Abstract
The abundant chromosome abnormalities in most carcinomas are probably a reflection of genomic instability present in the tumor, so the pattern and variability of chromosome abnormalities will reflect the mechanism of instability combined with the effects of selection. Chromosome rearrangement was investigated in 17 colorectal carcinoma-derived cell lines. Comparative genomic hybridization showed that the chromosome changes were representative of those found in primary tumors. Spectral karyotyping (SKY) showed that translocations were very varied and mostly unbalanced, with no translocation occurring in more than three lines. At least three karyotype patterns could be distinguished. Some lines had few chromosome abnormalities: they all showed microsatellite instability, the replication error (RER)+ phenotype. Most lines had many chromosome abnormalities: at least seven showed a surprisingly consistent pattern, characterized by multiple unbalanced translocations and intermetaphase variation, with chromosome numbers around triploid, 6–16 structural aberrations, and similarities in gains and losses. Almost all of these were RER−, but one, LS411, was RER+. The line HCA7 showed a novel pattern, suggesting a third kind of genomic instability: multiple reciprocal translocations, with little numerical change or variability. This line was also RER+. The coexistence in one tumor of two kinds of genomic instability is to be expected if the underlying defects are selected for in tumor evolution.
It has been known for a long time that many carcinomas have highly aneuploid karyotypes (1), suggesting that one of the steps selected for during tumor evolution results in genomic instability (2, 3). More recently, mismatch repair defects, with a replication error (RER)+ phenotype characterized by microsatellite instability (4, 5), have been described in a minority of tumors, for example, in about 15% of sporadic colorectal carcinoma. Intriguingly, most RER+ tumors have a stable near-diploid karyotype, whereas RER− tumors usually have stable microsatellites but unstable chromosome numbers and structure. The causes of this chromosomal instability are not yet clear, but may be various, because RER− tumors appear to be a heterogeneous group (6, 7). Mutation of p53 is one candidate, but some near-diploid RER+ tumors also have mutant p53 (8). Some aneuploid tumors may have defective mitotic checkpoint genes such as BUB1 (9), but the causal defects for the remainder are unknown. It has been suggested (2) that these instabilities are a byproduct of selection against apoptosis. Apoptosis after DNA damage, for example, can be abrogated by inactivation of either p53 or mismatch repair proteins (10–13). Clear definition of the different patterns of genomic instability in colorectal tumors therefore would be useful, as it may give clues to the nature of these undiscovered defects. Also, if genomic instability is a consequence of defects in apoptotic pathways, these patterns may prove predictive of response to therapy.
We have examined patterns of chromosome rearrangement and genomic instability in a series of 17 colorectal cancer cell lines, using chromosome painting methods. The lines were selected to include RER+ and RER− phenotypes, both with and without mutations in p53. Cell lines provide a source of tumor karyotypes that permits much more detailed analysis than primary material, but there has been doubt about how well they represent primary tumors. The patterns of genomic change in the cell lines were shown by comparative genomic hybridization (CGH) to reflect those in primary tumors. Karyotyping of the cell lines by multicolour chromosome painting—spectral karyotyping (SKY)—distinguished several patterns of chromosome abnormality and genomic instability, some of them not previously described in epithelial tumors.
Materials and Methods
The 17 human colorectal carcinoma cell lines were as described (4). CGH was essentially as described (14) using quips software (Vysis, Downer's Grove, IL) to calculate ratio profiles from 20 metaphases. SKY was as described (15). Briefly, whole chromosome paints for each chromosome labeled with different combinations of five fluorescent dyes were hybridized to cell line metaphases, and the fluorescence at each point in the image was analyzed with a spectrometer (Spectracube, Applied Spectral Imaging, Migdal HaEmek, Israel) to determine which chromosome was present. At least 10 metaphases were analyzed. Because SKY occasionally misidentifies small fragments of chromosome because of overlap between adjacent fluorescence signals, the identity of most translocated fragments was verified by conventional chromosome painting, using single fluorescent dyes for each chromosome (see Fig. 4, which is published as supplemental material on the PNAS web site, www.pnas.org) (15, 16).
Results
To demonstrate that the cell lines selected for karyotyping were representative of primary tumors in terms of their patterns of chromosomal abnormality, we analyzed the cell lines by CGH (Fig. 1) and compared the results with similar data reported by ourselves and others from primary, surgically removed tumors and first-pass xenografts (Fig. 2). Most RER+ tumors, both as cell lines and primary tumors, show too few chromosome changes to make valid comparisons, but in RER− lines and primary tumors, the same chromosome arms are subject to gains and losses (r = 0.7 for both tumors and xenografts). Loss of chromosome 6q, sometimes accompanied by loss of 6p, deviates from this pattern, suggesting either that there is selection pressure in vitro for loss of this chromosome, or that tumors with this defect more readily give rise to cell lines. The higher proportion of chromosome change in cell lines than primary tumors may reflect under-recording in primary tumors through contamination with normal stroma. The xenograft data support this hypothesis, because these transplanted tumors are free of human stroma and invariably more chromosomal changes are detected than in their parent primary tumors (6). Also, near-diploid RER− tumors may be under-represented in the lines and xenografts (6).
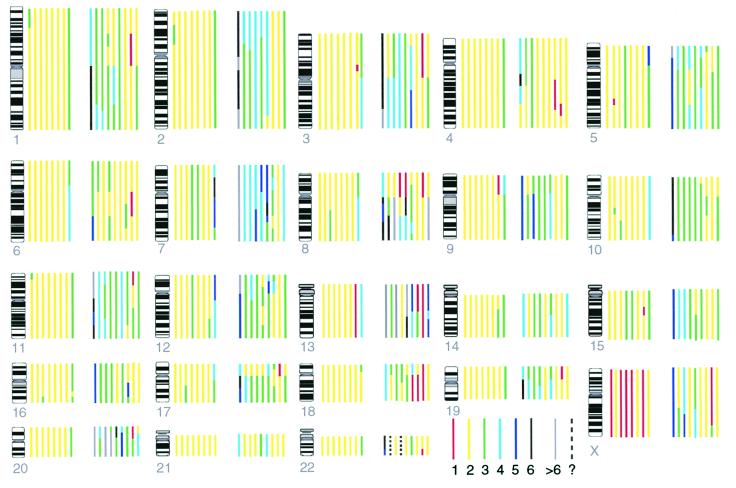
Figure 1.
Copy numbers of chromosome segments estimated by CGH, using ploidy information from the karyotype. Each colored bar represents the copy number of a chromosome in a particular cell line, different colors representing different copy numbers according to the key shown. For example, for chromosome 1, the bar nearest the ideogram represents DLD1 and shows that in DLD1 there are two copies (yellow) of most of chromosome 1, but three copies of distal 1p (green). Cell lines from left to right are: first group (RER+) DLD1 (nearest to chromosome ideogram), GP2d, HCT116, LoVo, LS174T, VACO5, HCA7, LS411; second group (RER−) C70, HT29, LIM1863, SW1417, SW403, SW480, SW620, SW837, VACO4A (furthest from chromosome ideogram).
Figure 2.
Comparison of CGH data between cell lines and surgical material (Left), xenografts (Right). CGH data are expressed as percentage of tumors, xenografts, or lines showing the change, and each point represents the gain or loss of an individual chromosome arm. (Left) Pooled data from primary tumors, unselected for RER status (28–32), compared with data from this study, combining the RER+ and RER− cell lines in the ratio 2:8 to mimic unselected surgical material. Linear regression analysis gave slope 1.4, r = 0.7. (Right) Data from RER−, first-pass xenografts, obtained in one of our laboratories (6, 33), compared with the RER− cell lines. Slope = 1.0, r = 0.7.
Karyotypes were obtained by SKY analysis, confirmed by single-dye chromosome painting (Figs. 3 and 4). The karyotypes are presented below as: modal number of chromosomes per metaphase (range), actual content of normal sex chromosomes, listing of all chromosomes with copy numbers, [number of metaphases with the given modal composition]. Additional distinct clones are separated by/followed by their differences from the main clone. Most of the RER− and atypical RER+ lines showed additional rearrangements found in only one metaphase, but because these could not be confirmed they are not shown. For the simple near-diploid lines, DLD1, GP2d, HCT116, LoVo, LS174T, and VACO5, apparently normal pairs of chromosomes are omitted. Structural abnormalities are described using ISCN 1995 nomenclature. For example del(2)(p21) indicates chromosome 2 deleted from p21 to p telomere. der(4)del(4)(q31q35)t(4;18)(?p15;?)×2 indicates derivative of chromosome 4 deleted between q31 and q35 and translocated with chromosome 18 at p15 on chromosome 4 (the query indicating uncertainty) and a position not determined on chromosome 18; modal number two copies in this clone. q14∼21 indicates break within region q14 to q21. Isochromosomes (abbreviated i), deletions (del), and duplications (dup) are reported where evident from size changes, or from both CGH and cytogenetic 4′,6-diamidino-2-phenylindole (DAPI) banding. Breakpoints given for C70, HT29, and HCA7 were judged by DAPI banding. * indicates apparently balanced translocation. † may be normal 1 in some metaphases.
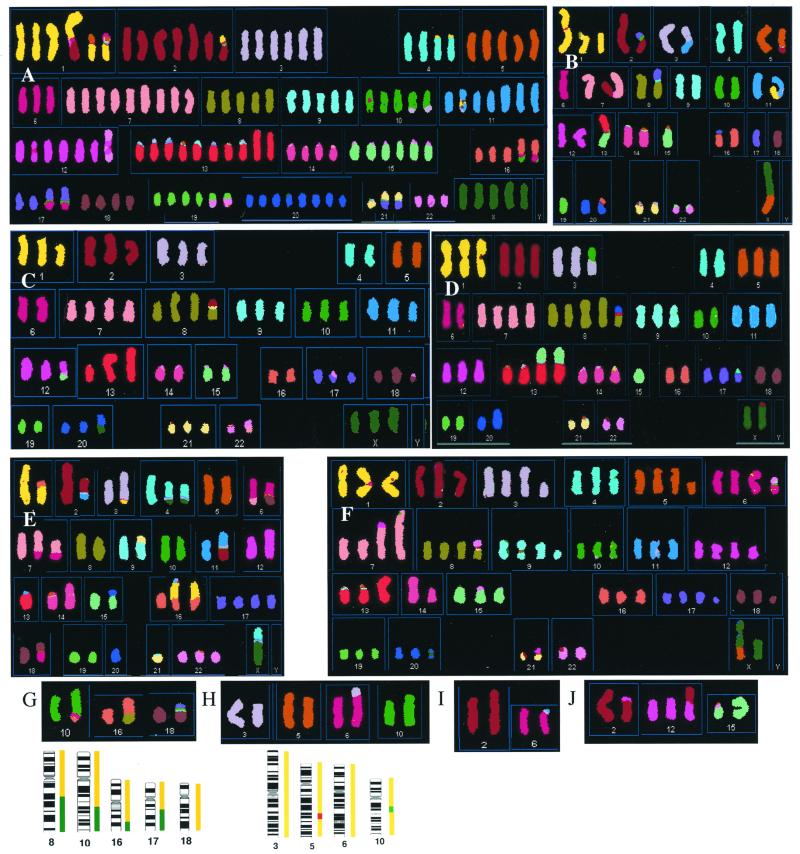
Figure 3.
Examples of karyotypes of the cell lines given by SKY analysis and their relation to CGH data. Lines not illustrated here are in Fig. 4 or have no abnormal chromosomes: Vaco5 and LS174T. (A–F) Images of complete metaphases. (A) C70. (B) SW837. (C) SW403. (D) VACO4A. (E) HCA7. (F) LS411. These are typical metaphases that may not show all of the chromosomes described in Results. The HCA7 metaphase is of the most complex clone. The metaphases for C70 and SW837 show rearranged chromosomes unique to that metaphase, respectively a reciprocal t(1;2) and a der(5)t(2;5). The chromosomes are shown in classification colors, i.e., each pixel is assigned a color representing the chromosome that the software has identified from the fluorescence at that position. Satellites at chromosomes 13, 14, 15, 21, and 22 as well as pericentromeric heterochromatin, e.g., of chromosome 1, hybridize nonspecifically so they are often miscolored. Some of the classifications are incorrect in detail, because of overlap of adjacent fluorescence colors, and their correct composition, determined by conventional fluorescence in situ hybridization with single dyes, is shown in Results. (G–J) Partial metaphases from near-diploid lines showing only the chromosomes with structural abnormalities and their normal counterparts for comparison. (G) HCT116. (H) GP2d. (I) DLD1. (J) LoVo. Underneath G and H the copy numbers of the corresponding chromosomes from CGH are shown as in Fig. 1.
Karyotypes of the colorectal carcinoma cell lines were:
RER−.
C70.
127 (115–130), XXXXX, 1×4, der(1)t(1;5)(p12;p13)×2, 2×1, del(2)(p21), del(2)×4, 3×6, 4×2, der(4)del(4)(q31q35)t(4;18)(?p15;?)×2, 5×4, 6×3, 7×9, 8×5, 9×5, 10×4, der(10)t(3;10)(?;q23∼24), 11×3, del(11)(?q23), der(11)t(11;11)×3, 12×6, der(12)t(12;22)t(12;22), 13×6, der(13)t(13;13)×2, 14×4, 15×6, 16×3, der(16)t(10;16)(q23∼24;q24)×2, 17×2, der(17)t(6;17)(?q23;q24∼25)×2, 18×4, 19×4, der(19)t(19;22)(p;q)×2, 20×9, 21×2, der(?)t(17;21)(q10;q10)×2, 22×3[12]/idem, +der(8)t(5;8)[2].
HT29.
70 (69), XX, del(X)(?p21), 1×3, 2×3, der(2)t(1;2)(q32;q11∼13), 3×3, 4×2, del(4)(?q31), 5×3, der(5)t(5;6)(p10;?), 6×2, 7×4, 8×2, hsr(8)(p22∼23), 9×2, der(?)t(6;9)(p10;q10), 10×3, 11×4, 12×3, 13×1, i(13)(q10), 14×2, 15×4, 16×3, 17×2, der(?)t(17;19)(q10;?p10), 18×2, del(18)(q12), 19×3, 20×3, del(20), 21×2, 22×3, der(22)t(17;22)(?;q12)[13]/idem, 11×3, der(?)t(9;11)[2]/idem, -11, +der(11)t(11;13), -13, +der(13)t(7;13)(?;q10), -16, +der(?)t(11;16)(q10;?p10)[3].
LIM1863.
80 (66), XXXX, 1×3, 2×3, 3×3, der(?)t(2;3)t(3;8), 4×3, 5×3, 6×2, 7×4, 8×2, del(8)(p?)×3, der(?)t(8;17), 9×3, 10×3, 11×3, der(?)t(1;11), 12×3, der(12)t(11;12), 13×0, i(13)(q10)×4, der(?)t(9;13), 14×3, 15×4, 16×3, 17×0, der(17)t(X;17)×2, 18×3, 19×3, 20×7, 21×2, 22×2[10]/idem, -der(17)t(X;17), +der(17)t(3;17)[2].
SW1417.
70 (66), XX, 1×1, del(1)(?)×2, 2×3, 3×4, 4×2, 5×1, del(5)(?)×2, der(5)t(5;17), der(5)t(5;20), 6×2, del(6)(q?), 7×1, dup(7)(q?), der(7)t(1;7)t(1;8)×2, 8×1, der(8)t(1;8), 9×1, del(9)(?)×2, 10×2, 11×4, 12×3, 13×2, 14×3, 15×1, i(15)(q10), 16×3, 17×2, 18×1, del(18)(?), dup(18)(?), 19×1, der(19)t(9;19)×2, 20×2, dup(20)(?)×2, 21×2, 22×4[13]/idem, -2, +del(2)(?), +der(2)t(2;3), +del(10) [2]/idem, der(2)t(2;20), -3, +del(3), +der(?)t(5;18), -12, +del(12)(q?), -17, +der(17)t(16;17)[3].
SW403.
64 (60), XXX, 1×2, del(1), 2×3, 3×3, 4×2, 5×3, 6×2, 7×4, 8×1, dup(8)(?), i(8)(q10), der(?)t(2;8), 9×3, 10×3, 11×3, 12×2, der(12)t(12;15), 13×1, dup(13)(q?)×2, 14×2, 15×2, 16×2, 17×1, del(17)(?), der(17)t(17;22), 18×1, dup(18)(q?), der(?)t(18;22)*, 19×3, 20×4, der(20)t(X;20), 21×3, 22×1, der(22)t(7;22)[9]/66, idem, +del(18) [3]/idem, -22, +der(22)t(18;22)* [2].
SW480–Clone 1.
58 (52), XX, Y×0, 1×1, der(1)t(1;9)*, 2×2, der(2)t(2;12), 3×2, del(3)(?), 4×2, 5×1, der(5)t(5;20)*, 6×2, 7×2, der(7)t(7;13), der(7)t(7;14), 8×1, der(?)t(8;19), 9×1, der(?)t(8;9), der(9)t(1;9)*, 10×1, der(10)t(10;12)(3;12), 11×3, 12×1, del(12)(?), 13×3, 14×2, 15×2, 16×2, 17×2, del(17)(q?), 18×1, del(18)(q?), 19×1, der(?)t(5;19)t(8;19), 20×2, der(20)t(5;20)*, 21×3, 22×2[9].
SW480–Clone 2.
90 (88), XX, Y×0, 1×4, 2×3, del(2)(?), der(2)t(2;12), der(2)t(2;18), 3×3, 4×3, 5×2, der(?)t(5;12), der(5)t(5;20)*×2, 6×3, 7×3, der(7)t(1;7), der(7)t(7;14)×2, 8×4, 9×3, 10×2, der(?)t(10;12)×2, der(10)t(10;15), 11×3, der(11)t(11;15)×2, 12×2, del(12)(?), der(12)t(12;14)*, 13×5, 14×1, der(14)t(12;14)*×2, 15×3, 16×3, 17×4, 18×1, del(18)(q?)×3, 19×2, der(?)t(5;19)t(8;19)×4, 20×4, der(20)t(5;20)*×3, 21×5, 22×4[13].
SW620.
48 (45), XX, Y×0, 1×2, 2×1, der(2)t(2;12), 3×1, del(3), 4×1, del(4), 5×1, der(5)t(5;20)*, 6×1, der(6)t(6;7)*, 7×2, del(7), der(7)t(6;7)*, 8×0, der(8)t(8;13), der(8)t(8;17), 9×2, 10×1, der(10)t(10;13), 11×3, 12×2, 13×1, 14×2, 15×2, 16×1, der(16)dup(16)t(3;16)t(6;16), 17×2, 18×1, der(?)t(5;18), 19×2, 20×2, der(20)t(5;20)*, 21×2, 22×2[11]/(46–47), idem, -X, +der(?)t(X;18), -der(?)t(5;18), +del(5)[3].
SW837.
38 (38), der(X)t(X;5), Y×0, 1×0, del(1)(?), der(1)t(1;8)×2, 2×2, 3×1, der(3)t(3;11), 4×2, 5×2, 6×1, del(6)(?), 7×1, der(7)t(7;19), 8×1, der(8)t(8;17), 9×2, 10×2, 11×1, der(11)t(1;11), 12×2, 13×0, der(?)t(13;15), 14×2, 15×1, 16×2, 17×1, 18×1, 19×1, 20×2, 21×2, 22×2[7]/idem, +1, -der(1)t(1;8), -2, + der(2)t(2;17), -der(7)t(7;19), +der(7)t(2;7), -16, +der(16)t(16;20)*, -20, +der(20)t(16;20)* [5].
VACO4A.
62 (61), XX, Y×0, 1×2, i(1)(q10), 2×3, 3×2, der(3)t(3;10)(q10;q10), 4×2, 5×3, 6×2, 7×4, 8×2, i(8)(q10)×2, der(?)t(8;20)(q10;?)×2, 9×3, 10×2, 11×3, 12×3, 13×2, der(13)dup(13)(q?)t(13;15)×2, 14×4, 15×1, 16×2, 17×2, der(?)t(10;17), 18×2, 19×2, 20×2, 21×2, 22×2[5]/idem, -der(?)t(10;17), +der(?)t(4;17)[4]/idem, -X, +der(X)t(X;2)[3]/idem, -9, +del(9)(q?)[2]/idem, -20, +dup(20) [2].
Typical RER+.
DLD1.
46 (43), XY, 1×1, dup(1)(p?)†, 2×1, dup(2)(p13p23), 6×1, der(6)t(6;11)[18].
GP2d.
46 (45), XX, 3×1, del(3), 5×1, del(5), 6×1 der(6)t(3;6), 10×1, dup(10)(q?)[12].
HCT116.
45 (43), X, Y×0, 10×1, der(10)dup(10)(q?)t(10;16), 16×1, der(16)t(8;16), 18×1, der(18)t(17;18)[18].
LoVo.
49 (48), XY, 2×1, der(2)t(2;12)*, 5×3, 7×3, 12×2, der(12)t(2;12)*, 15×1, i(15)(q10)[12].
LS174T.
47 (46), X, 7×3, 15×3 [14].
VACO5.
46 (43), XX[12]/idem, +del(7) [2]/idem, -21, +del(21)(p?)[4].
Atypical RER+.
HCA7.
43 (42), der(X)t(X;4)(p22;q25∼26)*, 1×1, der(1)del(1)(q)t(1;16)(p13;p11.2)*, 2×1, der(2)t(2;11)(q14∼21;q21)*, 3×0, del(3)(p13p21), der(3)t(1;3)(?p36;q27∼29)*, 4×1, der(4)t(X;4)(p22;q25∼26)*, 5×2, 6×0, der(6)t(6;7)(q21∼22;q31)*, der(6)t(6;18)(q13∼15;q11.2)*, 7×1, der(7)t(6;7)(q21∼22;q31)*, 8×2, 9×1, der(9)t(9;21)(p12∼13;q11.2), 10×2, 11×1, der(11)t(2;11)(q14∼21;q21)*, 12×1, dup(12)(q?), 13×1, 14×1, i(14)(q10), 15×1, del(15)(?q12q15), 16×1, der(16)t(1;16)(p13;p11.2)t(1;3)(p36;q27∼29)*, 17×2, 18×1, der(18)t(6;18)(q13∼15;q11.2)*, 19×2, 20×2, 21×1, 22×2[14]/44 (40), idem, +2, -der(2)t(2;11)(q14∼21;q21), -der(3)t(1;3)(?p36;q27∼29), +der(?)t(3;14), +6, -der(6)t(6;7)(q21∼22;q31), +7, -der(7)t(6;7)(q21∼22;q31), -10, +der(10)t(3;10), +11, -der(11)t(2;11)(q14∼21;q21), +14, -i(14)(q10), -der(16)t(1;16)(p13;p11.2)t(1;3)(p36;q27∼29), +der(16)t(1;16)(p13;p11.2)* [7].
LS411.
73 (70), X, der(X)dup(X)t(X;5), Y×0, 1×3, del(1), 2×3, 3×3, del(3), 4×3, 5×3, del(5), 6×2, 7×2, dup(7), der(7)dup(7)t(7;12), 8×3, der(?)t(8;22), 9×3, del(9), 10×3, 11×3, 12×3, del(12), 13×2, i(13)(q10), 14×1, i(14)(q10), 15×3, 16×3, 17×3, del(17), 18×2, del(18), 19×3, dup(19)(p?), 20×3, 21×1, der(?)t(12;21), 22×2, der(?)t(6;22)[8]/idem, -del(1), +del(6), +der(?)t(10;17), -dup(19)[3]/idem, -del(1), -der(?)t(6;22), +dup(6), +der(6)t(5;6), +del(11)(q?)[3].
These karyotypes were entirely consistent with the CGH data. Where chromosomes were found by SKY to be without rearrangement, the SKY karyotype independently confirmed the estimate of copy number obtained by CGH. In more complex karyotypes, estimates of copy-number changes derived from CGH frequently identified the fragments of chromosome arms that were involved in unbalanced translocations shown by SKY (17). Thus, for example, CGH identified gains of parts of 8q, 10q, 16q, and 17q in HCT116, corresponding to extra fragments of these chromosomes involved in translocations (Fig. 3G). Similarly, in GP2d, CGH showed loss around the APC gene locus at 5q21–22, corresponding to the short chromosome 5 in the karyotype (Fig. 3H).
SKY analysis (Figs. 3 and 4, tabulated above and summarized in Table 1) showed that all these RER− cell lines had multiple abnormalities in chromosome number, notably multiple trisomies, together with multiple chromosome rearrangements. They also showed substantial metaphase-to-metaphase variation within the same line, both in chromosome number (estimated by counting centromeres; Table 1) and by the presence of chromosome structural rearrangements that were present in only some of the metaphases. The great majority of translocations were nonreciprocal—no more than 13 of about 90 could be reciprocal (in the major clones), or no more than seven of 90 if the very unusual line, HCA7, is excluded, as discussed below. All chromosomes except Y were translocated in at least one line. Most translocations were observed no more than once in the entire series. The most frequent, unbalanced t(8;17), was present in only three lines, and it is not certain that the breakpoint was identical even in these. The CGH copy number profiles suggested that some breakpoints occurred repeatedly at approximately the same region in three or more lines—near 6q22, 8q22, 13q22, and 20p12, and near centromeres of 1, 3, 5, 6, 7, 8, 9, 17, 18 and 19—but the translocation partners were variable.
Table 1.
Genomic changes and variability of the cell lines
| Cell line | RER status | p53 status | Mode | Rearranged chromosomes | Variability of centromere number (%) |
|---|---|---|---|---|---|
| RER− | |||||
| C70 | − | ND | 127 | 14 | 29 |
| HT29 | − | Mut | 70 | 15 | 14 |
| LIM1863 | − | NF | 80 | 9 | 12 |
| SW1417 | − | NF | 70 | 22 | 13 |
| SW403 | − | Mut | 64 | 14 | 18 |
| SW480 | − | Mut | 58, 90 | 15, 16 | 16, 30 |
| SW620 | − | Mut | 48 | 15 | 10 |
| SW837 | − | Mut | 38 | 13 | 6 |
| VACO4A | − | NF | 62 | 10 | 6 |
| Typical RER+ | |||||
| DLD1 | + | Mut | 46 | 3 | 2 |
| GP2d | + | ND | 46 | 4 | 1 |
| HCT116 | + | Mut | 45 | 3 | 2 |
| LoVo | + | NF | 49 | 3 | 2 |
| LS174T | + | NF | 47 | 0 | 2 |
| VACO5 | + | Mut | 46 | 2 | 3 |
| Atypical RER+ | |||||
| HCA7 | + | Mut | 43 | 21 | 4 |
| LS411 | + | NF | 73 | 21 | 12 |
RER status is from ref. 4. p53 status: Mut, mutation detected; NF, no mutation found; ND, not done. Mutation screening was by chemical mismatch cleavage analysis of exons 3–9 and 11 (27). The variability of centromere numbers [adapted from Lengauer et al. (19)] was obtained by counting in each of 8–15 metaphases the number of copies of each centromere, whether in normal or rearranged copies of a chromosome; noting the percentage of metaphases that have deviations from the modal centromere number; and averaging over all centromeres. The two values for SW480 are for its two major clones. Our measurements of metaphase heterogeneity were slightly lower than Lengauer et al.'s (19), presumably because we examined cells in metaphase, whereas they studied interphase nuclei, and some of the cells they observed with altered chromosome numbers may not divide.
In contrast, six of the eight RER+ lines showed few or no breakpoints or abnormalities in chromosome number, and only minimal intermetaphase heterogeneity (Table 1). Of the six translocations observed in these six lines only one, the t(2;12) of LoVo, was found in any other tumor (the tumor that gave the two lines SW480 and SW620). (The 10;16 in HCT116 is not the same 10;16 as in C70.) A reciprocal translocation accounted for two of the three breakpoints in Lovo (Fig. 3J). The remaining two RER+ lines, however, showed strikingly different patterns of abnormality. LS411 had the features described above as characteristic of the majority of RER− cell lines: a near-triploid modal karyotype, multiple translocations, and pronounced intermetaphase heterogeneity (Table 1). We reconfirmed its RER+ phenotype on the cells that had been karyotyped. HCA7 had a near-diploid chromosome number, with very little variation in centromere number between metaphases, but had multiple breakpoints, the great majority reflecting reciprocal translocations. The modal karyotype contained six of these, but some individual metaphases showed additional reciprocal translocations.
Discussion
The results demonstrate a majority subtype among RER− cancers with surprisingly consistent features. The modal chromosome number tends to be near triploid, with many trisomies, showing relative excess of chromosome arms 7p, 7q, 8q, 13q, and 20q, and relative deficiency of 17p, 18q, and 8p. Invariably, there are multiple unbalanced translocations and pronounced intermetaphase variation. Six RER− lines are typical of this subtype, with modes between 58 and 80 (HT29, LIM1863, SW1417, SW403, VACO4A, and one clone of SW480). SW480 and SW620 originally were derived from the same patient, respectively from a primary tumor and metastasis, and they share several translocations, confirming that they shared a common founder cell (18). One SW480 clone falls at the lower end of the majority group with 58 chromosomes; the other clone has a mode of 90 with several duplicated translocations; whereas SW620 has a modal chromosome number of 48. The common founder may have had a near-diploid mode and the larger SW480 clone may have arisen through endoreduplication. The near-hexaploid line C70 probably is also derived from the majority type through endoreduplication, as it shows many duplicated translocations. SW837 appears distinct, with a subdiploid mode of 38. Dutrillaux (1) has suggested that most aneuploid breast and colon carcinomas evolve by losing chromosomes, because unbalanced translocations tend to result in net chromosome loss, often followed by endoreduplication to give a near-triploid mode with duplicated abnormalities. This pattern is clear in many breast cancer cell lines (1, 15), and SW837 is an example of net loss of chromosomes. However, the majority group of colorectal carcinoma lines described here do not obviously fit this pattern, being near-triploid but without the expected multiple duplicated abnormalities. Perhaps endoreduplication is a very early event, preceding most chromosome rearrangements, or another mechanism leads to gradual net chromosome gain. Alternatively, the near triploid pattern might result from grossly asymmetric chromosome partition at mitosis.
The consistent, pronounced intermetaphase heterogeneity in centromere numbers (Table 1) strongly suggests that errors in chromosome segregation continue to occur during growth, as shown formally by others (19, 20). The many examples of abnormal chromosomes that were detected in only one or two metaphases suggest that this instability also extends to structural changes. These structural changes must involve chromosome breakage, and the survival of colorectal cancer cells with this phenotype implies defective coupling between DNA breaks and apoptosis. There is good evidence that mutant p53 is associated with chromosome rearrangement and aneuploidy (e.g., ref. 21), and is probably permissive (22). However, three of the lines that were typically aneuploid had been chosen for study because they had no detectable p53 mutations, so there may be alternative mechanisms. Our data also confirm that p53 deficiency by itself is not sufficient for chromosomal instability (8).
Although six of the eight RER+ lines conformed to the now well-described near-diploid pattern, with few or no rearranged chromosomes, two, LS411 and HCA7, showed strikingly different karyotypes, with high numbers of altered chromosomes. Indeed, LS411 showed all of the features of the majority pattern in RER− tumors, including pronounced intermetaphase heterogeneity, although the evidence for mismatch repair deficiency in this line is incontrovertible: microsatellite instability with hMLH1 gene mutation and promoter hypermethylation (4). RER+ tumors with unstable, aneuploid genomes also have been observed among primary colorectal cancers (6, 7).
The most remarkable finding, however, was the observation of a near-diploid line with multiple reciprocal translocations: HCA7. The overwhelming majority of translocations reported in epithelial tumors are nonreciprocal (1, 15, 16), but HCA7 showed six reciprocal translocations in its modal karyotype. Moreover, these translocation events appear to be ongoing, because extra examples were found in some metaphases. Unlike LS411 and the RER− lines, HCA7's structural instability was not accompanied by significant numerical instability, supporting the suggestion that HCA7 has a defect that is distinct from that of the other groups. LoVo might be a second example of this pattern: although the sample of LoVo studied here showed a single reciprocal translocation t(2,12), the sample reported earlier by Soulie et al. (22) had two more, which almost certainly arose during passage in vitro. It may be significant that both HCA7 and LoVo are RER+, because cells defective in mismatch repair are prone to recombination repair between imperfectly matching, homeologous sequences (23, 24).
In conclusion, SKY, in combination with CGH, has demonstrated several distinctive patterns of karyotype and genomic instability in these 17 colorectal cancer cell lines. It has defined a typical colorectal tumor karyotype: near-triploid, with multiple trisomies, and chromosomally unstable. This type is usually RER−, but occasional tumors in this group also show the RER+, microsatellite instability, phenotype. This finding is consistent with the hypothesis that the defects that cause genomic instability are selected for in tumor evolution (probably because they protect against apoptosis; ref. 2), because it predicts that some tumors will acquire more than one kind of defect, independently. SKY also has revealed the existence of a type of instability characterized by multiple reciprocal translocation events, without evident variability of chromosome number. These instabilities all appear to give rise to repeated genomic alterations, presumably reflecting deficient pathways for recognition and removal of the altered cells. Because such pathways include apoptosis, and are therefore relevant to responses to therapy, it will be of interest to examine the relationships between drug sensitivity and these different patterns of genomic instability (25, 26).
Supplementary Material
Acknowledgments
We thank Joanne Davidson for advice on the SKY analysis and Despina Sanoudou and Ian Roberts for advice on CGH. This work was supported by the Egyptian Government (studentship for W.M.A.-R.), Kyoto Prefecture, Japan (support for K.K.), Imperial Cancer Research Fund, Isaac Newton Trust, Cambridge Fund for the Prevention of Disease, European Union, and the Cancer Research Campaign (grants to A.H.W., M.J.A., and P.A.W.E.).
Abbreviations
- CGH
comparative genomic hybridization
- RER−
RER+, replication error negative and positive, respectively
- SKY
spectral karyotyping
References
- 1.Dutrillaux B. Adv Cancer Res. 1995;67:59–82. doi: 10.1016/s0065-230x(08)60710-1. [DOI] [PubMed] [Google Scholar]
- 2.Tomlinson I P M, Novelli M R, Bodmer W F. Proc Natl Acad Sci USA. 1996;93:14800–14803. doi: 10.1073/pnas.93.25.14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 4.Wheeler J M, Beck N E, Kim H C, Tomlinson I P, Mortensen N J, Bodmer W F. Proc Natl Acad Sci USA. 1999;96:10296–10301. doi: 10.1073/pnas.96.18.10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leach F S, Polyak K, Burrell M, Johnson K A, Hill D, Dunlop M G, Wyllie A H, Peltomaki P, de la Chapelle A, Hamilton S R, et al. Cancer Res. 1996;56:235–240. [PubMed] [Google Scholar]
- 6.Georgiades I B, Curtis L J, Morris R G, Bird C C, Wyllie A H. Oncogene. 1999;18:7933–7940. doi: 10.1038/sj.onc.1203368. [DOI] [PubMed] [Google Scholar]
- 7.Yao J, Eu K W, Seow-Choen F, Vijayan V, Cheah P Y. Int J Cancer. 1999;80:667–670. doi: 10.1002/(sici)1097-0215(19990301)80:5<667::aid-ijc6>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 8.Eshleman J R, Casey G, Kochera M E, Sedwick W D, Swinler S E, Veigl M L, Willson J K V, Schwartz S, Markovitz S D. Oncogene. 1998;17:719–725. doi: 10.1038/sj.onc.1201986. [DOI] [PubMed] [Google Scholar]
- 9.Cahill D P, Lengauer C, Yu J, Riggins G J, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Nature (London) 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 10.Clarke A R, Gledhill S, Hooper M L, Bird C C, Wyllie A H. Oncogene. 1994;9:1767–1773. [PubMed] [Google Scholar]
- 11.Toft N J, Winton D J, Kelly J, Howard L A, Dekker M, Te Riele H, Arends M J, Wyllie A H, Margison G P, Clarke A R. Proc Natl Acad Sci USA. 1999;96:3911–3915. doi: 10.1073/pnas.96.7.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li R, Sutphin P D, Schwartz D, Matas D, Almog N, Wolkowicz R, Goldfinger N, Pei H, Prokocimer M, Rotter V. Oncogene. 1998;16:3269–3277. doi: 10.1038/sj.onc.1201867. [DOI] [PubMed] [Google Scholar]
- 13.Rich T, Allen R L, Wyllie A H. Nature (London) 2000;407:777–783. doi: 10.1038/35037717. [DOI] [PubMed] [Google Scholar]
- 14.Kallioniemi O P, Kallioniemi A, Piper J, Isola J, Waldman F M, Gray J W, Pinkel D. Genes Chromosomes Cancer. 1994;10:231–243. doi: 10.1002/gcc.2870100403. [DOI] [PubMed] [Google Scholar]
- 15.Davidson J M, Gorringe K L, Chin S-F, Orsetti B, Besret C, Courtay-Cahen C, Roberts I, Theillet C, Caldas C, Edwards P A W. Br J Cancer. 2000;83:1309–1317. doi: 10.1054/bjoc.2000.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Courtay-Cahen C, Morris J S, Edwards P A W. Genomics. 2000;66:15–25. doi: 10.1006/geno.2000.6178. [DOI] [PubMed] [Google Scholar]
- 17.Ghadimi B M, Sackett D L, Difilippantonio M J, Schrock E, Neumann T, Jauho A, Auer G, Ried T. Genes Chromosomes Cancer. 2000;27:183–190. [PMC free article] [PubMed] [Google Scholar]
- 18.Melcher R, Steinlein C, Feichtinger W, Muller C R, Menzel T, Luhrs H, Scheppach W, Schmid M. Cytogenet Cell Genet. 2000;88:145–152. doi: 10.1159/000015508. [DOI] [PubMed] [Google Scholar]
- 19.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 20.Bengtsson B O, Nabholz M, Kennett R, Bodmer W F, Povey S, Swallow D. Somatic Cell Genet. 1975;1:41–64. doi: 10.1007/BF01538731. [DOI] [PubMed] [Google Scholar]
- 21.Carder P J, Cripps K J, Morris R, Collins S, White S, Bird C C, Wyllie A H. Br J Cancer. 1995;71:215–218. doi: 10.1038/bjc.1995.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soulie P, Poupon M F, Remvikos Y, Dutrillaux B, Muleris M. Oncogene. 1999;18:775–781. doi: 10.1038/sj.onc.1202336. [DOI] [PubMed] [Google Scholar]
- 23.Schimenti J C. Am J Hum Genet. 1999;64:40–45. doi: 10.1086/302220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Wind N, Dekker M, Berns A, Radman M, te Riele H D. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 25.Eshleman J R, Markowitz S D. Curr Opin Oncol. 1995;7:83–89. [PubMed] [Google Scholar]
- 26.Bunz F, Hwang P M, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler K W, Vogelstein B. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigues N R, Rowan A, Smith M E, Kerr I B, Bodmer W F, Gannon J V, Lane D P. Proc Natl Acad Sci USA. 1990;87:7555–7559. doi: 10.1073/pnas.87.19.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Mulla F, Keith W N, Pickford I R, Going J J, Birnie G D. Genes Chromosomes Cancer. 1999;24:306–314. doi: 10.1002/(sici)1098-2264(199904)24:4<306::aid-gcc3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 29.De Angelis P M, Clausen O P F, Schjolberg A, Stokke T. Br J Cancer. 1999;80:526–535. doi: 10.1038/sj.bjc.6690388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meijer G A, Hermsen M A J A, Baak J P A, van Diest P J, Meuwissen S G M, Beliën J A M, Hoovers J M N, Joenje H, Snijders P J F, Walboomers J M M. J Clin Pathol. 1998;51:901–909. doi: 10.1136/jcp.51.12.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paredes-Zaglul A, Kang J J, Essig Y P, Mao W, Irby R, Wloch M, Yeatman T J. Clin Cancer Res. 1998;4:879–886. [PubMed] [Google Scholar]
- 32.Ried T, Knutzen R, Steinbeck R, Blegen H, Schröck E, Heselmeyer K, du Manoir S, Auer G. Genes Chromosomes Cancer. 1996;15:234–245. doi: 10.1002/(SICI)1098-2264(199604)15:4<234::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 33.Curtis L J, Georgiades I B, White S, Bird C C, Harrison D J, Wyllie A H. J Pathol. 2000;192:440–445. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH761>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.