Abstract
Glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) and its paralogues were implicated in late-onset Alzheimer’s disease (LOAD), although the strength and direction of association have not been consistent. We genotyped three previously reported SNPs (rs3741916-GAPDH 5’UTR, rs2029721-pGAPD and rs4806173-GAPDHS) in three case-control series (2112 cases and 3808 controls). Rs3741916 showed the strongest LOAD association (p=0.003). The minor allele of rs3741916 showed a protective effect in our combined series (OR=0.87, 95% confidence interval (CI)=0.79–0.96). This is consistent with results from the two published follow-up studies and in opposite direction of the original report. Meta-analysis of the published series with ours suggests presence of heterogeneity (Breslow-Day p<0.0001). Meta-analysis of only the follow-up series including ours revealed a significant protective effect for the minor allele of rs3741916 (OR=0.85, 95% CI=0.76–0.96, p=0.009). Our results support the presence of LOAD variants and heterogeneity at the GAPDH locus. The most promising rs3741916 variant is unlikely to be functional given opposing effects in different series. Identification of functional variant(s) in this region likely awaits deep sequencing.
Search Terms: Alzheimer's disease, Association studies in genetics, Case control studies
Introduction
Late Onset Alzheimer’s disease (LOAD) is a complex disease with an estimated 80% genetic component(Gatz, et al., 2006). Until recently only the APOE4 allele showed consistent, reproducible association with LOAD (reviewed in(Ertekin-Taner, 2007)). The large LOAD GWAS published in the past two years have identified five novel LOAD genes with genome-wide significance(Carrasquillo, et al., 2009,Harold, et al., 2009,Lambert, et al., 2009,Reiman, et al., 2007). Three (CLU, PICALM, CR1) of these genes achieved genome-wide significance in the first stage of the two largest LOAD GWAS to date(Harold, et al., 2009,Lambert, et al., 2009). The two remaining genes (PCDH11X and GAB2) reached this level of significance in the combined Stage 1 and 2 analyses(Carrasquillo, et al., 2009,Reiman, et al., 2007). An additional 500+ LOAD candidate genes and alleles have been published(Bertram, et al., 2007) but most failed to show consistent replication. One such example is the GAPDH locus on chromosome 12p. GAPDH encodes an glyceraldehyde 3-phosphate dehydrogenase, most commonly known for its role in glycolysis, but which has also been recently implicated in neuronal apoptosis and transcriptional activation(Colell, et al., 2007,Nakajima, et al., 2009).. GAPDH is located on chromosome 12p proximal to a LOAD linkage peak described in multiple studies(12–15), making it both a positional and functional candidate LOAD gene.
In 2004, Li et al.(Li, et al., 2004) reported replicable association of multiple SNPs at the GAPDH locus and its paralogues, GAPDHS (on 19q) and pGAPD (on 12q), with LOAD in up to four Caucasian case-control series. This study focused on rs3741916 in the 5’UTR of GAPDH, rs4806173 in intron 1 of GAPDHS and rs2029721, a missense mutation, in pGAPD. All three SNPs were also analyzed in follow-up studies by Lin et al. (9), who analyzed a Caucasian case-control and a Caucasian family-based series. GAPDH SNP rs3741916 was also assessed by Lee et all (10), who analyzed one Caucasian and one Carribbean-Hispanic family-based series in addition to a Caucasian case-control series.
In the initial study of Li et al. (8), the minor allele of rs3741916 was significantly associated with increased risk of LOAD but in both follow up studies (9, 10) it was significantly associated with decreased risk These results suggest that the association of rs3741916 with LOAD may be influenced by genetic and/or environmental factors that vary among the populations studied. Significant series to series heterogeneity of this sort, with increased risk in some studies and decreased risk in others, is relatively common in genetic association studies of LOAD and other genetically complex diseases. Given the multiple, independent LOAD associations reported for SNPs in GAPDH and its paralogues, it seemed likely to us that variants in these genes could have a complicated effect on LOAD pathogenesis. To investigate this possibility further, we genotyped the three SNPs that were previously reported (8–10) to show significant association (rs3741916, rs2029721, and rs4806173) in three additional case-control series with a combined total of 5920 subjects (2112 cases and 3808 controls). We then analyzed these SNPs using models identical to those employed in the previous studies to assess the same stratified sets of subjects that were analyzed in those studies. To characterize the association at these loci more fully, we assessed an additional 22 SNPs in these genes.
Materials and Methods
Patient samples
Two independent clinically diagnosed series of late-onset AD (LOAD) cases (age of diagnosis > 60) and elderly controls (age at evaluation >60) were collected at Mayo Clinic Jacksonville (JS series; 882 cases and 986 controls) and Mayo Clinic Rochester (RS series; 640 cases and 2460 controls), in addition to an autopsy confirmed series of elderly AD cases maintained at the Brain Bank at Mayo Clinic Jacksonville (AUT; 590 cases and 362 controls, age at death >60). These three series combined have 2112 cases and 3808 controls; details of which can be found in Table 1.
Table 1.
Demographic details of three “Mayo Clinic case-control series” and the subset strata used for analysis models.
| Series | Strata | Cs N | Cn N | Total N | Mean Age | Males (%) | ApoE4+ (%) |
|---|---|---|---|---|---|---|---|
| JS | All Ages | 882 | 986 | 1868 | 77.1 | 745 (40%) | 837 (45%) |
| >78 | 452 | 418 | 870 | 71.9 | 347 (40%) | 353 (41%) | |
| <78 | 430 | 568 | 998 | 83.1 | 398 (40%) | 484 (49%) | |
| ApoE4− | 321 | 710 | 1031 | 77.7 | 421 (41%) | na | |
| ApoE4+ | 561 | 276 | 837 | 76.4 | 324 (39%) | na | |
| RS | All Ages | 640 | 2460 | 3100 | 78.7 | 1363 (44%) | 916 (30%) |
| >78 | 383 | 1223 | 1606 | 83.4 | 642 (40%) | 467 (29%) | |
| <78 | 233 | 1211 | 1444 | 73.4 | 721 (50%) | 449 (31%) | |
| ApoE4− | 278 | 1856 | 2134 | 78.9 | 963 (45%) | na | |
| ApoE4+ | 338 | 578 | 916 | 78.1 | 400 (44%) | na | |
| AUT | All Ages | 590 | 362 | 952 | 79.1 | 452 (47%) | 437 (46%) |
| >78 | 379 | 130 | 509 | 85.7 | 200 (39%) | 257 (51%) | |
| <78 | 211 | 232 | 443 | 71.5 | 252 (57%) | 180 (41%) | |
| ApoE4− | 233 | 282 | 515 | 78.5 | 254 (49%) | na | |
| ApoE4+ | 357 | 80 | 437 | 79.7 | 198 (45%) | na | |
Cs = LOAD Case, Cn = LOAD Control. JS = Jacksonville Series, RS = Rochester Series, AUT = Autopsy confirmed series. <78 Age of Diagnosis/Examination/Death is less than 78 years, >78 Age of Diagnosis/Examination/Death is greater than 78 years.
All subjects from the JS and RS series were diagnosed by a Mayo Clinic neurologist. The neurologist confirmed a Clinical Dementia Rating score of 0 for all subjects enrolled as controls; cases had diagnoses of possible or probable AD made according to NINCDS-ADRDA criteria(McKhann G, et al., 1984). In the autopsy-confirmed series all brains were evaluated by the neuropathologist, Dr. Dennis Dickson where diagnosis of definite AD was also made according to NINCDS-ADRDA criteria. This study was approved by the appropriate institutional review board and appropriate informed consent was obtained from all participants. These series have previously been used in our studies including the recent Mayo Clinic late-onset AD genome-wide association study(Carrasquillo, et al., 2009).
SNP selection
Three SNPs reported in the initial study (rs3741916, rs2029721, rs4806173)(Li, et al., 2004) were genotyped in our complete case-control series with ages at diagnosis/evaluation/death above 60 years. An additional 22 SNPs were successfully assessed in the 60–78 age group. These SNPs were selected according to the criteria outlined in the Supplementary Text.
Genotyping
Three platforms were used for genotyping; Taqman, Sequenom and Illumina. The details of genotyping methods are in the Supplementary Text.
Statistical analysis
Single SNP association analysis
Each SNP was assessed individually for association with LOAD by multivariate logistic regression analysis using an allelic dosage model, adjusted for the following covariates: presence of an APOE4 allele (0,1), age at diagnosis/evaluation/death and gender.
In order to accurately replicate the tests from the published studies we analyzed the 3 key SNPs using the “best model” described by each prior study(Li, et al., 2004)-(Lee, et al., 2008). Furthermore we tested each of these three SNPs for difference in effect based on the following covariate strata: APOE4 allele +/−, Age > vs. ≤ 78 years and male vs. female gender. Breslow-Day tests for each of these strata did not identify significant differences in effect for each of the 3 key SNPs (Breslow-Day p-value > 0.3).
Meta-Analysis
To perform meta-analysis of all published rs3741916 allelic associations and our series, allelic counts were calculated from the reported allelic frequency and sample size information, when available(Lee, et al., 2008,Li, et al., 2004). Breslow-Day test for non-compatibility was used to test for series heterogeneity. Test statistics are reported for each series and pooled test statistics are reported using the random effects model (DerSimonian-Laird).
Linkage Disequilibrium Analysis
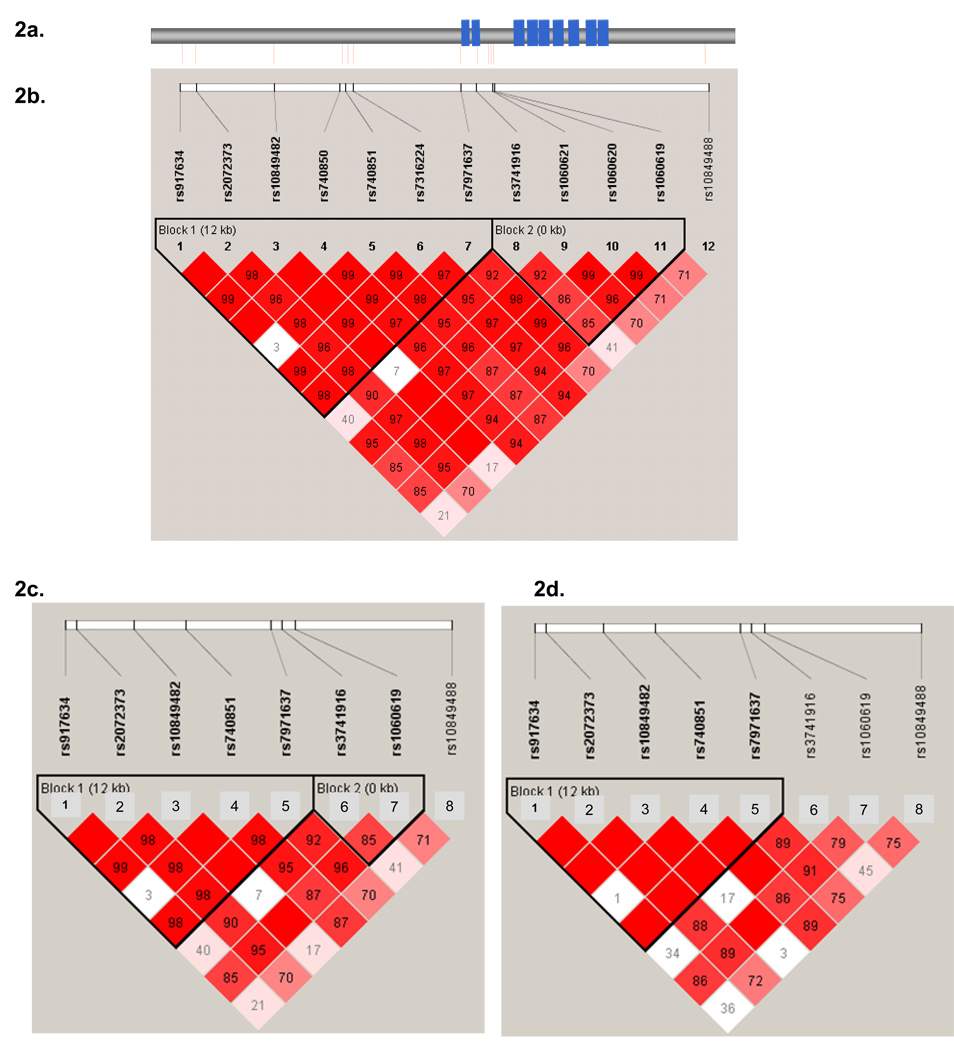
Analysis (solid spine of LD) by HaploView(Barrett, et al., 2005) of the genotypes for all 25 SNPs (3 key SNPs initially genotyped and 22 subsequent SNPs) in our 60–78 year age group was used to identify LD blocks in GAPDH (Figures 2 a–b) and GAPDHS (Figure A1–2). To compare the LD surrounding the rs3741916 SNP with other Caucasian subjects, we analyzed downloaded HapMap data according to genome build 36 and assessed it in Haploview. Only a subset of our genotyped GAPDH SNPs were available in HapMap. Figure 2c depicts the LD plot of this subset in our series and figure 2d is that in the HapMap Caucasian (CEU) series.
Figure 2.
Figure 2a–d: Linkage disequilibrium in the combined Mayo Clinic series at the GAPDH locus for all genotyped SNPs (a–b), the subset of HapMap SNPs in our series (c) and HapMap Caucasian subjects (d). SNP = Single nucleotide Polymorphism. 2a: Exons are represented with blue boxes and SNPs are represented with red lines. 2b–d: LD was estimated and haplotype blocks were defined using the “Solid Spine” method implemented in HAPLOVIEW.
Darker shades of red indicate increasing strength of LD (D’). 2d. HapMap SNP data is based on their Caucasian (CEU) subjects and genome build 36 downloaded from the HapMap website.
Results
Replication analysis of SNPs previously reported by others
The demographics of the three case-control series that we analyzed are summarized in Table 1. Table 2 compares the results from previous studies of rs3741916, rs2029721, and rs4806173 with the results we obtained for each case-control series and for the three series combined.
Table 2.
Replication analysis of previous reports
| Allelic | Logistic Regression (Dominant Model) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP (Locus) |
Chr | Base Position |
Strata | Series | Cs N (MAF) | Cn N (MAF) | OR (95%CI) | p-value | OR (95%CI) | p-value |
| A rs3741916 (GAPDH) aka rs1136666 | 12 | 6,514,252 | ApoE4- (Li et al, 2004) | Li et al W/UC/UK | (0.297) | (0.247) | 1.27(1.06:1.53) | 0.008 | nr | nr |
| JS | 311 (0.25) | 702 (0.27) | 0.92 (0.74: 1.15) | 0.478 | 0.95 (0.75: 1.28) | 0.874 | ||||
| RS | 276 (0.28) | 1838 (0.28) | 1.00 (0.82: 1.23) | 0.960 | 1.12 (0.86: 1.45) | 0.406 | ||||
| AUT | 274 (0.22) | 232 (0.28) | 0.73 (0.54: 0.99) | 0.042 | 0.67 (0.46: 0.97) | 0.034 | ||||
| JS/RS/AUT | 819 (0.25) | 2814 (0.28) | 0.88 (0.78: 1.00) | 0.051 | 0.92 (0.79: 1.08) | 0.313 | ||||
| B rs3741916 (GAPDH) aka rs1136666 | 12 | 6,514,252 | AAE/D*< mean (Lin et al, 2006) | Lin et al C-C series | nr | nr | nr | nr | 0.39 (0.21:0.70) | 0.002 |
| JS | 417 (0.27) | 563 (0.28) | 0.95 (0.78: 1.17) | 0.683 | 0.98 (0.74: 1.29) | 0.870 | ||||
| RS | 229 (0.28) | 1200 (0.29) | 0.93 (0.74: 1.16) | 0.537 | 0.95 (0.70: 1.30) | 0.765 | ||||
| AUT | 209 (0.24) | 225 (0.27) | 0.83 (0.60: 1.13) | 0.243 | 0.85 (0.56: 1.28) | 0.435 | ||||
| JS/RS/AUT | 855 (0.26) | 1988 (0.29) | 0.89 (0.79: 1.02) | 0.089 | 0.92 (0.77: 1.10) | 0.349 | ||||
| C rs3741916 (GAPDH) aka rs1136666 | 12 | 6,514,252 | All (Lee et al, 2008) | Lee et al C-C series | (0.14) | (0.21) | nr | 0.027 | nr | 0.054 |
| JS | 859 (0.26) | 974 (0.28) | 0.90 (0.78: 1.05) | 0.178 | 0.92 (0.75: 1.12) | 0.399 | ||||
| RS | 611 (0.27) | 2411 (0.28) | 0.91 (0.79: 1.05) | 0.212 | 0.95 (0.78: 1.14) | 0.554 | ||||
| AUT | 585 (0.23) | 354 (0.27) | 0.81 (0.65: 1.00) | 0.053 | 0.77 (0.57: 1.04) | 0.086 | ||||
| JS/RS/AUT | 2055 (0.25) | 3739 (0.28) | 0.86 (0.79: 0.94) | 8.0E-04 | 0.88 (0.78: 0.99) | 0.033 | ||||
| D rs2029721 (pGAPD) | 12 | 61,435,611 | AAE/D* > mean(Li et al, 2004 & Lin et al 2006) | Li et al W/UC/UK | (0.308) | (0.352) | 0.80 (0.68:0.97) | 0.018 | nr | nr |
| Lin et al C-C series | nr | nr | nr | 0.004 | 0.55 (0.32:0.96) | 0.036 | ||||
| JS | 320 (0.39) | 309 (0.36) | 1.13 (0.89: 1.42) | 0.322 | 1.16 (0.82: 1.63) | 0.404 | ||||
| RS | 336 (0.36) | 719 (0.38) | 0.94 (0.78: 1.14) | 0.530 | 0.96 (0.73: 1.27) | 0.766 | ||||
| AUT | 369 (0.36) | 130 (0.34) | 0.95 (0.70: 1.31) | 0.759 | 1.31 (0.84: 2.05) | 0.237 | ||||
| JS/RS/AUT | 1025 (0.37) | 1158 (0.37) | 1.00 (0.88:1.13 | 0.999 | 1.05 (0.87: 1.26) | 0.642 | ||||
| E rs4806173 (GAPDHS) | 19 | 40,716,765 | AAE/D* < mean (Li et al, 2004) | Li et al W/UC/UK | (0.326) | (0.42) | 0.66 (0.55:0.80) | 3.0E-04 | nr | nr |
| JS | 270 (0.39) | 330 (0.40) | 0.97 (0.76: 1.23) | 0.812 | 0.95 (0.65: 1.38) | 0.785 | ||||
| RS | 220 (0.39) | 644 (0.38) | 1.05 (0.83: 1.32) | 0.691 | 1.00 (0.70: 1.41) | 0.977 | ||||
| AUT | 209 (0.38) | 229 (0.36) | 1.09 (0.82: 1.45) | 0.575 | 1.16 (0.77: 1.75) | 0.490 | ||||
| JS/RS/AUT | 699 (0.39) | 1203 (0.38) | 1.03 (0.89: 1.18) | 0.729 | 1.00 (0.81: 1.23) | 0.960 | ||||
Chr = Chromosome, Cs = AD case, Cn = Control subject. N = number of subjects in series that have genotype data. MAF = minor allele frequency. OR = Odds Ratio, CI = Confidence Interval.
Allelic association tested using chi-squared test with no covariates. Logistic regression uses Age*, Gender and presence of an APOE4 allele as covariates, *Age and AAE/D refers to Age at Diagnosis, Examination or Death. The mean age differs between the different publications. The mean age is 78 in our combined series. Nominally significant p-values (<0.05) are highlighted in bold, nr = Not reported. .Panels A and E: Li et al., 2004 models of analysis. Panel B: Lin et al., 2006 model. Panel C: Lee et al., 2008 model. Panel D: Li et al., 2004 and Lin et al., 2006 models.
GAPDH SNP rs3741916
In the original study of rs3741916, Li et al.(8) showed strongest association in the APOE4- group. Using an allelic association model to analyze the APOE4- group in their combined series (Table 2A), they found that the minor allele of rs3741916 was associated with (p=0.008) increased risk of LOAD (OR=1.27, 95%CI=1.06–1.53). When we used the same model to analyze the APOE4- subjects in our AUT series (Table 2A), we found that the minor allele of rs3741916 was associated with (p=0.042) decreased risk of LOAD (OR=0.73, 95%CI = 0.54–0.99). There was, however, no evidence of association when we analyzed the APOE4- group in our JS, RS, or combined series (Table 2A).
In a follow up study of rs3741916, Lin et al. found strongest evidence of association in their young age group with onset below the series mean. Using logistic regression under a dominant model to analyze the young age group (Table 2B), they found that the minor allele of rs3741916 was associated with (p=0.002) decreased risk of LOAD (OR=0.39, 95%CI=0.21–0.70). When we used the same model to analyze the young age group with age at diagnosis/evaluation below our mean of 78 years, none of our series showed significant association. (Table 2B).
In another follow-up study, Lee et al. analyzed their Northern European case control series using an allelic association model. This analysis (Table 2C) showed that the minor allele of rs3741916 was associated with (p=0.027) decreased risk of LOAD. When we analyzed our combined series in the same way (Table 2C), we also found that the minor allele of rs3741916 was associated with (p=8×10−4) decreased risk of LOAD (OR=0.86, 95%CI=0.79–0.94). Moreover, each of our 3 series trended toward decreased risk with ORs ranging from 0.81–0.91 and p values ranging from 0.053–0.212.
To evaluate rs3741916 for series to series heterogeneity based on APOE4, age and gender, we stratified our combined series by APOE4 (+ vs. −), age (> vs ≤ 78 years) and gender (male vs. female) and did not find any significant difference between the groups (Breslow-Day test P > 0.32).
pGAPD SNP rs2029721
In the initial study of rs2029721, Li et al.(8) showed strongest association in the older age group with onset above the series mean. Using an allelic association model to analyze this group (Table 2D), they found that the minor allele of rs2029721 was associated with (p=0.018) decreased risk of LOAD (OR=0.80, 95%CI=0.68–0.97). In follow-up, using the same model to analyze their older group, Lin et al.(Lin, et al., 2006) also found that the minor allele was associated with (p=0.004) decreased risk of LOAD (Table 2D). When they analyzed their entire series, Lin et al. also found that the minor allele of rs2029721 was associated with significantly decreased risk (data not shown). Using the same allelic association model, we did not identify any significant association of this SNP with LOAD in the older age group of our individual or combined series (Table 2D). Analysis of all subjects in each series or in the combined series also yielded no significant association (data not shown).
GAPDHS SNP rs4806173
In the initial study of rs4806173, Li et al.(8) showed strongest association in the young age group with onset below the series mean. Using an allelic association model to analyze this group (Table 2E), they found that the minor allele of rs4806173 was associated with (p=0.0003) decreased risk of LOAD (OR=0.66, 95%CI=0.55–0.80). When we used the same model to analyze the young age group in our individual or combined series, we did not observe any significant association with the minor allele of this SNP (Table 2E).
Logistic regression using an additive model with covariates
To explore a conventional additive model while controlling for covariates, we analyzed the three key SNPs by logistic regression with covariates age at diagnosis/evaluation/death, gender and presence of an APOE4 allele in our 3 series, individually and combined. We also analyzed each of these series after stratifying by mean age at diagnosis/evaluation/death (≥ or ≤ 78 years), gender and presence or absence of an APOE4 allele.
As expected from our replication analyses, GAPDH SNP rs3741916 was the only SNP that showed significant association in any of our series (Table 3). Using the additive model with covariates, rs3741916 showed association in the combined series (p=0.003), where the minor allele was associated with decreased risk of LOAD (OR= 0.87, 95%CI=0.79–0.96). Each of the 3 series had ORs associated with reduced risk of LOAD, and the AUT series achieved significance (p=0.047). When corrected for the 3 original SNPs tested in this study, the rs3741916 association in our combined series would still be significant (p=3×0.003=0.009). However, when the stringent Bonferroni correction is applied for all 25 SNPs tested in this study, this overall association becomes marginal (p=25×0.003=0.075).
Table 3.
The results of logistic regression analysis under an additive model in the Mayo Clinic Series.
| Logistic Regression Additive Model |
||||||||
|---|---|---|---|---|---|---|---|---|
| SNP (Locus) | Chr | Base Position |
Strata | Series | Cs N (MAF) | Cn N(MAF) | OR (95%CI) | p-value |
| rs3741916 (GAPDH) aka rs1136666 | 12 | 6,514,252 | All | JS | 859 (0.26) | 974 (0.28) | 0.90 (0.77: 1.05) | 0.159 |
| RS | 611 (0.27) | 2411 (0.28) | 0.93 (0.81: 1.08) | 0.361 | ||||
| AUT | 585 (0.23) | 354 (0.27) | 0.78 (0.61: 1.00) | 0.047 | ||||
| JS/RS/AUT | 2055 (0.25) | 3739 (0.28) | 0.87 (0.79: 0.96) | 0.003 | ||||
| rs2029721 (pGAPD) | 12 | 61,435,611 | AAE/D ≤78 | JS | 590 (0.40) | 637 (0.36) | 1.39 (1.07: 1.81) | 0.013 |
| RS | 556 (0.37) | 1356 (0.38) | 0.94 (0.74: 1.21) | 0.638 | ||||
| AUT | 574 (0.37) | 354 (0.36) | 1.20 (0.90: 1.60) | 0.225 | ||||
| JS/RS/AUT | 1720 (0.38) | 2347 (0.37) | 1.14 (0.98: 1.31) | 0.090 | ||||
| rs4806173 (GAPDHS) | 19 | 40,716,765 | All | JS | 591 (0.39) | 642 (0.38) | 1.03 (0.87: 1.23) | 0.732 |
| RS | 556 (0.39) | 1372 (0.38) | 0.99 (0.86: 1.15) | 0.923 | ||||
| AUT | 586 (0.38) | 358 (0.38) | 0.87 (0.70: 1.09) | 0.222 | ||||
| JS/RS/AUT | 1733 (0.38) | 2372 (0.38) | 0.98 (0.89: 1.08) | 0.688 | ||||
Chr = Chromosome Cs = AD case, Cn = Control subject. N = number of subjects in series that have genotype data. MAF = minor allele frequency. OR = Odds Ratio, CI = Confidence Interval. Logistic regression includes Age*, Gender and presence of an ApoE4 allele included as covariates, *Age and AAE/D refers to Age at Diagnosis, Examination or Death. Nominally significant p-values (<0.05) are highlighted in bold.
The other 2 SNPs did not reach significance in the combined series, under this model, although rs2029721 was significantly risky in the JS series (OR=1.39, 95%CI=1.07–1.81) and also had a risky trend in the combined series (OR=1.14, 95%CI=0.98–1.31). The stratum that yielded the most significant results in the combined group using the additive logistic regression approach is shown in Table 3.
Meta-analysis
We performed a meta-analysis of GAPDH SNP rs3741916 (Figure 1) because it had the strongest evidence of association in our series and showed association with AD in all previously published series. Allele counts for rs3741916 were calculated for the previously reported case control series where allele frequencies and series sizes were available. In Figure 1a, the “Wash”, “UCSD”, “Linkage” and “UK” series are from the initial publication (Li, et al., 2004) and the “NE” series is from Lee et al. There was insufficient data to calculate the allele counts for the Lin et al. study (Lin, et al., 2006). As shown Figure 1a, three of the four series from the initial study are opposite in direction to all of the follow-up studies, leading to significant series-to-series heterogeneity for rs3741916 (Breslow-Day p value < 0.0001). When the first, exploratory series (WashU) from the initial study (Li et al) was removed from the meta analysis (Figure 1b), the pooled OR estimate for the minor allele of rs3741916 was 0.95 (95%CI=0.81–1.10); and there was still evidence for significant series-to-series heterogeneity for rs3741916 (Breslow-Day p value = 0.0003).
Figure 1.
Figure 1 a. Meta-Analysis of all series with reported counts and frequencies. Breslow-Day p-value <0.0001. * indicates series reported in the original study (Li et al).
Figure 1 b. Meta-Analysis of all series with reported counts and frequencies except first series from the original study. Breslow-Day p-value =0.0003. * indicates series reported in the original study (Li et al).
Figure 1.c Meta-Analysis of all follow-up series. Breslow Day p-value = 0.1967, Combined series p-value = 0.0094
When the four series from the inital study (Li et al) were removed and the four follow-up series (NE, JS, RS, and AUT) were analyzed, meta-analysis yielded a pooled OR estimate of 0.85 (95%CI=0.76–0.96) for the minor allele G of rs3741916 (random effects p=0.0094) (Figure 1.c). We expect that addition of the Lin et al series to the meta-analysis would further improve this estimate, since that series reported significant association in the same direction as the four other follow-up series.
We assessed LD amongst our 12 GAPDH SNPs (Figures 2a–b). We also downloaded HapMap data of this region and determined that 8 of our 12 SNPs had data in the Caucasian HapMap subjects (CEU), where we assessed LD (Figure 2d) in comparison to that in our study population (Figure 2c). We determined that the extent of LD between rs3741916 and its surrounding SNPs was slightly different between these two datasets, which may be one potential source of heterogeneity, though the extent of heterogeneity in LD between this SNP and rarer functional variants in this region may not be possible to fully appreciate based on the LD plots for these available SNP.
Discussion
GAPDH is an excellent LOAD candidate gene given the genetic linkage(Mayeux, et al., 2002,Myers, et al., 2002,Rogaeva E, et al., 1998,Scott WK, et al., 2000) and association(Lee, et al., 2008,Li, et al., 2004,Lin, et al., 2006) findings reported at this locus and functional evidence for its role in neurodegeneration(Colell, et al., 2007,Nakajima, et al., 2009). Three previous studies analyzed SNPs at the GAPDH locus and/or its paralogues in a total of 6 case-control and 3 family-based series. Using models identical to those employed in the previous studies, we evaluated three SNPs previously reported to show significant association, rs3741916, rs2029721, and rs4806173. We also analyzed 22 additional SNPs (Supplementary Text) in GAPDH and GAPDHS.
The only SNP that showed significant association in our study was rs3741916. This SNP yielded nominally significant associations with LOAD in each of the three previous studies, but the minor allele was associated with increased risk of LOAD in the initial study and with decreased risk in the two Caucasian follow-up studies. In our combined series, rs3741916 was significantly associated with decreased risk of LOAD and trended toward association with decreased risk in each of the 3 individual series. Meta-analysis of our 3 case-control series and a series from a published follow-up study with sufficient data for analysis(Lee, et al., 2008) showed significant association with decreased risk and no evidence of heterogeneity. However, meta-analysis that included the initial series as well as these follow-up series showed highly significant heterogeneity. Thus, in the studies performed to date, the minor allele of rs3741916 was associated with decreased risk of LOAD in the majority of the series. There is, however, marked heterogeneity because in 3 series from the original study(Lee, et al., 2008,Li, et al., 2004,Lin, et al., 2006), the minor allele was associated with significantly increased risk.
Heterogeneity for candidate LOAD loci is a common problem(Newton-Cheh and Hirschhorn, 2005). Small sample size can lead to false positive results in initial studies as well as false negative results in underpowered follow-up studies. Although small sample size can explain lack of replication, it is unlikely to account for results like those for rs3741916 where there is significant association with increased risk in some studies and decreased risk in others. Lin et al.(Lin, et al., 2007) investigated the reasons for this “flip-flop” of the GAPDH rs3741916 locus in their series(9) vs. the initial report(8) and concluded that differences in the correlation of this SNP with APOE could account for the different effects in the two series. Specifically, in subjects of younger age at onset, the minor G allele of rs3741916 was inversely correlated with APOE4 in the follow-up Lin et al. study(Lin, et al., 2006), offering an explanation as to why this allele associated with reduced risk of LOAD in their study. The authors observed that the strongest association in the initial study was obtained in those subjects who lacked APOE4, where the minor allele of rs3741916 was associated with increased risk. Thus, the differential correlation between APOE4 and the rs3741916 G allele might account for the opposite effects of this GAPDH SNP allele in the two studies. The authors also concluded that differences in the ages-of-onset of the study subjects might contribute to the “flip-flop” that was observed.
In our series, we did not see a difference in the association of rs3741916 with LOAD when neither our APOE4+ and – subjects nor our old and young subjects were analyzed separately. Furthermore, in analyses where we controlled for age, gender and presence of APOE4 allele, we still found that the minor allele of rs3741916 was associated with decreased risk of LOAD. This does not invalidate the hypothesis previously suggested(11) to account for the difference between the studies by Li et al.(8) and Lin et al.(9), but it does suggest that there may be heterogeneity unrelated to APOE4 or age that accounts for the opposite effects of rs3741916 in different series.
It is possible that multiple alleles with weak effects and/or environmental factors have an important influence on rs3741916/LOAD association and that these factors vary enough among series that there is significantly increased risk in some series and significantly decreased risk in others. Another possible explanation is that the major and/or minor alleles of rs3741916 are in LD with rarer functional SNP(s) that have relatively strong effect(s) on AD risk. If LD and/or functional allele frequency varied substantially from series to series, the minor allele of rs3741916 could be associated with increased risk of LOAD in some series and decreased risk in others.
Our results support a role for the chromosome 12p locus in LOAD, where our most significant association is reported for rs3741916 in the 5’UTR of GAPDH using all subjects from all 3 series. More work is needed to determine if this association is the result of GAPDH variants or those at other loci which are in LD with the GAPDH variants. It may be that multiple variants in multiple genes within this chromosome 12p region contribute to the findings reported at this locus. This possibility may explain the heterogeneity observed for rs3741916 and likely requires deep sequencing to uncover the true functional variants accounting for the association with LOAD.
Supplementary Material
Acknowledgments
Funding:
Support for this research was provided by the National Institutes of Health grants: National Institute on Aging [R01 AG018023 to N.R.G-R and S.G.Y.]; Mayo Alzheimer’s Disease Research Center: [P50 AG016574 to R.C.P, D.W.D, N.R.G-R, and S.G.Y]; Mayo Alzheimer’s Disease Patient Registry: [U01 AG006576 to R.C.P]; National Institute on Aging [AG025711, AG017216, AG003949 to D.W.D]. This project was also generously supported by the Robert and Clarice Smith and Abigail Van Buren Alzheimer's Disease Research Program [to R.C.P., D.W.D.,N.R.G-R; S.G.Y] and by the Palumbo Professorship in Alzheimer’s Disease Research [to S.G.Y.]. N. E-T is the recipient of National Institute on Aging [F32 AG20903], National Institutes of Health [KL2 RR024151], Johnnie B. Byrd and Siragusa Foundation grants. OB thanks the Alzheimer’s Research Trust (ART) for funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data Information: Supplementary Text, Figure A and Tables AC.
Disclosures:
M. Allen reports no disclosures.
C. Cox reports no disclosures.
O. Belbin reports no disclosures.
L. Ma reports no disclosures.
G.D. Bisceglio reports no disclosures.
S.L. Wilcox reports no disclosures.
C.C. Howell reports no disclosures.
T. A. Hunter reports no disclosures.
O. Culley reports no disclosures.
L.P. Walker reports no disclosures.
M.M. Carrasquillo, Ph.D. reports no disclosures.
D.W. Dickson, MD reports no disclosures.
R.C. Petersen, MD, PhD has been a consultant to GE Healthcare and Elan Pharmaceuticals, has served on a data safety monitoring board in a clinical trial sponsored by Elan Pharmaceuticals, and a safety monitoring board for Wyeth Pharmaceuticals.
N. Graff-Radford, MD has served as a consultant to Codman and received grant support from Elan Pharmaceutical Research, Pfizer Pharmaceuticals, Medivation and Forrest.
S.G. Younkin, MD, PhD reports no disclosures.
N. Ertekin-Taner, MD, PhD reports no disclosures.
References
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, Younkin SG, Younkin CS, Younkin LH, Bisceglio GD, Ertekin-Taner N, Crook JE, Dickson DW, Petersen RC, Graff-Radford NR, Younkin SG. Genetic variation in PCDH11X is associated with susceptibility to lateonset Alzheimer's disease. Nat Genet. 2009;41(2):192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colell A, Ricci JE, Tait S, Milasta S, Maurer U, Bouchier-Hayes L, Fitzgerald P, Guio-Carrion A, Waterhouse NJ, Li CW, Mari B, Barbry P, Newmeyer DD, Beere HM, Green DR. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129(5):983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- Ertekin-Taner N. Genetics of Alzheimer's disease: a centennial review. Neurol Clin. 2007;25(3):611–667. doi: 10.1016/j.ncl.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63(2):168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O'Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009 doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009 doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Rogaeva E, Meng Y, Stern Y, Santana V, Lantigua R, Medrano M, Jimenez-Velazquez IZ, Farrer LA, St George-Hyslop P, Mayeux R. Further examination of the candidate genes in chromosome 12p13 locus for late-onset Alzheimer disease. Neurogenetics. 2008;9(2):127–138. doi: 10.1007/s10048-008-0122-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Nowotny P, Holmans P, Smemo S, Kauwe JS, Hinrichs AL, Tacey K, Doil L, van Luchene R, Garcia V, Rowland C, Schrodi S, Leong D, Gogic G, Chan J, Cravchik A, Ross D, Lau K, Kwok S, Chang SY, Catanese J, Sninsky J, White TJ, Hardy J, Powell J, Lovestone S, Morris JC, Thal L, Owen M, Williams J, Goate A, Grupe A. Association of late-onset Alzheimer's disease with genetic variation in multiple members of the GAPD gene family. Proc Natl Acad Sci U S A. 2004;101(44):15688–15693. doi: 10.1073/pnas.0403535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PI, Martin ER, Bronson PG, Browning-Large C, Small GW, Schmechel DE, Welsh-Bohmer KA, Haines JL, Gilbert JR, Pericak-Vance MA. Exploring the association of glyceraldehyde-3-phosphate dehydrogenase gene and Alzheimer disease. Neurology. 2006;67(1):64–68. doi: 10.1212/01.wnl.0000223438.90113.4e. [DOI] [PubMed] [Google Scholar]
- Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. Am J Hum Genet. 2007;80(3):531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeux R, Lee JH, Romas SN, Mayo D, Santana V, Williamson J, Ciappa A, Rondon HZ, Estevez P, Lantigua R, Medrano M, Torres M, Stern Y, Tycko B, Knowles JA. Chromosome-12 mapping of late-onset Alzheimer disease among Caribbean Hispanics. Am J Hum Genet. 2002;70(1):237–243. doi: 10.1086/324773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Myers A, Wavrant De-Vrieze F, Holmans P, Hamshere M, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Knowles H, Morris JC, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, Rice F, Giles P, Tunstall N, Jones L, Lovestone S, Williams J, Owen MJ, Hardy J, Goate A. Full genome screen for Alzheimer disease: stage II analysis. Am J Med Genet. 2002;114(2):235–244. doi: 10.1002/ajmg.10183. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Amano W, Kubo T, Fukuhara A, Ihara H, Azuma YT, Tajima H, Inui T, Sawa A, Takeuchi T. Glyceraldehyde-3-phosphate dehydrogenase aggregate formation participates in oxidative stress-induced cell death. J Biol Chem. 2009;284(49):34331–34341. doi: 10.1074/jbc.M109.027698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Cheh C, Hirschhorn JN. Genetic association studies of complex traits: design and analysis issues. Mutat Res. 2005;573(1–2):54–69. doi: 10.1016/j.mrfmmm.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Huentelman MJ, Craig DW, Coon KD, Liang WS, Herbert RH, Beach T, Rohrer KC, Zhao AS, Leung D, Bryden L, Marlowe L, Kaleem M, Mastroeni D, Grover A, Heward CB, Ravid R, Rogers J, Hutton ML, Melquist S, Petersen RC, Alexander GE, Caselli RJ, Kukull W, Papassotiropoulos A, Stephan DA. GAB2 alleles modify Alzheimer's risk in APOE epsilon4 carriers. Neuron. 2007;54(5):713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogaeva E, Premkumar S, Song Y, Sorbi S, Brindle N, Paterson A, Duara R, Levesque G, Yu G, Nishimura M, Ikeda M, O'Toole C, Kawarai T, Jorge R, Vilarino D, Bruni AC, Farrer LA, St George-Hyslop PH. Evidence for an Alzheimer disease susceptibility locus on chromosome 12 and for further locus heterogeneity. JAMA. 1998;280(7):614–618. doi: 10.1001/jama.280.7.614. [DOI] [PubMed] [Google Scholar]
- Scott WK, Grubber JM, Conneally PM, Small GW, Hulette CM, Rosenberg CK, Saunders AM, Roses AD, Haines JL, MA P-V. Fine mapping of the chromosome 12 late-onset Alzheimer disease locus: potential genetic and phenotypic heterogeneity. Am J Hum Genet. 2000;66(3):922–932. doi: 10.1086/302828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.