP arrays versus IgG control-IP arrays.
Identification of a novel GR-mRNA binding motif and of putative GR targets bearing the motif
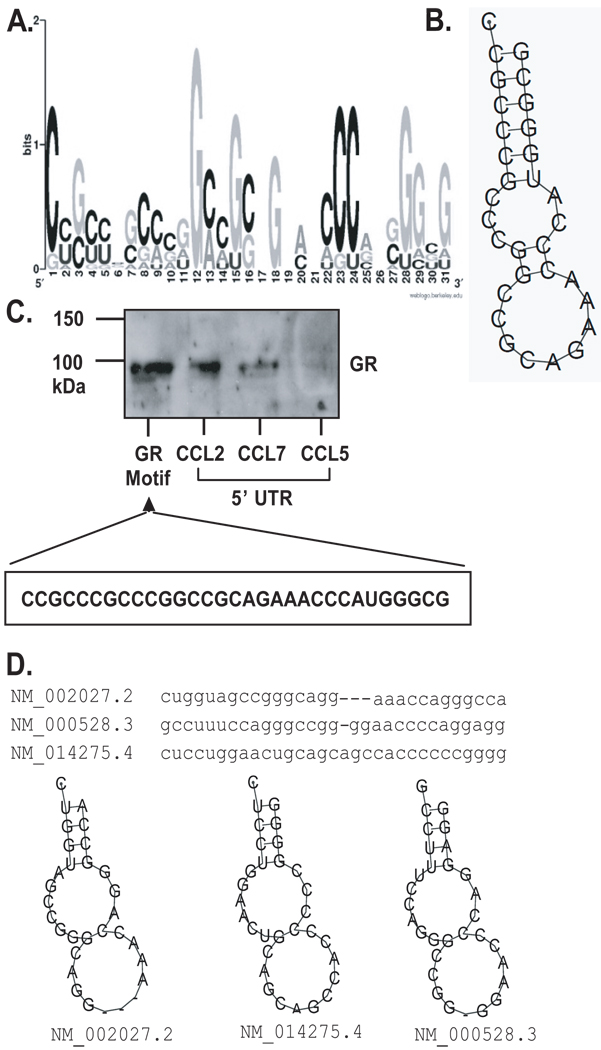
To investigate the presence of an RNA signature motif on the transcript for GR, the ~500 transcripts found to be specifically associated with GR were used as the experimental dataset for the computational analysis. The analysis searched for a common mRNA recognition motif among the transcript sequences of the experimentally determined genes based on both primary sequences and secondary structures, and then scanned the entire human transcriptome sequences (i.e. the UniGene database) for the putative target genes of the identified RNA signature motif of GA (see Materials and Methods). One motif spanning 27–31 nucleotides displayed the highest frequency of hits per kb in the experimental dataset when compared to the entire UniGene transcript database. Evaluation of the primary and secondary structure of the motif indicates a sequence highly enriched in G and C nucleotides (78% of the signature motif), forming two contiguous loops on a short stem (Fig. 7A and B). Association of GR with this motif was demonstrated via biotin pulldown assay using a biotinylated oligonucleotide corresponding to the motif, which was incubated with cytoplasmic lysate from unstimulated BEAS-2B cells. GR associated with the motif sequence, the 5’UTR of CCL2 and CCL7, but not the CCL5 5’UTR (Fig. 7C).
Figure 7. Primary sequence and secondary structure of the predicted GR mRNA motif.
A. Graphic logo representing the probability matrix of the GR motif, showing the relative frequency of each nucleotide for each position within the motif sequence. The motif is originated from the experimental data set from the array study. B. Secondary structure of the GR motif comprising the nucleotides with highest frequency for each position within the motif shown in A. C. Biotin pull-down assay showing association of GR from unstimulated BEAS-2B cell lysates with the GR motif shown in B., compared to GR association with the full-length 5’UTR of CCL2, CCL7 and CCL5 mRNA (the latter as negative control). D. Sequence and secondary structure of the GR motif in three mRNAs from the UniGene list of putative GR targets; the corresponding RefSeq accession numbers and names are shown.
Based on the structure of this motif, we conducted a search against the entire UniGene databases for identifying putative mRNA targets of GR. We firstly searched the 5’UTR sequences of UniGene transcripts, given that the identification of GR is associated with the 5’UTR of CCL2, and identified 7889 transcripts bearing the motif in their 5’UTR (45% of the entire database). We also searched full-length transcripts and found a total of 25672 mRNAs bearing the GR motif (21% of the entire database). Table 2 reports a selection of the sequences, listed in full in the Supplemental Table 3.
Table 2.
Putative GR mRNA targets bearing the GC-rich motif (full list in Supplemental Table 3).
| Unigene ID | Gene Name | Symbol | Highest Score * |
No. of hits |
|---|---|---|---|---|
| Hs#S2332203 | Pur-gamma A-form | PURG | 9.1 | 13 |
| Hs#S1726769 | AF4/FMR2 family, member 2 | AFF2 | 8.77 | 13 |
| Hs#S1731710 | nuclear receptor subfamily 2, group F, member 1 | NR2F1 | 8.13 | 12 |
| Hs#S3294 | c-sis/platelet-derived growth factor 2 | SIS/PDGF2 | 9.77 | 11 |
| Hs#S226081 | B-cell CLL/lymphoma 7A | BCL7A | 9.54 | 10 |
| Hs#S2332607 | APP beta-secretase mRNA | BACE1 | 9.45 | 10 |
| Hs#S24303175 | diphosphomevalonate decarboxylase | MVD | 9.26 | 10 |
| Hs#S16761125 | solute carrier family 8 member 3 | SLC8A3 | 8.62 | 9 |
| Hs#S4435678 | signal peptide peptidase 3 | SPPL3 | 8.85 | 8 |
| Hs#S21224286 | activity-dependent neuroprotector homeobox | ADNP | 8.74 | 7 |
| Hs#S25049596 | retrotransposon gag domain containing 4 | RGAG4 | 8.7 | 7 |
| Hs#S21504120 | family with sequence similarity 150, member B | FAM150B | 8.57 | 7 |
| Hs#S2139673 | vasohibin 1 | VASH1 | 8.13 | 7 |
| Hs#S1727370 | paired-like homeodomain 1 | PITX1 | 8.93 | 6 |
| Hs#S4838504 | glycogen synthase kinase 3 beta | GSK3B | 8.89 | 6 |
| Hs#S15970748 | Notch-regulated ankyrin repeat protein | NRARP | 8.61 | 6 |
| Hs#S1729405 | transmembrane protein 187 | TMEM187 | 8.36 | 6 |
| Hs#S2140082 | potassium channel tetramerisation domain containing 3 | KCTD3 | 8.01 | 6 |
| Hs#S1731173 | LIM domain binding 1 | LDB1 | 9.77 | 5 |
| Hs#S2293641 | GATA zinc finger domain containing 2A | GATAD2A | 9.77 | 5 |
| Hs#S3940069 | F-box protein 9 | FBXO9 | 8.71 | 5 |
| Hs#S15631530 | notum pectinacetylesterase homolog (Drosophila) | NOTUM | 8.67 | 5 |
| Hs#S3335589 | FERM-containing protein | CG1 | 8.57 | 5 |
| Hs#S3219251 | neuromedin B | NMB | 8.49 | 5 |
| Hs#S1731193 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 | PLOD3 | 8.43 | 5 |
| Hs#S3219580 | H2.0-like homeobox | HLX | 8.41 | 5 |
| Hs#S1732363 | Fas (TNFRSF6) associated factor 1 | FAF1 | 8.29 | 5 |
| Hs#S1726650 | dipeptidyl-peptidase 4 | DPP4 | 8.28 | 5 |
| Hs#S4285033 | roundabout, axon guidance receptor, homolog 1 | ROBO1 | 8.09 | 5 |
| Hs#S6664690 | chromodomain protein, Y-like | CDYL | 8.06 | 5 |
| Hs#S5860310 | leucine rich repeat neuronal 1 | LRRN1 | 8.02 | 5 |
The value of the score reflects how each motif matches the GR motif model (shown Figure 7.)
Discussion
Cytoplasmic function for GR: acceleration of mRNA decay
Our data support a novel role for the GR within the cytoplasmic cell compartment as an RNA-binding protein. We found that in GC-treated human airway epithelial cells, GR associates with CCL2 and CCL7 mRNA and is necessary for GC-induced acceleration of the chemokine mRNA decay. Moreover, we discovered the ability of cytoplasmic GR to associate with a discrete subset of mRNA and we identified, through a well-tested bioinformatic analysis of the newly identified GR targets, a novel GC-rich RNA motif mediating GR association with mRNA.
It is well established that the GRα, generated from the human GR gene by alternative splicing together with the GRβ isoform, acts as a ligand-dependent transcription factor and conveys, by transcriptional control of gene expression, the metabolic functions of the endogenous GC hormone and of the synthetic anti-inflammatory corticosteroids. Like many transcription factors and hormone receptors, the hGRα is subject to nucleocytoplasmic shuttling: in the absence of ligand, it resides mostly in the cytoplasm as part of a heterooligomeric complex containing heat shock proteins (HSPs) 90, 70 and 50, immunophilins, and other proteins (41). Ligand binding induces conformational changes of the protein complex leading to disassociation and nuclear translocation of GC, where GR dimerizes and regulates gene transcription (5). Similarly to the GR, many transcription factors and hormone receptors are subject to nucleocytoplasmic shuttling, induced by cell activation or ligand binding. Importantly, several of them carry out additional roles in gene regulation while stationing in the cytoplasmic compartment. The zinc finger protein NF-90 regulates transcription of IL-2 by binding to the NFAT site as well as directing IL-2 RNA export and localization (26). Moreover, NF-90 modulates mRNA stability and translation of a specific pool of transcripts via binding to a recently identified AU-rich motif in their 3'UTR (36, 42). Similar examples are the zinc-finger proteins YY1 and tristetraprolin (TTP) as well as nucleolin, all of which regulate both transcriptional and post-transcriptional events by association with DNA and transcription factors in the nucleus, and with RNA and other components of the mRNA decay and translation while in the cytoplasm (22, 40, 43–45).
In support of potential cytoplasmic functions for GR, earlier work reported that the GR is indeed able to bind to RNA (46, 47), specifically to the transfer RNA (tRNA) (48). More recently, Dhawan et al. described in rat smooth muscle cells, using biochemical approaches also used in our study - such as cell-free decay assay and RNP-IP - that the rat GR associated with CCL2 mRNA and that its presence in the RNP complex was necessary for GC-induced increase in CCL2 mRNA degradation (27). Our data build upon this initial observation, providing evidence of a specific GR interaction with CCL2 mRNA using multiple, established assays (RNP-IP, biotin pull-down, EMSA, UV-crosslinking, Figures 3 and 4). The differences in CCL2 mRNA enrichment in the GR-IP after GC or TNFα indicate that this is a dynamic process, and while further studies to define quantitatively the affinity of GR to transcripts and to fully characterize the RBP function of GR are warranted, the present data strongly support the existence of an as yet uncharted mechanism of GR-mediated posttranscriptional control that could integrate with its known role in transcriptional gene regulation, in order to deliver the full GC anti-inflammatory action.
GC-mediated chemokine mRNA decay is dependent upon GR association with discrete, non-ARE regions of the transcripts
Posttranscriptional control of gene expression by GC has been so far mainly attributed to its promotion of ARE-mediated decay by induction of genes critical for this function, such as MAP Kinase phosphatase-1 (MKP-1) and the RNA-binding protein TTP (15, 25, 49, 50). However, our data support the hypothesis of an additional, direct effect of GC on RNA turnover. Previous studies have described GC action on mRNA stability independent from the presence of ARE. For rat CCL2, the presence of a 224-long sequence including the 5’ UTR was necessary to convey GC-mediated increased mRNA decay using a cell-free mRNA decay assay, while the isolated ARE-bearing 3’UTR was dispensable (31). In agreement with these published findings, we mapped GR association for human CCL2 and CCL7 within the 5’UTR of the mRNA. In the case of CCL2 mRNA, the region necessary for the GR association is composed by 16 nucleotides in position 44–60 – however, it is possible that larger sequences neighboring this site are needed for the proper assembly of the functional RNP ultimately conveying the effect on mRNA decay in vivo. In fact, several lines of evidence indicate that the interaction of GR with its target transcripts occurs in the context of a larger RNP complex. First, the GR-mediated acceleration of CCL2 and CCL7 mRNA decay occurs only after GC treatment of intact cells; second, GC treatment induces an upward mobility shift in the RNP complex detected in the cell-free decay assay. These data suggest that the ligand-activated GR may function either by associating with cytoplasmic components involved in mRNA decay and functionally activating this process, or by transcriptional activation of a necessary RNA-binding partner (potentially TTP), which is subsequently recruited to the CCL2 RNP complex.
Association of GR with a subset of endogenous mRNAs
Regulation of mRNA decay is coordinated by RNA-binding proteins in an operon-like fashion, through their binding to common regulatory elements present in a discrete subset of transcripts (18). Array-based analysis of gene expression in cytoplasmic RNA was obtained by RNP-IP, which is an efficient and well-tested method to recover endogenous RNP complexes (39). Using a GR-specific antibody, we identified almost 500 transcripts as significantly enriched in the GR IP, a number comparable to that observed for other RBP-bound transcript pools (32, 36). In line with the numerous studies that have utilized the standard RNP-IP for ribonomic analysis, and reinforced by the agreement with the other assays showing GR interaction with chemokine mRNAs, the RNP-IP array data indicate that the association with CCL2 and CCL7 mRNA is part of a broader ability of the GR to interact with cytoplasmic mRNA.
The GR-associated transcripts, according to GO analysis, are mostly involved in cell metabolism and signal transduction, which are biological processes well known to be targeted by GC as part of their homeostatic functions (51). The relative lack of immune and inflammatory transcripts in the GR-associated transcript pool is not surprising, as these targets are underrepresented in unstimulated airway epithelial cells. The target profile of RBP can change upon cell stimulation (52), reflecting the induction of a different pattern of gene expression, and it will be relevant to investigate whether the GR-associated mRNA pool in cells activated by an inflammatory stimulus, such as TNFα, is enriched in proinflammatory genes known to be antagonized by GC treatment, and if co-treatment with GC triggers GR-mediated mRNA decay for those targets.
Identification of GC-rich sequence on mRNA as putative GR binding motif
Using biotin pull-down assay, we confirmed that the GR associates with the GC-rich motif identified computationally, suggesting that the putative targets bearing this motif are likely to be regulated by the cytoplasmic function of GR. It remains to be tested if the functional role of this motif is indeed to convey acceleration of mRNA decay, as documented for CCL2 and CCL7, and whether other GR motifs can be found in genes expressed upon inflammatory cell activation. Multiple motifs have been documented for the RBP TIAR, which binds to U-rich motifs within the 3’UTR of its targets with high affinity when conveying translational repression, while its low-affinity binding to a C-rich element decreases upon cellular stress (32). It is very likely that GC treatment, as well as a host of intra- and extracellular triggers could modulate the GR binding to the RNA motifs in many ways. For example, this process could be modified by stimulus-induced changes in the RBP and microRNA potentially associated with GR in the RNP complex, and/or by action upon the association of GR-bound RNA with cytoplasmic foci of regulated mRNA decay and translation, such as P bodies and stress granules, where these functions are critically regulated during inflammation (17).
The GR lacks a conserved RNA-binding domain (RBD), present in other transcription factors like NF-90 and in nucleolin (53, 54). Future analysis of the GR domain interfacing with the GC-rich motif is warranted. In addition to potential RNA-binding through the GR RBD, GR could associate to other RBP components that bind directly to RNA. Along these lines, the GR has been shown to engage in protein-protein interaction with other shuttling RBPs, such as nucleolin (55).
The identification of GR-mediated acceleration of chemokine mRNA decay and of a novel function of GR in the cytoplasmic compartment opens a new paradigm in the GC mechanism of action, so far centered on its control of gene transcription. It will be critical to fully characterize this newly identified process, in order to understand its biological relevance in the antiinflammatory action of GC.
Supplementary Material
Acknowledgments
We thank Kevin Becker and William Wood at the NIH Genomics core for their assistance with analysis of array data and deposition of results in the GEO online repository.
Funding: This work was supported by NIH grant R01 AI060990-01A1 (Dr. Stellato) and by the American Academy of Allergy, Asthma and Immunology/Glaxo-Smith Kline Fellows Career Development Award (Dr. Ishmael). Drs. Abdelmohsen., Zhan and Gorospe are supported by the National Institute on Aging - Intramural Research Program, National Institutes of Health.
Footnotes
Abbreviations: Act D, actinomycin D; ARE, adenylate-uridylate rich region; ARE-BP, ARE-binding protein; EMSA, electrophoretic mobility shift assay; GC, glucocorticoid; GR, glucocorticoid receptor; GRE: glucocorticoid recognition element; IP, immunoprecipitation; PTR, post-transcriptional gene regulation, RBD, RNA-binding domain; RBP, RNA-binding protein; RNP, ribonucleoprotein complex; RNP-IP, immunoprecipitation of messenger ribonucleoprotein complexes; TTP, tristetraprolin; USER, untranslated sequence elements for regulation; UTR, untranslated region.
References
- 1.Barnes PJ. How corticosteroids control inflammation: Quintiles Prize Lecture 2005. Br J Pharmacol. 2006;148:245–254. doi: 10.1038/sj.bjp.0706736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stellato C. Glucocorticoid actions on airway epithelial responses in immunity: functional outcomes and molecular targets. J Allergy Clin Immunol. 2007;120:1247–1263. doi: 10.1016/j.jaci.2007.10.041. quiz 1264-1245. [DOI] [PubMed] [Google Scholar]
- 3.Adcock IM, Ito K, Barnes PJ. Glucocorticoids: Effects on Gene Transcription. Proc Am Thorac Soc. 2004;1:247–254. doi: 10.1513/pats.200402-001MS. [DOI] [PubMed] [Google Scholar]
- 4.Bhavsar P, Hew M, Khorasani N, Torrego A, Barnes PJ, Adcock I, Chung KF. Relative corticosteroid insensitivity of alveolar macrophages in severe asthma compared with non-severe asthma. Thorax. 2008;63:784–790. doi: 10.1136/thx.2007.090027. [DOI] [PubMed] [Google Scholar]
- 5.Rhen T, Cidlowski JA. Antiinflammatory Action of Glucocorticoids -- New Mechanisms for Old Drugs. N Engl J Med. 2005;353:1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- 6.Clark AR. Anti-inflammatory functions of glucocorticoid-induced genes. Mol Cell Endocrinol. 2007;275:79–97. doi: 10.1016/j.mce.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 7.Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immunol. 2010;10:24–35. doi: 10.1038/nri2685. [DOI] [PubMed] [Google Scholar]
- 8.Anderson P, Phillips K, Stoecklin G, Kedersha N. Post-transcriptional regulation of proinflammatory proteins. J Leukoc Biol. 2004;76:42–47. doi: 10.1189/jlb.1103536. [DOI] [PubMed] [Google Scholar]
- 9.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends in Genetics. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Fan J, Heller NM, Gorospe M, Atasoy U, Stellato C. The role of post-transcriptional regulation in chemokine gene expression in inflammation and allergy. Eur Respir J. 2005;26:933–947. doi: 10.1183/09031936.05.00120204. [DOI] [PubMed] [Google Scholar]
- 12.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ysla RM, Wilson GM, Brewer G, Lynne EM, Megerditch K. Methods in Enzymology. Academic Press; 2008. Chapter 3 Assays of Adenylate Uridylate-Rich Element-Mediated mRNA Decay in Cells; pp. 47–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stellato C. Post-transcriptional and nongenomic effects of glucocorticoids. Proceedings of the American Thoracic Society. 2004;1:255–263. doi: 10.1513/pats.200402-015MS. [DOI] [PubMed] [Google Scholar]
- 15.Smoak K, Cidlowski JA. Glucocorticoids Regulate Tristetraprolin Synthesis and Posttranscriptionally Regulate Tumor Necrosis Factor Alpha Inflammatory Signaling. Mol. Cell. Biol. 2006;26:9126–9135. doi: 10.1128/MCB.00679-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stellato C, Matsukura S, Fal A, White J, Beck LA, Proud D, Schleimer RP. Differential regulation of epithelial-derived C-C chemokine expression by IL-4 and the glucocorticoid budesonide. J Immunol. 1999;163:5624–5632. [PubMed] [Google Scholar]
- 17.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 18.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 19.Shyu AB, Wilkinson MF. The double lives of shuttling mRNA binding proteins. Cell. 2000;102:135–138. doi: 10.1016/s0092-8674(00)00018-0. [DOI] [PubMed] [Google Scholar]
- 20.Vavassori S, Covey LR. Post-transcriptional regulation in lymphocytes: the case of CD154. RNA Biol. 2009;6:259–265. doi: 10.4161/rna.6.3.8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 22.Chen CY, Gherzi R, Andersen JS, Gaietta G, Jurchott K, Royer HD, Mann M, Karin M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14:1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 23.Yarovinsky To, Butler NS, Monick MM, Hunninghake GW. Early Exposure to IL-4 Stabilizes IL-4 mRNA in CD4+ T Cells via RNA-Binding Protein HuR. J Immunol. 2006;177:4426–4435. doi: 10.4049/jimmunol.177.7.4426. [DOI] [PubMed] [Google Scholar]
- 24.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 25.Ishmael FT, Fang X, Galdiero MR, Atasoy U, Rigby WF, Gorospe M, Cheadle C, Stellato C. Role of the RNA-binding protein tristetraprolin in glucocorticoid-mediated gene regulation. J Immunol. 2008;180:8342–8353. doi: 10.4049/jimmunol.180.12.8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi L, Godfrey WR, Lin J, Zhao G, Kao PN. NF90 regulates inducible IL-2 gene expression in T cells. J Exp Med. 2007;204:971–977. doi: 10.1084/jem.20052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhawan L, Liu B, Blaxall BC, Taubman MB. A Novel Role for the Glucocorticoid Receptor in the Regulation of Monocyte Chemoattractant Protein-1 mRNA Stability. J. Biol. Chem. 2007;282:10146–10152. doi: 10.1074/jbc.M605925200. [DOI] [PubMed] [Google Scholar]
- 28.Reddel RR, Salghetti SE, Willey JC, Ohnuki Y, Ke Y, Gerwin BI, Lechner JF, Harris CC. Development of tumorigenicity in simian virus 40-immortalized human bronchial epithelial cell lines. Cancer Res. 1993;53:985–991. [PubMed] [Google Scholar]
- 29.Atasoy U, Curry SL, López de Silanes I, Shyu AB, Casolaro V, Gorospe M, Stellato C. Regulation of Eotaxin Gene Expression by TNF and IL-4 Through Messenger RNA Stabilization: Involvement of the RNA-binding protein HuR. J Immunol. 2003;171:4369–4378. doi: 10.4049/jimmunol.171.8.4369. [DOI] [PubMed] [Google Scholar]
- 30.Casolaro V, Fang X, Tancowny B, Fan J, Wu F, Srikantan S, Asaki SY, De Fanis U, Huang SK, Gorospe M, Atasoy UX, Stellato C. Posttranscriptional regulation of IL-13 in T cells: role of the RNA-binding protein HuR. J Allergy Clin Immunol. 2008;121:853–859. doi: 10.1016/j.jaci.2007.12.1166. e854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poon M, Liu B, Taubman MB. Identification of a novel dexamethasone-sensitive RNA-destabilizing region on rat monocyte chemoattractant protein 1 mRNA. Mol Cell Biol. 1999;19:6471–6478. doi: 10.1128/mcb.19.10.6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HS, Kuwano Y, Zhan M, Pullmann R, Jr, Mazan-Mamczarz K, Li H, Kedersha N, Anderson P, Wilce MC, Gorospe M, Wilce JA. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol Cell Biol. 2007;27:6806–6817. doi: 10.1128/MCB.01036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez de Silanes I, Galban S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furuta GT, Liacouras CA, Collins MH, Gupta SK, Justinich C, Putnam PE, Bonis P, Hassall E, Straumann A, Rothenberg ME. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–1363. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 35.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci U S A. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuwano Y, Pullmann R, Jr, Marasa BS, Abdelmohsen K, Lee EK, Yang X, Martindale JL, Zhan M, Gorospe M. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucl. Acids Res. 2010;38:225–238. doi: 10.1093/nar/gkp861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorodkin J, Heyer LJ, Stormo GD. Finding the most significant common sequence and structure motifs in a set of RNA sequences. Nucleic Acids Res. 1997;25:3724–3732. doi: 10.1093/nar/25.18.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eddy S, Durbin R. RNA sequence analysis using covariance models. Nucl. Acids. Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Helou EF, Simonson J, Arora AS. 3-yr-follow-up of topical corticosteroid treatment for eosinophilic esophagitis in adults. Am J Gastroenterol. 2008;103:2194–2199. doi: 10.1111/j.1572-0241.2008.01989.x. [DOI] [PubMed] [Google Scholar]
- 40.Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J. Biol. Chem. 2008 doi: 10.1074/jbc.M709657200. M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolaides NC, Galata Z, Kino T, Chrousos GP, Charmandari E. The human glucocorticoid receptor: Molecular basis of biologic function. Steroids. 2010;75:1–12. doi: 10.1016/j.steroids.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Jr, Martindale JL, Yang X, Gorospe M. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol Cell Biol. 2008;28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh K, Laughlin J, Kosinski PA, Covey LR. Nucleolin Is a Second Component of the CD154 mRNA Stability Complex That Regulates mRNA Turnover in Activated T Cells. J Immunol. 2004;173:976–985. doi: 10.4049/jimmunol.173.2.976. [DOI] [PubMed] [Google Scholar]
- 44.Mongelard F, Bouvet P. Nucleolin: a multiFACeTed protein. Trends in Cell Biology. 2007;17:80–86. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 45.Liang J, Lei T, Song Y, Yanes N, Qi Y, Fu M. RNA-destabilizing factor tristetraprolin negatively regulates NF-kappaB signaling. J Biol Chem. 2009;284:29383–29390. doi: 10.1074/jbc.M109.024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossini GP. Glucocorticoid receptors are associated with particles containing DNA and RNA in vivo. Biochem Biophys Res Commun. 1987;147:1188–1193. doi: 10.1016/s0006-291x(87)80195-x. [DOI] [PubMed] [Google Scholar]
- 47.Rossini GP, Wikstrom AC, Gustafsson JA. Glucocorticoid-receptor complexes are associated with small RNA in vitro. J Steroid Biochem. 1989;32:633–642. doi: 10.1016/0022-4731(89)90507-4. [DOI] [PubMed] [Google Scholar]
- 48.Ali M, Vedeckis WV. The glucocorticoid receptor protein binds to transfer RNA. Science. 1987;235:467–470. doi: 10.1126/science.3798121. [DOI] [PubMed] [Google Scholar]
- 49.Lasa M, Abraham SM, Boucheron C, Saklatvala J, Clark AR. Dexamethasone causes sustained expression of mitogen-activated protein kinase (MAPK) phosphatase 1 and phosphatase-mediated inhibition of MAPK p38. Mol Cell Biol. 2002;22:7802–7811. doi: 10.1128/MCB.22.22.7802-7811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abraham SM, Lawrence T, Kleiman A, Warden P, Medghalchi M, Tuckermann J, Saklatvala J, Clark AR. Antiinflammatory effects of dexamethasone are partly dependent on induction of dual specificity phosphatase 1. J Exp Med. 2006;203:1883–1889. doi: 10.1084/jem.20060336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.George PC, Tomoshige K. Glucocorticoid Signaling in the Cell. Annals of the New York Academy of Sciences. 2009;1179:153–166. doi: 10.1111/j.1749-6632.2009.04988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukherjee N, Lager PJ, Friedersdorf MB, Thompson MA, Keene JD. Coordinated posttranscriptional mRNA population dynamics during T-cell activation. Mol Syst Biol. 2009;5:288. doi: 10.1038/msb.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112(Pt 6):761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 54.Shi L, Zhao G, Qiu D, Godfrey WR, Vogel H, Rando TA, Hu H, Kao PN. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3'-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J Biol Chem. 2005;280:18981–18989. doi: 10.1074/jbc.M411034200. [DOI] [PubMed] [Google Scholar]
- 55.Schulz M, Schneider S, Lottspeich F, Renkawitz R, Eggert M. Identification of nucleolin as a glucocorticoid receptor interacting protein. Biochem Biophys Res Commun. 2001;280:476–480. doi: 10.1006/bbrc.2000.4141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.